Abstract
Peptides and proteins possess an inherent propensity to self-assemble into generic fibrillar nanostructures known as amyloid fibrils, some of which are involved in medical conditions such as Alzheimer disease. In certain cases, such structures can self-propagate in living systems as prions and transmit characteristic traits to the host organism. The mechanisms that allow certain amyloid species but not others to function as prions are not fully understood. Much progress in understanding the prion phenomenon has been achieved through the study of prions in yeast as this system has proved to be experimentally highly tractable; but quantitative understanding of the biophysics and kinetics of the assembly process has remained challenging. Here, we explore the assembly of two closely related homologues of the Ure2p protein from Saccharomyces cerevisiae and Saccharomyces paradoxus, and by using a combination of kinetic theory with solution and biosensor assays, we are able to compare the rates of the individual microscopic steps of prion fibril assembly. We find that for these proteins the fragmentation rate is encoded in the structure of the seed fibrils, whereas the elongation rate is principally determined by the nature of the soluble precursor protein. Our results further reveal that fibrils that elongate faster but fracture less frequently can lose their ability to propagate as prions. These findings illuminate the connections between the in vitro aggregation of proteins and the in vivo proliferation of prions, and provide a framework for the quantitative understanding of the parameters governing the behavior of amyloid fibrils in normal and aberrant biological pathways.
Keywords: Amyloid, Kinetics, Prions, Protein assembly, Protein Folding, Ure2p, Biosensor, Fragmentation, Transmissibility
Introduction
Many proteins exhibit a tendency to form organized aggregates such as the amyloid fibrils associated with pathological conditions including Alzheimer disease (1). In certain cases, such aggregates can self-propagate in living systems as prions and transmit information encoded in their conformations to other living organisms without the need for storing this information in coding nucleic acids. The prion phenomenon was first discovered in the context of transmissible spongiform encephalopathies (TSEs) (2–3), but has now been identified not only in pathological conditions but also as an epigenetic phenomenon in fungi (4–6). In particular, yeast prions have proved to be a powerful model for probing and understanding the complex functional and structural aspects characteristic of prion biology (4–5, 7–13).
The non-Mendelian elements [URE3] and [PSI+] of Saccharomyces cerevisiae have been shown to be prion forms of the cellular proteins Ure2 and Sup35 (14–15) and in recent years further yeast prions have been identified (4–5). Ure2p is one of the best characterized prions in yeast (16). Its activity is connected with the regulation of nitrogen catabolism: it interacts with the transcription factor Gln3p, and when a good source of nitrogen is available, it can repress the genes that code for the enzymes and transporters needed for using poor nitrogen sources (17). The N-terminal region of Ure2p is required for its prion properties in vivo (18) as well as for amyloid formation in vitro (19–21); this region is therefore known as the prion domain (PrD).3 The presence of the flexible N-terminal domain is not required for its regulatory function in vivo (22) or for its enzymatic activity in vitro (23–25) reinforcing the idea that the role of the PrD is related to controlling the prion behavior. The Ure2 protein exists in the closely related hemiascomycetous yeasts S. cerevisiae and S. paradoxus in the form of ScUre2p and SpUre2p, respectively. The C-terminal domains of ScUre2p and SpUre2p are identical, and the PrDs of ScUre2p and SpUre2p differ by only a few residues (26), see Fig. 1. Unlike ScUre2p, however, SpUre2p has a low propensity to function as a prion even when overexpressed in S. paradoxus (27). Further in vivo and in vitro studies have demonstrated that SpUre2p does in fact readily form fibrils in vitro, and have identified less efficient fragmentation as a likely origin of its reduced ability to propagate transmissible prions in vivo (28).
FIGURE 1.
Sequence alignment of Ure2p from S. cerevisiae and S. paradoxus. The differences in amino acid sequence between the two homologues all lie within the N/Q-rich N-terminal prion domain (boxed area), while the C-domains are identical.
Though the ability to switch to an aggregated state from a soluble one is a feature common to prion proteins, it is important to note that only a small fraction of amyloidogenic proteins have been shown to act as prions. The successful propagation of a prion depends on the capacity of the soluble protein to undergo conversion to the fibrillar form, to promote the multiplication of prion seeds, and to transmit these seeds to other cells. If any step in this process is impaired, the ability to propagate as a prion will be reduced or even lost. The sensitivity of fibrils to shearing forces or agents promoting fragmentation, which can greatly affect the generation of prion seeds, have been identified as indications that the breakage rate of amyloid fibrils is a likely key determinant of the physiological impact for both prion and non-prion amyloid (12). Consequently much effort has focused on characterizing the kinetic parameters governing fibril growth, either theoretically (12, 29–31) or experimentally (10, 12); many experimental techniques have been developed to monitor the growth of amyloid fibrils, including: dye binding assays (9, 32), dynamic light scattering (DLS) (33), atomic force microscopy (AFM) (12, 34–35), and quartz crystal microbalance (QCM) sensors (36–37).
In this study, we use a combination of theoretical analysis based on explicit solutions (38) to the master equations (10, 12, 39–42) describing fibrillar growth, together with a range of experimental solution and biosensor assays monitoring the rates of fibril growth of ScUre2p and SpUre2p, to shed light on the kinetics of prion assembly in a quantitative manner. By varying in a controlled way the system parameters, such as the protein seed concentration, we have been able to extract and compare the rate constants characterizing the growth of amyloid fibrils for these two Ure2p homologues. Our results reveal that the fibrils of SpUre2p elongate faster but break less frequently than those formed from ScUre2p; as the propensity of the latter fibrils to function as prions is substantially higher than that of the former, these results provide quantitative support for the idea that breakage rates play a determining role in prion propagation (10, 12). Our results in addition allow a quantitative comparison of the kinetics of the individual steps underlying prion assembly and reveal that for the proteins studied here, the fragmentation rates are transmitted to daughter fibrils from the structure of seed fibrils used to initiate the growth reaction, whereas the elongation rate is influenced primarily by the ability of the soluble protein molecules to attach themselves to the fibril ends rather than by the nature of the seed structures.
EXPERIMENTAL PROCEDURES
Chemicals
Thioflavin T (ThT), Tris, and other chemicals were obtained from Sigma. ScUre2p and SpUre2p proteins were expressed in Escherichia coli with an N-terminal His tag and purified under native conditions as described previously (28, 43) except that a French press was used to disrupt E. coli cells instead of sonication.
ThT Assay
The kinetics of Ure2 fibril formation were monitored using an assay based on binding of the fluorescence dye ThT, as described previously (44–45). At regular time intervals during incubation, 10-μl aliquots were removed from the incubated mixture and assayed for ThT binding. For each sample, the intensity of ThT fluorescence at 485 nm after excitation at 450 nm was measured on a Hitachi F-4500 spectrofluorimeter.
Quantification of the Amount of Ure2 in Fibrillar Form
The concentration of protein in the fibrils was calculated as the difference in the initial total protein concentration and the final protein concentration in the supernatant fraction after centrifugation to pellet the fibrils; protein concentration was determined by Bradford assay.
Seed Preparation
To induce fibril formation, Ure2p was incubated at a concentration of 40 μm at 4 °C without shaking in 50 mm Tris-HCl, pH 8.4, 200 mm NaCl for at least 7 days, by which time fibril formation was complete as measured by ThT assay. Mature fibrils were then sonicated using a probe sonicator (Sonics and Materials VCX750) for 5 s (22% of the maximal sonication power) to produce the seeds used in the solution seeding experiments.
Cross-seeding Experiments
20 μm ScUre2p or SpUre2p protein was incubated with a series of concentrations (1–10%) of preprepared Sc or Sp seeds at 4 °C without shaking in 50 mm Tris-HCl, pH 8.4, 200 mm NaCl. The kinetics of fibril formation were monitored by ThT assay. Protein solutions with no addition of pre-formed seed fibrils at the start of the experiment were monitored as controls to confirm that no significant degree of spontaneous nucleation occurred during the time course of the seeded experiments.
Breakage and Elongation Kinetics
When formation of fibrils reaches the plateau phase, the concentration of soluble Ure2p is less than 3% of Ure2p present in solution initially and the conversion from the soluble to fibrillar form is therefore very efficient. To convert the measured ThT fluorescence values at intermediate times into concentrations of protein in fibrillar form, we used a linear scaling between the initial fluorescence value and the final reading (see “Results”). Data were fitted globally to an explicit analytical solution to the master equation of fibrillar growth derived previously (38) and outlined below. In the presence of seed fibrils, the fraction of protein in fibrillar form is given as a double exponential form as shown in Equation 1,
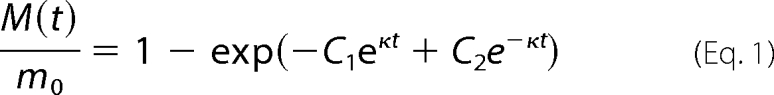 |
where M(t) is the concentration of protein in fibrillar form at time t, m0 is the initial concentration of soluble Ure2p and the constants, C1 and C2, are fixed by the initial conditions.
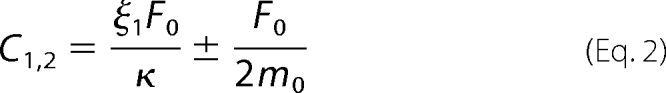 |
F0 = P0L0 is the mass concentration of seed fibrils at t = 0, where P0 is the number of seed fibrils and L0 is the average number of molecular units in a seed fibril. The single effective rate constant κ describes the rate of growth of the fibril population through the concerted action of fragmentation and elongation.
 |
This functional form (Equations 1–3), therefore, essentially contains only two free global parameters,
 |
which can be obtained through fitting the kinetics of fibril formation.
In a cross-seeding experiment, the seeds added to ScUre2p and SpUre2p protein solutions are the same (i.e. have the same value of L0). Therefore, the ratio of the elongation rates between SpUre2p and ScUre2p (r+) and the ratio of breakage rates between SpUre2p and ScUre2p (r−) can be evaluated for a given type and batch of seed as shown in Equations 6 and 7,
 |
 |
where the superscript indicates the protein in solution. A value of r+ or r− = 1 indicates that the nature of the soluble protein does not influence the rate constants for elongation (+) or breakage (−), respectively. Finally, to compare the effects associated with the nature of the seed, we introduce the ratio shown in Equation 8,
 |
where the subscript denotes the protein used as the seed. A value of η+ or η− = 1 indicates that the nature of the seed does not affect the rate constants for elongation (+) or breakage (−), respectively.
Electron Microscopy
A 10-μl aliquot of sample was taken and drop cast onto a copper grid coated with a Formvar film and allowed to adsorb for 1 min. The grid was then rinsed briefly with a droplet of water, stained with 10 μl of 2% uranyl acetate and observed with a Philips Tecnai 20 electron microscope operating at 100 kV.
Quartz Crystal Microbalance (QCM)
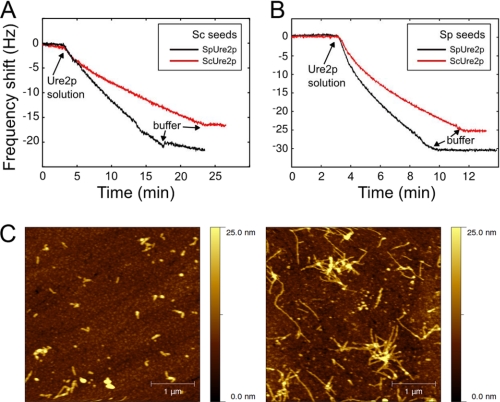
Seed fibril fragments from ScUre2p or SpUre2p, prepared as described above, were covalently coupled to a modified QCM sensor (Q-sense, QSX 301). The gold surface of the QCM sensor was immersed in 10 mm MUA (mercaptoundecanoic acid) in ethanol for 10 h to allow the assembly of an ordered monolayer to take place. The MUA-modified QCM sensors were subsequently rinsed with distilled water before activation of the carboxylic acid groups by immersion in 0.3 mg/ml 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and 0.5 mg/ml N-hydroxysuccinimide (NHS) for 1.5 h. The sensor was rinsed with distilled water and dried. Finally a 100-μl aliquot of a suspension containing 0.5 μm Sc or Sp seed fibrils was added on to the activated sensor and left to react for 30 min. The sensor was rinsed with buffer (50 mm Tris-HCl, pH 8.4, 200 mm NaCl) before passivation of the activated groups by incubating the surface for 1 h with a solution of 500 mm of ethanolamine at pH 10.0. After a final rinsing step, again with distilled water, the functionalized sensors were inserted into the microbalance flow cell (Q-sense, E4). After equilibration for 12 h to achieve a stable frequency baseline, protein solution was introduced into the flow cell and the increase in mass of the surface-bound seed fibrils resulting from their elongation was monitored in real time via the shift in the resonant frequency of the quartz crystal (36). The fundamental resonant frequency of the crystal, together with six overtones were monitored simultaneously; in Fig. 6 the overtone with n = 3 is shown.
FIGURE 6.
Measurement of fibril growth kinetics using QCM. Cross-seeding experiments were performed using (A) Sc seeds or (B) Sp seeds immobilized on the QCM Chip in 50 mm Tris-HCl, pH 8.4, 200 mm NaCl. The linear decline in frequency of the piezoelectric quartz crystal was detected when solutions of ScUre2p (red line) or SpUre2p (black line) were introduced into the flow cell. C, AFM images of QCM chips before and after fibril growth. The seeds immobilized on the QCM sensor (left) and the fibrils obtained after the growth experiment (right) were observed by tapping mode AFM. A 4-μm square area was scanned in each case.
Atomic Force Microscopy (AFM)
The seed fibrils were coupled as described above to two identical modified QCM sensors in each case. Then one of the sensors was dried immediately and imaged in tapping mode using a PicoPlus AFM (Molecular Imaging, Tempe, AZ). The other one was imaged after the measurement in the microbalance.
RESULTS
The Concentration of Ure2 Protein in Fibrillar Form Shows a Linear Relationship with ThT Fluorescence
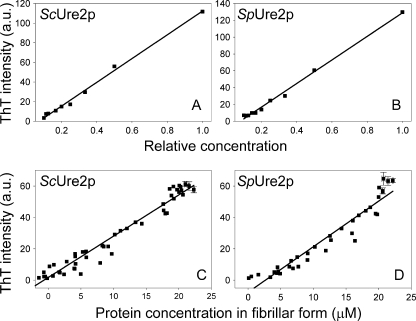
The ThT assay is commonly used to monitor fibril formation as ThT fluoresces when bound to fibrils and the intensity of fluorescence can reflect the amount of protein in amyloid form (46). However, this method does not necessarily show a strict quantitative relationship between the extent of fibril formation and fluorescence for a given protein (47). To test whether ThT can be used to quantify the amount of Ure2p fibrils, different concentrations of fibrils were produced by serial dilution of mature fibrils (with or without first sonicating the fibrils). Plotting the ThT values against their relative concentrations (i.e. setting the original concentration of mature fibrils as 1), we obtained a linear relationship between the ThT value and the quantity of Ure2p fibrils (Fig. 2, A and B). We also monitored this relationship during fibril formation. At different time points of fibril formation, the ThT value of the complete sample and the protein concentration in the pellet fraction of the sample after centrifugation, were measured at the same time point (see “Experimental Procedures”). The quantity of Ure2p fibrils formed at each stage of the time course also shows a good linear relationship with the ThT value (Fig. 2, C and D). The ThT assay therefore directly reflects the quantity of Ure2p fibrils, and so can be used to monitor the kinetic properties of the fibrils.
FIGURE 2.
Linear relationship between ThT intensity and protein concentration in fibrillar form. A and B, different concentrations of seeds were produced by serial dilution of (A) ScUre2p fibrils and (B) SpUre2p fibrils, and the ThT intensity was plotted against the relative seed concentration. C and D, relationship of ThT intensity and concentration of Ure2p in the pellet fraction at different stages throughout the process of fibril formation for (C) ScUre2p and (D) SpUre2p. The data from 5 growth curves were included for each protein. The points within the plateau region (where the error in determining the protein concentration is largest) were averaged for each individual growth curve, and the error bars represent the standard error of the mean (n = 3 to 6).
Comparing Kinetic Parameters Describing Fibril Growth for Different Protein Homologues in Solution
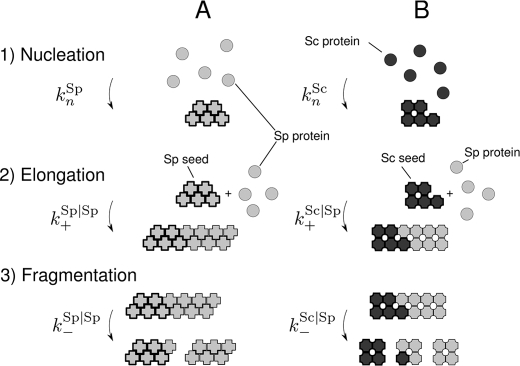
In this study we analyzed fibril growth measurements using a master equation which includes the three key processes governing amyloid growth (Fig. 3): nucleation, elongation, and breakage (10, 12, 34, 48). Underlying this picture, there are three kinetic constants: the nucleation rate constant kn, the elongation rate constant k+ and the breakage rate constant k−. In the presence of seed fibrils, soluble Ure2p will attach to the ends of existing seeds with a rate of k+, which is generally faster than spontaneous formation of nuclei (48). If this condition is met, nucleation is circumvented and only elongation and breakage will contribute to fibril growth.
FIGURE 3.
Mechanism of amyloid fibril formation. For seeded growth, when nucleation kn can be neglected, there are two kinetic constants defining the growth kinetics: the elongation rate constant (k+) and the breakage rate constant (k−). A, spontaneous fibril formation of SpUre2p. B, fibril formation of SpUre2p under the induction of Sc seeds.
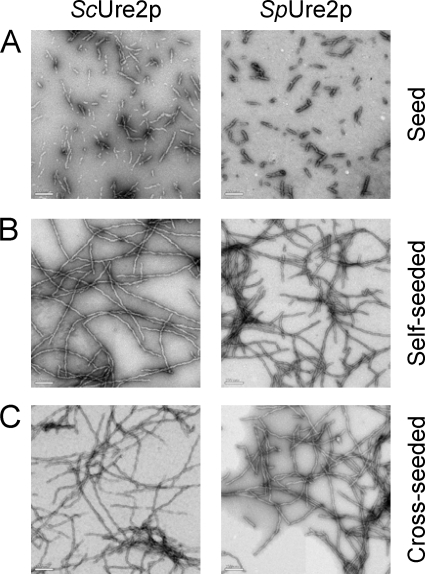
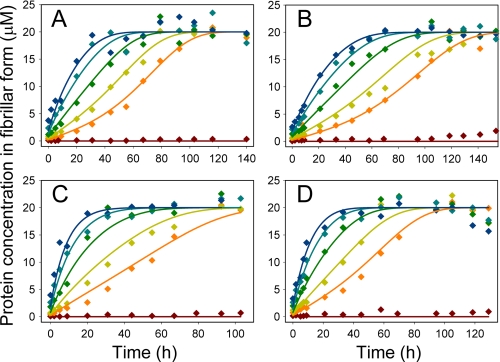
Seed fibrils of the Ure2p homologues Sc and Sp were produced by sonication (Fig. 4). Under the conditions used here for fibril growth experiments, even in the presence of a small concentration of preformed fibrils, the aggregation reaction reaches completion before measurable fibril formation takes place in samples lacking added seeds (Fig. 5). Therefore, in the presence of seed fibrils, the contribution to the overall reaction of spontaneous nucleation events can be neglected and only the two constants (k+, k−) characteristic of the system are left.
FIGURE 4.
Electron microscopy images of Sc seeds and Sp seeds, and the fibrils produced in the cross-seeding experiments. The images show (A) freshly prepared seeds, (B) self-seeded fibrils, and (C) cross-seeded fibrils grown in a solution of 20 μm ScUre2p (left column) or SpUre2p (right column) in 50 mm Tris-HCl, pH 8.4, 200 mm NaCl, as described in the “Experimental Procedures.” The bars represent 200 nm.
FIGURE 5.
Time courses of fibril formation in cross-seeding experiments monitored by ThT fluorescence. Solutions (20 μm) of ScUre2p (A and B) or SpUre2p (C and D) were incubated with a series of concentrations of Sc seeds (A and C) or Sp seeds (B and D): 0% seed (brown), 1% seed (orange), 2% seed (ochre), 4% seed (green), 7% seed (teal), 10% seed (blue), in 50 mm Tris-HCl, pH 8.4, 200 mm NaCl. The data were fitted globally to obtain the ratios of the rate constants for elongation and breakage, as described under “Experimental Procedures.”
The measured ThT fluorescence values were converted into absolute concentrations of protein present in fibrillar form (see above) and the entire dataset describing the fibril formation rates at different seed concentrations was fitted to obtain the two global parameters, ξ1 = k+/L0 and ξ2 = k+ k−, derived from the explicit solutions (38) to the elongation-fragmentation growth equations (see “Experimental Procedures”). The entire experiment was repeated several times for each combination of the two protein homologues as seed or soluble protein, using different batches of soluble protein and fibril seeds, and generally an excellent fit of the experimental data was obtained in each case (Fig. 5 and data not shown). However, the absolute values obtained, particularly of ξ1, were found to vary by a factor of around 5-fold for a given combination of seed and soluble protein (Table 1). This principally reflects the fact that although the Sc and Sp seed fibrils were produced under identical conditions, it is in general very difficult to ensure that the number of fibril ends in different batches of seeds are identical. In addition, there is no method available to determine accurately the value of L0 (the average number of molecular units in a seed fibril). Therefore, instead of attempting to estimate the absolute values of k+ and k− for each batch of every seed/protein combination, we instead expressed the data as ratios of elongation or breakage rate constants for the two protein homologues, where each ratio was obtained for a single batch of seed. In addition to circumventing the need to estimate the absolute value of L0, a further advantage of the ratio method is that the effect on the rate constants of any experimental errors that are constant within a single parallel cross-seeding experiment but vary between separate experimental runs (e.g. incubation temperature) will tend to cancel out. Indeed, the ratio values thus obtained were found to be highly reproducible among independent repetitions of the same experiment. The mean values obtained for each of the ratios are displayed in Table 2.
TABLE 1.
Parameters related to elongation and breakage rates determined by fitting of solution cross-seeding experiments
The values shown were derived from the global fit (see “Experimental Procedures”) of the data of cross-seeding experiments (as displayed in Fig. 5) and the errors shown are the standard error of the global fit of each experiment. ξ1 = k+/L0 and ξ2 = k+ k−, where k+ and k− are the rate constants for elongation and breakage, respectively, and L0 is the average number of molecular units in a seed fibril, which will be constant for a given batch of seed.
| Seed |
ScUre2p |
|||
|---|---|---|---|---|
| Soluble protein |
ScUre2p |
SpUre2p |
||
| Parameter | ξ1 | ξ2 | ξ1 | ξ2 |
| s−1·m−1 | 10−6·s−2·m−2 | s−1·m−1 | 10−6·s−2·m−2 | |
| Batch | ||||
| I | 3.4 ± 0.2 | 2.4 ± 0.3 | 8.0 ± 0.5 | 1.2 ± 0.3 |
| II | 6.7 ± 0.4 | 2.7 ± 0.2 | 14.6 ± 0.6 | 2.5 ± 0.2 |
| III | 5.7 ± 0.2 | 2.1 ± 0.2 | 10.3 ± 0.5 | 1.1 ± 0.2 |
| Seed |
SpUre2p |
|||
|---|---|---|---|---|
| Soluble protein |
ScUre2p |
SpUre2p |
||
| Parameter | ξ1 | ξ2 | ξ1 | ξ2 |
| s−1·m−1 | 10−6·s−2·m−2 | s−1·m−1 | 10−6·s−2·m−2 | |
| Batch | ||||
| I | 2.4 ± 0.1 | 1.7 ± 0.1 | 6.3 ± 0.3 | 2.2 ± 0.4 |
| II | 11.7 ± 0.5 | 3.9 ± 0.6 | 27.3 ± 1.3 | 7.9 ± 2.9 |
| III | 4.9 ± 0.2 | 5.4 ± 0.3 | 11.2 ± 0.5 | 6.5 ± 0.7 |
TABLE 2.
Ratios of elongation and breakage rate constants of fibrils formed from ScUre2p and SpUre2p determined from cross-seeding experiments
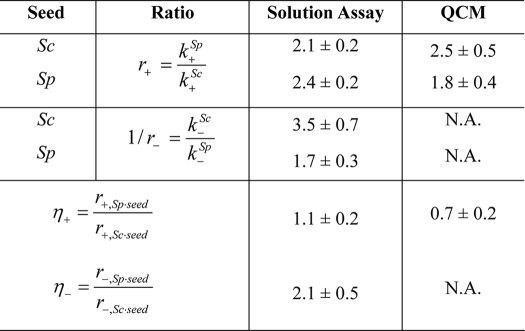
The values shown represent the mean and S.E. derived from three independent experiments. The values were determined as described in “Experimental Procedures,” where r is the ratio of rate constants for the two proteins in solution, for a given type and batch of seed; and η is the ratio of the ratios (see Eq. 6–8). A value of r+ > 1 means that the elongation rate for SpUre2p is greater than for ScUre2p; a value of 1/r− > 1 means that the breakage rate for ScUre2p is greater than for SpUre2p; a value of η+ or η− that deviates from unity indicates that the nature of the protein seed influences the elongation or breakage rate, respectively. N.A., not applicable.
These results reveal that in each case, the values of r+ were observed to be >1 (Table 2), indicating that fibrils grown in a solution of SpUre2p possess a higher elongation rate than those grown in a solution of ScUre2p, regardless of which protein was used as the seed. However, in each case the values of 1/r− were observed to be >1 (Table 2), indicating that SpUre2p fibrils fracture less frequently than ScUre2p fibrils, once again regardless of which protein was used as the seed. Interestingly, the ratios of elongation rates were found to be largely independent of the nature of the seed fibrils (η+ of the order of unity in Table 2), indicating that the elongation reaction is in each case predominantly governed by the nature of the soluble species. On the other hand, the nature of the protein seed was observed to have around a 2-fold effect on the ratios of breakage rates (η− in Table 2); this finding suggests that the mechanical frangibility of fibrils can be encoded into the seed fibrils and can be transmitted through the self-templating action characteristic of the growth process to proteins of a related but differing sequence.
Comparing Kinetic Parameters Describing Fibril Growth for Different Protein Homologues by Biosensor Assay
In solution assays of amyloid growth, measurements are sensitive to different microscopic processes such as fibril elongation and fragmentation, the relative contributions of which have to be deconvoluted from the data describing the bulk reaction. Recently, however, it has become possible to monitor directly individual steps that underlie amyloid growth by the use of biosensors, which allow precise control of the number of growth sites (49). Therefore, in addition to the solution assay, we also probed the elongation of fibrils using a QCM sensor (Fig. 6).
Fibril seeds of ScUre2p or SpUre2p were covalently attached to the surface of the sensor. As in the solution assays, both self-seeded and cross-seeded reactions were monitored. When the protein solution is allowed to flow across the sensor surface, the changes in the resonant frequency of the crystal then accurately report on the nanogram-scale mass changes resulting from the addition of protein molecules to the ends of the fibrils, and the slope of the frequency shift reflects the elongation rate of the protein fibril (Fig. 6, A and B). In addition, the length of the seed fibrils before and after growth can be observed by AFM (Fig. 6C), confirming the origin of the mass increase as the supra-molecular assembly reaction. A comparison of the rates of fibril elongation from ScUre2p and SpUre2p proteins by QCM shows that similar trends are observed to those indicated by the fits to the bulk reactions in solution (Table 2). This validates our solution cross-seeding analysis as able to separate out and compare the individual steps of fibril elongation and breakage for the two protein homologues studied here.
DISCUSSION
Although much progress has been made in understanding the basic molecular steps which underlie protein aggregation and prion propagation, the quantitative characterization of the rates of these individual events has been challenging to achieve. In this report we have addressed this problem with a combination of solution state aggregation assays and biosensor-based measurements. The complexity of prion propagation stems in part from the fact that the three distinct microscopic processes of nucleation, elongation, and fragmentation all contribute to the growth of prion fibrils, and their relative importance for the propagation of prions is not straightforward to evaluate from in vitro bulk kinetic measurements. Mathematical modeling has emerged as a vital tool to extract from kinetic measurements information about the processes of initiation and proliferation of prion-related peptide and protein aggregates, and master equations describing different processes contributing to the overall growth process have been proposed and analyzed (12, 29–30). It has been shown from such analysis that fibril breakage is an important process for the proliferation of prion aggregates (12, 30) and the prediction of the average length is consistent with experimental data only if breakage is incorporated into the mechanism of fibril growth (34). By analyzing the results from the growth assays using an analytical solution (38) of the master equation describing prion fibril growth we have determined the individual contributions of elongation and breakage to Ure2p fibril growth; the results represent a significant advance by providing direct quantitative evidence corroborating the determining role of fragmentation for prion propagation.
The different behavior of ScUre2p and SpUre2p both in vitro and in vivo provide an opportunity to analyze the mechanisms that allow fibril formation to be transmissible. Whereas ScUre2p has been shown to function effectively as a prion in a range of systems, the prion propensity of SpUre2p has remained more elusive and there are contradictory results pertaining to it (26–27, 50–52), an indication that the prion character of SpUre2p is less stable than that of ScUre2p. Our measurements reveal that the differences in the breakage and elongation rates between ScUre2p and SpUre2p vary by only about a factor of two; this finding could signify that SpUre2p is at the limit between non-prion and prion amyloid behavior and its prion propagation therefore can be greatly affected by the environment, such as the expression level of the protein (51). In previous work (28) we have demonstrated that ScUre2p and SpUre2p have similar fibril growth behavior under quiescent conditions. In the presence of agitation, however, the lag time for SpUre2p fibril formation can be reduced more dramatically than for ScUre2p. As agitation serves to increase the rate of fibril breakage, these results suggest differences in the mechanical strength of ScUre2p and SpUre2p aggregates (28). In the present study, we have found that ScUre2p not only showed a higher breakage rate than SpUre2p, but also a lower elongation rate than SpUre2p.
The physical properties of a fibril can depend both on the nature of the seed used as a template and the characteristics of the soluble protein from which it is grown. In some cases, the templating action of the seeds has a dominant role (12, 53–55) whereas in other cases the properties of the protein from which the fibrils are grown emerges as being more important (56). We probed the effects of Sp/Sc cross-seeding and, intriguingly, found that SpUre2p elongates faster and breaks more slowly than ScUre2p under the induction of both Sc and Sp seeds. This result suggests a mechanistic explanation of why no [URE3] clone could be detected in S. paradoxus despite strong overexpression of the ScUre2p PrD (51), as while the ScUre2p PrD may provide additional seeds, the properties of the aggregates formed will still display the growth and fragmentation properties of the host protein.
Comparing the ratios of elongation rates or breakage rates under the induction of different seeds (Table 2) confirms that the ratios of the breakage rates differ when the growth is induced by different seeds, but the ratios of elongation rates are similar under the induction of different seeds. From a kinetic point of view, the strain type of a prion is determined by a combination of the division rate and the growth rate (12). Stable and strong prion strains prevail in given regions of parameter space where proliferation is faster than clearance. Although ScUre2p fibrils elongate more slowly than SpUre2p, their greater breakage rate gives the former the advantage for propagation, probably representing the key determinant for transmissibility. On the other hand, the faster elongation rate but slower breakage rate of SpUre2p results in a low multiplication rate and lower propensity to be transmitted to the next generation, leading to a reduced ability to behave as a prion.
In this study, we have succeeded in comparing the rates of the individual microscopic steps involved in fibril formation for two homologues of Ure2p, allowing the relative contributions of the elongation and breakage rates to the overall rate of fibril growth to be distinguished. The results in the present report highlight the importance of fibril fragmentation in contributing to prion propensity and open up opportunities for understanding from a kinetic point of view the non-transmissible versus transmissible nature of amyloid diseases.
Acknowledgments
We thank Dr. Laura Itzhaki for access to facilities at the Hutchison/MRC Research Centre, Cambridge. We thank the Institute of Biophysics Electron Microscopy Centre for assistance with electron microscopy experiments.
This work was supported in part by grants from the National Natural Science Foundation of China (30620130109, 30670428, 30870482, 31070656), the Chinese Ministry of Science and Technology (2006CB500703, 2006CB910903) and the Chinese Academy of Sciences (KSCX2-YW-R-119, KSCX2-YW-R-256) (to S. P.), as well as support from the IRC in Nanotechnology (to M. E. W. and A. K. B.), the Wellcome and Leverhulme Trusts (to C. M. D.), and St John's College, Cambridge (to T. P. J. K.). The initial stages of this work were supported by a “UK China Partners in Science II”' fund award from the Science and Innovation Section of the Foreign & Commonwealth Office (to T. P. J. K.). This work was facilitated by a Royal Society International Joint Project Grant (to Laura Itzhaki and S. P.).
- PrD
- prion domain
- Sc
- Saccharomyces cerevisiae
- Sp
- Saccharomyces paradoxus
- ThT
- Thioflavin T
- AFM
- atomic force microscopy
- QCM
- quartz crystal microbalance.
REFERENCES
- 1. Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 2. Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aguzzi A., Haass C. (2003) Science 302, 814–818 [DOI] [PubMed] [Google Scholar]
- 4. Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T. (2007) Nat. Rev. Microbiol. 5, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tessier P. M., Lindquist S. (2009) Nat. Struct. Mol. Biol. 16, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saupe S. J. (2007) Prion 1, 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chien P., Weissman J. S. (2001) Nature 410, 223–227 [DOI] [PubMed] [Google Scholar]
- 8. DePace A. H., Weissman J. S. (2002) Nat. Struct. Biol. 9, 389–396 [DOI] [PubMed] [Google Scholar]
- 9. Fay N., Inoue Y., Bousset L., Taguchi H., Melki R. (2003) J. Biol. Chem. 278, 30199–30205 [DOI] [PubMed] [Google Scholar]
- 10. Collins S. R., Douglass A., Vale R. D., Weissman J. S. (2004) PLoS Biol. 2, e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krishnan R., Lindquist S. L. (2005) Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 13. Perrett S., Jones G. W. (2008) Curr. Opin. Struct. Biol. 18, 52–59 [DOI] [PubMed] [Google Scholar]
- 14. Wickner R. B. (1994) Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 15. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. (1995) Science 268, 880–884 [DOI] [PubMed] [Google Scholar]
- 16. Lian H. Y., Jiang Y., Zhang H., Jones G. W., Perrett S. (2006) Biochim. Biophys. Acta 1764, 535–545 [DOI] [PubMed] [Google Scholar]
- 17. Cooper T. G. (2002) FEMS Microbiol. Rev. 26, 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masison D. C., Wickner R. B. (1995) Science 270, 93–95 [DOI] [PubMed] [Google Scholar]
- 19. Taylor K. L., Cheng N., Williams R. W., Steven A. C., Wickner R. B. (1999) Science 283, 1339–1343 [DOI] [PubMed] [Google Scholar]
- 20. Thual C., Bousset L., Komar A. A., Walter S., Buchner J., Cullin C., Melki R. (2001) Biochemistry 40, 1764–1773 [DOI] [PubMed] [Google Scholar]
- 21. Jiang Y., Li H., Zhu L., Zhou J. M., Perrett S. (2004) J. Biol. Chem. 279, 3361–3369 [DOI] [PubMed] [Google Scholar]
- 22. Coschigano P. W., Magasanik B. (1991) Mol. Cell. Biol. 11, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai M., Zhou J. M., Perrett S. (2004) J. Biol. Chem. 279, 50025–50030 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Z. R., Bai M., Wang X. Y., Zhou J. M., Perrett S. (2008) J. Mol. Biol. 384, 641–651 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Z. R., Perrett S. (2009) J. Biol. Chem. 284, 14058–14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baudin-Baillieu A., Fernandez-Bellot E., Reine F., Coissac E., Cullin C. (2003) Mol. Biol. Cell 14, 3449–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Talarek N., Maillet L., Cullin C., Aigle M. (2005) Genetics 171, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Immel F., Jiang Y., Wang Y. Q., Marchal C., Maillet L., Perrett S., Cullin C. (2007) J. Biol. Chem. 282, 7912–7920 [DOI] [PubMed] [Google Scholar]
- 29. Masel J., Jansen V. A. (2004) Biophys. J. 87, 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hall D., Edskes H. (2004) J. Mol. Biol. 336, 775–786 [DOI] [PubMed] [Google Scholar]
- 31. Watzky M. A., Morris A. M., Ross E. D., Finke R. G. (2008) Biochemistry 47, 10790–10800 [DOI] [PubMed] [Google Scholar]
- 32. Stöhr J., Weinmann N., Wille H., Kaimann T., Nagel-Steger L., Birkmann E., Panza G., Prusiner S. B., Eigen M., Riesner D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2409–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lomakin A., Chung D. S., Benedek G. B., Kirschner D. A., Teplow D. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith J. F., Knowles T. P., Dobson C. M., Macphee C. E., Welland M. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Knowles T. P., Fitzpatrick A. W., Meehan S., Mott H. R., Vendruscolo M., Dobson C. M., Welland M. E. (2007) Science 318, 1900–1903 [DOI] [PubMed] [Google Scholar]
- 36. Knowles T. P., Shu W., Devlin G. L., Meehan S., Auer S., Dobson C. M., Welland M. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hovgaard M. B., Dong M., Otzen D. E., Besenbacher F. (2007) Biophys. J. 93, 2162–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knowles T. P., Waudby C. A., Devlin G. L., Cohen S. I., Aguzzi A., Vendruscolo M., Terentjev E. M., Welland M. E., Dobson C. M. (2009) Science 326, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 39. Carulla N., Caddy G. L., Hall D. R., Zurdo J., Gairi M., Feliz M., Giralt E., Robinson C. V., Dobson C. M. (2005) Nature 436, 554–558 [DOI] [PubMed] [Google Scholar]
- 40. Xue W. F., Homans S. W., Radford S. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pöschel T., Brilliantov N. V., Frömmel C. (2003) Biophys. J. 85, 3460–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kunes K. C., Cox D. L., Singh R. R. (2005) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72, 051915 [DOI] [PubMed] [Google Scholar]
- 43. Perrett S., Freeman S. J., Butler P. J., Fersht A. R. (1999) J. Mol. Biol. 290, 331–345 [DOI] [PubMed] [Google Scholar]
- 44. Zhu L., Zhang X. J., Wang L. Y., Zhou J. M., Perrett S. (2003) J. Mol. Biol. 328, 235–254 [DOI] [PubMed] [Google Scholar]
- 45. Schlumpberger M., Wille H., Baldwin M. A., Butler D. A., Herskowitz I., Prusiner S. B. (2000) Protein Sci. 9, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naiki H., Higuchi K., Hosokawa M., Takeda T. (1989) Anal. Biochem. 177, 244–249 [DOI] [PubMed] [Google Scholar]
- 47. Nilsson M. R. (2004) Methods 34, 151–160 [DOI] [PubMed] [Google Scholar]
- 48. Jarrett J. T., Lansbury P. T., Jr. (1993) Cell 73, 1055–1058 [DOI] [PubMed] [Google Scholar]
- 49. White D. A., Buell A. K., Dobson C. M., Welland M. E., Knowles T. P. (2009) FEBS Lett. 583, 2587–2592 [DOI] [PubMed] [Google Scholar]
- 50. Edskes H. K., Wickner R. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, Suppl. 4, 16384–16391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Crapeau M., Marchal C., Cullin C., Maillet L. (2009) Mol. Biol. Cell 20, 2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edskes H. K., McCann L. M., Hebert A. M., Wickner R. B. (2009) Genetics 181, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. (2005) Science 307, 262–265 [DOI] [PubMed] [Google Scholar]
- 54. Jones E. M., Surewicz W. K. (2005) Cell 121, 63–72 [DOI] [PubMed] [Google Scholar]
- 55. Collinge J., Clarke A. R. (2007) Science 318, 930–936 [DOI] [PubMed] [Google Scholar]
- 56. Devlin G. L., Knowles T. P., Squires A., McCammon M. G., Gras S. L., Nilsson M. R., Robinson C. V., Dobson C. M., MacPhee C. E. (2006) J. Mol. Biol. 360, 497–509 [DOI] [PubMed] [Google Scholar]