Abstract
Pneumolysin (PLY) is a cholesterol-binding, pore-forming protein toxin. It is an important virulence factor of Streptococcus pneumoniae and a key vaccine target against pneumococcal disease. We report a systematic structure-driven approach that solves a long-standing problem for vaccine development in this field: detoxification of PLY with retention of its antigenic integrity. Using three conformational restraint techniques, we rationally designed variants of PLY that lack hemolytic activity and yet induce neutralizing antibodies against the wild-type toxin. These results represent a key milestone toward a broad-spectrum protein-based pneumococcal vaccine and illustrate the value of structural knowledge in formulating effective strategies for antigen optimization.
Keywords: Antigen, Bacterial Toxins, Computer Modeling, Immunology, Membrane Proteins, Protein Structure, Site-directed Mutagenesis, Molecular Design, Streptococcus pneumoniae, Cholesterol-dependent Cytolysin
Introduction
Infections by Streptococcus pneumoniae cause an estimated 1 million deaths worldwide, and the development of an accessible and universal vaccine against pneumococcal disease is a global health priority (1). Several efforts are currently under way to design and develop vaccines based on conserved S. pneumoniae protein antigens (2). In contrast to successes with small-molecule drugs, the structure-based design of vaccines is still in its infancy (3). This is understandable given that vaccine antigens are generally complex biological macromolecules. The role of structure-driven methods in vaccinology is poised to expand, however, owing to recent advances in experimental and computational structural biology (3). The vast wealth of information currently available in bioinformatics databases can be mined and applied to the prediction of unknown three-dimensional protein structures as well as the design of new proteins with novel functionality (4–6). Furthermore, emerging molecular dynamics simulation techniques have made it possible to venture beyond static conformational snapshots and into computational modeling of the functionally relevant motions of proteins in their natural environments (7, 8). As proof of concept that these tools are now sophisticated enough to enable vaccine antigen design, we have engineered the major cholesterol-dependent cytolysin of S. pneumoniae into a detoxified form that retains its protective antigenicity.
To date, vaccination strategies against S. pneumoniae have focused on its capsular polysaccharide as the source of protective antigen in vaccine formulations (9). However, this approach covers only a limited number of serotypes, and production issues make polysaccharide conjugates inaccessible in sufficient quantity to the population most in need in developing countries (10). As a result, research efforts are now being directed toward recombinant protein vaccines, which are cheaper, more robust, and easier to manufacture in comparison with conjugates and also have the potential to provide coverage against all serotypes (2, 10).
A leading candidate for such a protein-based vaccine is pneumolysin (PLY)3: it is an important virulence factor of S. pneumoniae (11, 12), is highly conserved across all serotypes (13), is immunogenic in humans (14, 15), and has shown protection in numerous animal models of infection (16–18). However, PLY is a toxin that oligomerizes to form large pores in the membranes of target host cells (19). The native protein is therefore undesirable for use as a human vaccine antigen. Less toxic mutant forms have been described but have generally retained some degree of hemolytic activity (20). Historically, the best characterized of these is PdB (PLY(W433F)), which possesses ∼1% of the hemolytic activity of native PLY (21). More recently, completely inactive PLY mutants have been discovered by scanning mutagenesis (22); however, the mechanism of their attenuation is not well understood.
The objective of this study was to identify and introduce amino acid modifications that completely detoxify PLY by arresting its transitioning to the pore-forming state without compromising its ability to elicit neutralizing antibodies against the native toxin. To achieve this goal, we applied a targeted genetic detoxification approach based on insights into the toxin's putative three-dimensional structure and functional mechanics. The first step in the computational design process was the prediction of atomic resolution structures of PLY in its water-soluble, prepore, and pore-forming states. Contact maps and dynamic domain analyses were then used to formulate strategies for stabilization and detoxification. The resulting protein designs were cloned, purified, and validated experimentally by their ability to elicit antibodies that neutralized the wild-type toxin.
EXPERIMENTAL PROCEDURES
Construction of Structural Models
A model of soluble wild-type PLY was constructed by comparative modeling using a published x-ray crystallographic three-dimensional structure of perfringolysin O (PFO) as the template (Protein Data Bank code 1PFO) (23). Template coordinates were obtained from the RSCB Protein Data Bank (24). Models of the prepore and pore-forming conformations were based on Cα traces of PFO fit into cryo-electron microscopy maps of prepore and pore-forming states of PLY (Protein Data Bank codes 2BK1 and 2BK2) (25). Computational modeling was performed using Discovery Studio® 2.1 (Accelrys). Sequence alignments were generated using the align multiple sequences protocol, and homology models were constructed with MODELLER (26). Twenty-five models of each conformation (soluble, prepore, and pore-forming) were predicted, and the best models of each category were selected based on the probability density function energy scores.
Dynamic Domain Analyses
Dynamic domain analyses were carried out using the Hingefind algorithm (27).
Contact Map Calculation
Contact matrices for the soluble and pore-forming conformations were independently generated using Discovery Studio® 2.1 with the Cα-to-Cα cutoff distance set at 7 Å. A superposition of the two contact maps was then obtained by plotting on common axes.
Cloning
The ply gene was PCR-cloned from the genome of S. pneumoniae strain R36A into the NdeI/XhoI sites of pET-28a (Novagen) for expression of His-tagged protein. Expression constructs for PLY mutants were created from this plasmid template by site-directed mutagenesis using the QuikChange Multi-Site-directed mutagenesis kit (Stratagene). All mutations were verified by sequencing.
Protein Purification
Recombinant His-tagged proteins were expressed in Escherichia coli BL21(DE3) grown in Overnight Express Autoinduction System 1 medium (Novagen) at 25 °C for 24 h. Protein from the soluble fraction was purified using Ni2+-nitrilotriacetic acid-agarose (Qiagen) according to the manufacturer's instructions. Protein purity was assessed by SDS-PAGE (≥85% in all cases).
Differential Scanning Calorimetry (DSC)
DSC measurements were performed on a VP-Capillary DSC microcalorimeter with an autosampler (MicroCal, Northampton, MA). Protein samples were loaded onto a 96-well plate and then loaded automatically and sequentially into a 0.325-ml analysis cell. Samples were heated at a scan rate of 1 °C/min (temperature range of 20–80 °C) at a constant pressure of 42–44 p.s.i. The heat capacity profiles were recorded using VP Viewer Version 1.2 software. The transition temperature and enthalpy of transitions were determined using MicroCal OriginTM Version 7.0 software.
CD Spectroscopy
Samples were analyzed using a Jasco Model 810 CD spectropolarimeter. The CD spectra were recorded in the range of 170–280 nm using a 0.1-cm path length quartz cell. Spectra were obtained by scanning at 50 nm/min. Data were converted to molar ellipticity [θ] expressed in degrees cm2/dmol and plotted using the instrument-specific software.
Hemolysis Assay
Membranolytic activity was measured using an in vitro hemolysis assay. Purified PLY mutants were serially diluted (2-fold) in a round-bottom microtiter plate in Hanks' buffered saline (Invitrogen) supplemented with 0.5% bovine serum albumin (Sigma). Washed sheep red blood cells (Rockland Immunochemicals) were added to each well (1.5% (v/v) final concentration), and the plate was incubated for 30 min at 37 °C. Intact cells were removed by centrifugation, and supernatants were transferred to a flat-bottom plate to measure hemoglobin release by absorbance at 540 nm. Percent lysis values were graphed versus protein concentration (milligrams/milliliter), and specific activity was taken as the inverse of the concentration at which 50% erythrocyte lysis was achieved.
Antiserum Generation
Antisera were generated in rats using protocols approved by an internal Animal Care Committee. Animal handling adhered to all guidelines outlined by the Canadian Council on Animal Care. Antibody titers were assessed by ELISA using wild-type PLY as the coating antigen.
Neutralization Assay
Toxin neutralization was tested using an in vitro hemolysis inhibition assay. Because free cholesterol inhibits PLY activity (28), antisera were first depleted of cholesterol by precipitation with dextran sulfate (Dextralip® 50, Sigma). Antisera were then serially diluted (2-fold) across a round-bottom microtiter plate in Hanks' buffered saline supplemented with 0.5% bovine serum albumin. Wild-type PLY was added to each well (10 ng), and the plate was incubated for 30 min at 37 °C to allow antibody binding. Washed sheep red blood cells were then added to each well (0.75% final concentration), and the plate was incubated for another 30 min at 37 °C. Intact cells were removed by centrifugation, and supernatants were transferred to a flat-bottom plate to measure hemoglobin release by absorbance at 540 nm. Percent lysis values were graphed versus antiserum dilution, and the neutralizing titer was taken as the inverse of the dilution at which 50% inhibition of lysis was achieved.
RESULTS
Prediction of PLY Three-dimensional Structures
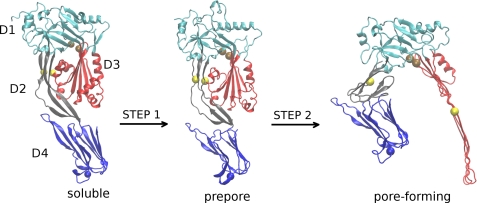
Distinct conformations of PLY were modeled to analyze the series of inter- and intradomain rearrangements that drive its transformation from the water-soluble state to the membrane-associated prepore state and ultimately the membrane-inserted pore-forming state (29). Both low-resolution structures of PLY and high-resolution coordinates of its homolog, PFO, were used as templates to guide model construction (25, 30, 31). These models represent snapshots of key conformational states sampled by a molecule of PLY as it converts from a soluble monomer to an oligomeric transmembrane pore-forming structure (Fig. 1). The transition from the water-soluble monomer to the membrane-bound prepore oligomer is characterized by significant structural reorganization (25). One of the initial steps may involve modest deformation of the D2 domain and the twisting and bending of the D4 domain relative to D1–D3 (Fig. 1, STEP 1). The most remarkable internal structural rearrangements are those associated with the transformation of the prepore state to the membrane-inserted pore-forming conformation (Fig. 1, STEP 2). These rearrangements include crumpling of the D2 β-sheet, separation of D3 from D2, and uncoiling of two helical regions in D3 into an extended membrane-traversing β-sheet that constitutes the interior lining of the pore. Forty-four oligomeric partners, contributing a total of 176 β-strands, assemble into a pore measuring ∼260 Å in diameter (25).
FIGURE 1.
Homology models of three distinct conformations of PLY illustrate a stepwise mechanistic model for the transformation of PLY from the water-soluble to the pore-forming conformation. The structural domains of PLY are labeled D1, D2, D3, and D4, and colored cyan, gray, red, and blue, respectively. For clarity, oligomeric partners and the host cell membrane are excluded from the rendering of the prepore and pore-forming conformations. The locations of some amino acids pairs that were linked via disulfide bridging are shown as spheres. The yellow spheres represent amino acid pair Glu-159 and Thr-348, and the brown spheres represent Thr-65 and Gly-293. The blue sphere represents Cys-428. Rendering was done using VMD (47).
Analyses of Structural Models and Design of Detoxified Mutants
The computationally modeled structures of various conformations of PLY were used to develop strategies for its detoxification. The hypothesis was that PLY could be rendered inactive by trapping it in the soluble or prepore conformation. The three main approaches adopted to prevent structural rearrangement of the toxin to the pore-forming state were the following: (a) introduction of conformation-trapping intramolecular disulfide bridges, (b) engineering of rigidity-conferring mutations at mechanical hinge regions, and (c) introduction of steric hindrance at conserved Gly residue positions.
Conformational Trapping Using Disulfide Linkages
This approach has been previously applied to structural studies on the related toxin PFO (32). It was shown that a mutant form of PFO featuring a disulfide linkage between domains 2 and 3 was able to assemble into a prepore complex but did not convert to the pore-forming state. Similarly, it was demonstrated that disulfide cross-linking of two β-stands in domain 3 of PFO prevented prepore oligomerization and membrane insertion even though membrane binding was not significantly affected (33).
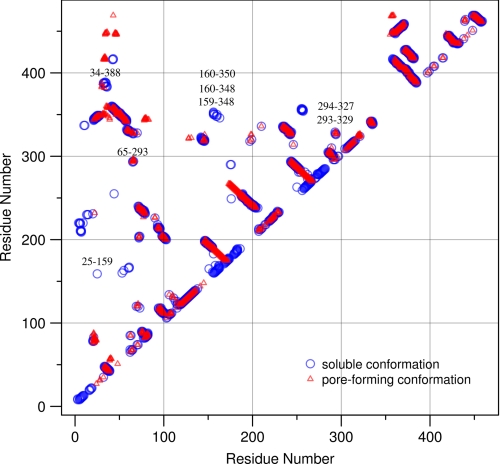
The typical Cα-Cα distance for disulfide-bridged cysteine residues in proteins is reported to be ∼6.5 Å (34, 35). To accommodate inherent protein flexibility, we used a slightly higher Cα-Cα value of 7 Å to calculate contact matrices for both the soluble and pore-forming conformations of PLY (Fig. 2). These matrices enabled the identification of residue pairs that could potentially be linked by disulfide bridges. The suitability of residue pairs as candidates for disulfide-trapping mutations was further assessed by visual examination of the structural models. Particular attention was paid to residue pairs at interdomain boundaries based on the expectation that the introduction of interfacial disulfide bridges would prevent the relative displacement of domains. An important consideration, besides the magnitude of Cα-Cα separation, was the directionality of Cα-Cβ bond vectors. Residue pairs fulfilling one of the following criteria were targeted for mutation to Cys: (i) within the 7-Å Cα-Cα cutoff distance and with side chains suitably oriented for disulfide bonding in the initial soluble state but spatially too distant for disulfide linking in the final pore-forming conformation or (ii) within the disulfide cutoff distance in both the soluble and pore-forming conformations but with side chain orientations suitable for disulfide bridging only in the soluble state. To prevent formation of unintended intermolecular disulfide linkages, the sole free Cys at position 428 of native PLY was mutated to Ala (known to have no effect on toxin activity (36)). Designs 1, 2, and 6–9 in Table 1 fulfilled the first criterion defined above, whereas Designs 3–5 fulfilled the second criterion. It is evident from Table 1 that, in some cases, Cα-Cα separations in the soluble form differ from corresponding distances in the pore-forming state by as much as 50 Å. Such dramatic disparities suggest that disulfide trapping could effectively prevent the structural rearrangements required for pore formation, and hence toxicity, as hypothesized.
FIGURE 2.
Overlay of contact maps of the soluble and pore-forming conformations of PLY. Points shown as blue circles represent residue pairs whose Cα atoms are within 7 Å of each other in the three-dimensional structural model of the soluble form. Red triangles represent residues in similarly close proximity in the pore-forming conformation. Off-diagonal points are particularly noteworthy because they represent close three-dimensional proximity between residues that are distant in the amino acid linear sequence. Isolated blue or red points represent contacts present in only one of the two conformations. The positions of cysteine substitutions in the disulfide-trapped mutants designed in this study are labeled.
TABLE 1.
List of mutants designed in this study
For each design, the amino acid modifications made and the rationale behind the changes are specified.
| Design no. | Mutation(s) | Modified domains | Cα-Cα (Å) |
Rationale | |
|---|---|---|---|---|---|
| Soluble | Pore-forming | ||||
| 1 | G25C, E159C, C428A | D2–D3 | 5.7 | 50.4 | Constrained via disulfide bond |
| 2 | E159C, T348C, C428A | D3–D2 | 6.8 | 52.0 | Constrained via disulfide bond |
| 3 | G294C, L327C, C428A | D3–D1 | 4.9 | 4.2 | Constrained via disulfide bond |
| 4 | G293C, I329C, C428A | D3–D1 | 6.4 | 5.7 | Constrained via disulfide bond |
| 5 | T65C, G293C, C428A | D1–D3 | 5.7 | 4.6 | Constrained via disulfide bond |
| 6 | Q160C, Y350C, C428A | D3–D2 | 6.6 | 56.7 | Constrained via disulfide bond |
| 7 | Q160C, T348C, C428A | D3–D2 | 6.7 | 55.6 | Constrained via disulfide bond |
| 8 | G25C, Q160C, C428A | D2–D3 | 7.7 | 54.0 | Constrained via disulfide bond |
| 9 | K34C G388C | D2–D4 | 4.1 | 18.2 | Constrained via disulfide bond |
| 10 | E29P | D2 | Hinge stiffening | ||
| 11 | E352P | D2 | Hinge stiffening | ||
| 12 | E29P, E352P | D2 | Hinge stiffening | ||
| 13 | G243V | D1 | Introduce steric hindrance at conserved Gly positions | ||
| 14 | G293A | D3 | Introduce steric hindrance at conserved Gly positions | ||
| 15 | G293T | D3 | Introduce steric hindrance at conserved Gly positions | ||
| 16 | G293V | D3 | Introduce steric hindrance at conserved Gly positions | ||
| 17 | G294P | D3 | Introduce steric hindrance at conserved Gly positions | ||
| 18 | G293C | D3 | Introduce steric hindrance at conserved Gly positions | ||
Rigidity-conferring Mutations at Mechanical Hinge Regions
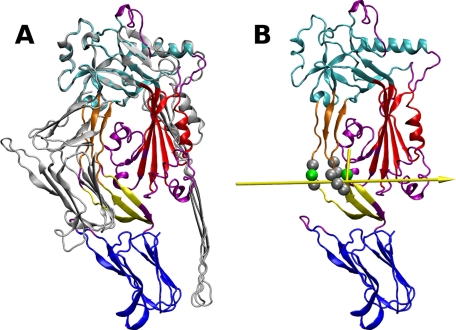
Protein domains that move relative to each other during pore formation and the associated hinge regions were identified using the Hingefind algorithm (27). Coordinates of the soluble and pore-forming conformations were provided as input. Through least-squares superposition of the two configurations, PLY was partitioned into geometrically preserved domains that move as rigid bodies relative to each other (Fig. 3A). Hinge axes between the dynamic domains were then determined. Considering movements between physically connected regions only, the domains colored yellow and orange in Fig. 3 undergo the largest relative displacement. The 117° effective rotation between them reflects the extensive deformation of D2 during pore formation. The interface between these two regions was thus chosen as the focal point for the introduction of hinge-stiffening mutations. A pictorial view of the Hingefind result showing the axis of rotation of the yellow domain relative to the orange domain is presented in Fig. 3B. When designing hinge-stiffening mutants of PLY, candidate amino acids for mutation were selected by taking into account the following: (i) proximity to the hinge axis, (ii) the magnitude of change the backbone dihedral angles undergo in transitioning from the soluble to the pore-forming conformation, and (iii) the compatibility of backbone dihedrals in the soluble state with ϕ and ψ angles typical of Pro. The range of ϕ and ψ angles accessible to Pro is limited by its pentagonal ring that connects the Cα atom to the backbone nitrogen atom. As a result, the ϕ angle is restricted to approximately −60°, whereas the ψ angle clusters around two regions in the Ramachandran plot, −35° (α-region) and +150° (β-region) (37). Listed in Table 2 are backbone dihedral angles of nine residues in closest proximity to the rotation axis described in Fig. 3B. Table 2 shows that Glu-29, Glu-50, and Glu-352 best match the previously stated selection criteria for Pro substitution. Three variants of PLY were thus designed based on the result of the Hingefind analysis: PLY(E29P), PLY(E352P), and PLY(E29P,E352P). These are designated as Designs 10–12 in Table 1. In all three, the modified positions are located in D2, where the backbone flexibility restrictions imposed by Pro are expected to prevent the twisting and bending of the β-sheet linking D1 to D4, which appears to be necessary for the conformational transition from the soluble to the pore-forming state (see Figs. 1 and 3).
FIGURE 3.
Structural transitions of PLY. A, structural alignment of the soluble and pore-forming conformations of PLY. The largest dynamic domain (colored cyan in the soluble conformation) was used as the reference for superposition. Domain partitioning was done using the Hingefind algorithm with a tolerance of 5.0 Å (27). Other dynamic domains identified are shown in blue, red, orange, and yellow, whereas regions that could not be portioned into a dynamic domain are purple. B, the soluble conformation is used to illustrate the movement of the orange domain relative to the yellow domain. The axis of rotation is shown as a yellow arrow, with its orientation representing a left-handed 117° rotation about the axis. The pivot point in the middle of the arrow is connected to domain centroids to illustrate the rotation angle. Residues in close proximity to the axis are rendered as spheres, with the position of the two hinge residues modified to prevent interdomain rotation (Glu-29 and Glu-352) highlighted in green. The images were produced with VMD (47).
TABLE 2.
Mechanical hinges of PLY identified by the Hingefind algorithm
Comparisons of backbone ϕ and ψ dihedral angles (in degrees) for each hinge residue in the soluble and pore-forming conformation are shown.
| Hinge residue |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ile-28 | Glu-29 | Asn-30 | Arg-48 | Lys-49 | Lys-50 | Tyr-350 | Val-351 | Glu-352 | |
| ϕ | |||||||||
| Soluble | −104 | −68 | −112 | −115 | −127 | −79 | −110 | −89 | −82 |
| Pore | −137 | −77 | −56 | −179 | −78 | −144 | −149 | −156 | −41 |
| ψ | |||||||||
| Soluble | 122 | −47 | 136 | 134 | 132 | 116 | 152 | 105 | 110 |
| Pore | 171 | −111 | 120 | −166 | −51 | 106 | −117 | −169 | −43 |
Introduction of Steric Hindrance at Conserved Glycines
Two recent studies have revealed the existence of structural similarity between the bacterial cholesterol-dependent cytolysin family of proteins (to which PLY belongs) and proteins containing membrane attack complex/perforin (MACPF) domains (38, 39). Despite an overall lack of sequence similarity between cholesterol-dependent cytolysin toxins and the MACPF domain, some core structural elements and three Gly residues are conserved (supplemental Fig. S1). In PLY, the conserved glycines correspond to Gly-243, Gly-293, and Gly-294. The structural and functional similarities between these two distinct classes of pore-forming proteins suggest that they share a common mechanism and that the three conserved glycines may play an important role in pore formation (39). These Gly residues were therefore targeted for substitution with amino acids of varying side chain sizes, with the goal of sterically hindering the conformational transition from the soluble to the pore-forming state. Substitution of a small residue like Gly with a larger one was predicted to increase the energy barrier to conformational transition from the soluble to the pore state by reducing backbone dihedral space and restricting the torsional rotation of nearby side chains of surrounding residues. The mutations evaluated for this strategy are listed in Table 1 (Designs 13–18), with the rationale summarized briefly as follows. Because Gly-243 is located at a bend in a long β-strand that connects D1 and D3 (supplemental Fig. S1), it was substituted with Val, a larger amino acid that is also typically a β-strand former and hence likely to preserve local secondary structure (40). On the other hand, Gly-293 and Gly-294 are located in a loop connecting two short β-strands in D3. Several amino acids (Ala, Thr, Val, and Cys) were substituted for Gly-293 to explore the effect of side chain size. The G294P mutation was inspired by previous studies with unrelated proteins demonstrating that Pro residues in loop regions increase conformational stability by enhancing the rigidity of the loop (41, 42).
Generation of Recombinant Proteins
Mutant proteins were expressed and purified using standard techniques (see “Experimental Procedures”). Initially, the quality of the mutant proteins was assessed by comparing their DSC profiles with those obtained using the wild-type active protein (supplemental Table S1). In the case of disulfide-constrained mutants, it was also possible to show the maintenance of correct overall conformation by reducing the disulfide constraints and demonstrating the reacquisition of hemolytic activity (supplemental Fig. S2). Ultimately, the ability to elicit antibodies that neutralized native PLY was the critical test of mutant protein quality (discussed below).
Toxin Activity of Mutant PLY Proteins
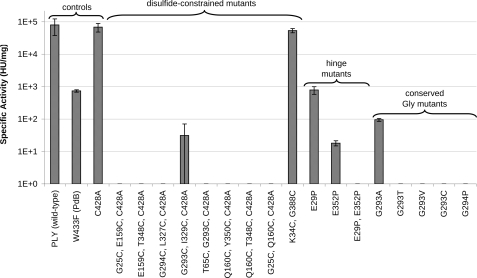
The toxin activity of the PLY mutants was measured using a hemolysis assay designed to monitor the pore-induced release of hemoglobin from red blood cells by absorbance spectroscopy (see “Experimental Procedures”). Specific hemolytic activities are summarized in Fig. 4, with results grouped according to class of mutation. In agreement with published data (21), PdB was 100-fold less active than wild-type PLY, and the mutation C428A (36) did not affect toxin activity. Consequently, PdB hemolytic activity was used as the benchmark for improvement, and the PLY(C428A) substitution was incorporated into the disulfide-constrained designs to prevent the unwanted formation of disulfides with the endogenous Cys. Most of the novel PLY mutants were less hemolytic than PdB, and many were completely inactive (≥100,000-fold less active than wild-type PLY, below the assay limit of detection) (Fig. 4).
FIGURE 4.
Specific activity of engineered PLY mutants under nonreducing conditions. An absence of a gray bar indicates undetectable levels of activity. For activity under reducing conditions, see supplemental Fig. S2. HU, hemolytic units.
Antigenic Integrity of Mutant PLY Proteins
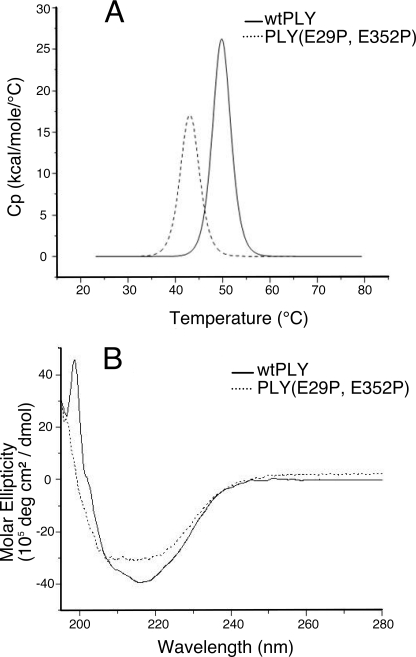
To test whether the attenuated mutants were still able to elicit neutralizing antibodies, antisera were generated against these proteins. Immunogenicity of the mutant proteins was confirmed by determination of PLY-specific IgG titers and by testing their ability to neutralize the wild-type toxin (see “Experimental Procedures”). All mutants elicited IgG titers similar to wild-type PLY (supplemental Table S2). Neutralization titers of antisera were similar to that of the control serum raised against wild-type PLY with the exception of two mutants; the neutralization titers of anti-PLY(E352P) and anti-PLY(E29P,E352P) antisera were 4- and 64-fold less effective than that of the control serum, respectively (supplemental Table S2). This observation suggests that PLY(E352P) and PLY(E29P,E352P) may have adopted non-native conformations compared with wild-type PLY and would thus be less effective vaccine antigens. This interpretation is supported by thermal stability data obtained by DSC: the thermal transition midpoint temperature (Tm) of PLY(E29P,E352P) was nearly 7 °C lower than that of wild-type PLY (Tm = 43.0 °C versus 49.8 °C for wild-type PLY) (Fig. 5A). In addition, the profile of the PLY(E29P,E352P) thermal transition (Fig. 5A) suggests that the protein would be at least partially unfolded at the body temperature of the immunized rats. This is further supported by CD spectroscopy experiments that showed PLY(E29P,E352P) had a different secondary structural profile compared with wild-type PLY (Fig. 5B). Based on these results, PLY(E352P) and PLY(E29P,E352P) were deemed to be structurally compromised and unsuitable as vaccine candidates.
FIGURE 5.
A, overlay of DSC thermograms for wild-type PLY (solid line) and mutant PLY(E29P,E352P) (dashed line). Tm values were 49.8 °C and 43.0 °C, respectively. Cp, specific heat capacity at constant pressure. B, overlay of CD spectra for wild-type PLY (solid line) and mutant PLY(E29P,E352P) (dashed line). deg, degrees.
DISCUSSION
Using a systematic structure-driven approach, we have solved a long-standing problem in pneumococcal vaccine development: detoxification of the key virulence factor PLY with retention of its antigenic integrity. Our objective was to identify and introduce modifications into PLY that could prevent its transition from the soluble to the pore-forming state while retaining its ability to elicit neutralizing antibodies. In all, 11 of our designs met these criteria.
The premise that Gly-293 would be critical to PLY activity was inspired by the recently demonstrated commonality of structural folds between cholesterol-dependent cytolysins and MACPF domain proteins and the observation that specific Gly residues also appeared to be conserved between these protein families (38, 39). This hypothesis was validated by experiments in which we substituted Gly-293 and Gly-294 with side chains larger than Ala to generate mutants that lost hemolytic activity (Fig. 4). Both Gly residues are well conserved in cholesterol-dependent cytolysins and MACPF domain proteins (38, 39). Therefore, for this class of mutants, we propose a mechanism of inactivation in which large side chains introduce steric interference. This would impede the conformational progression of PLY to the competent pore-forming state. The structural model of PLY indicates that large amino acid residues at position 293 are subject to steric hindrance from three neighboring side chains: Thr-65, Asn-66, and Ile-329. Extrapolating our findings, we predict that the pore-forming activity of MACPF proteins would be similarly impeded by the introduction of large residues at the positions equivalent to Gly-293 and Gly-294 of PLY (e.g. Gly-270 and Gly-271 in Photorhabdus luminescens MACPF).
Computer models of the multiple PLY conformational states were instrumental to our antigen-engineering efforts involving disulfide bonds. Of nine in silico designs featuring disulfide constraints, seven were rendered completely inactive, another had its hemolytic activity reduced by 3 orders of magnitude relative to wild-type PLY, and one mutant (PLY(K34C,G388C)) was not attenuated as intended (Fig. 4). Exposure of these mutants to reducing conditions led to the recovery of hemolytic activity where expected (supplemental Fig. S2). We conclude from these data that the mechanism of detoxification involves the disulfide trapping of PLY in conformations that are incapable of proceeding through one or more steps of association and assembly into oligomeric membrane-spanning pores. Of particular note is the disulfide-constrained mutant PLY(T65C,G293C,C428A), which seems to be attenuated via a dual mechanism, one aspect of which depends on disulfide trapping and the other of which depends on alteration of the conserved Gly-293 residue. Such redundancy in detoxification mechanisms is highly attractive from a vaccine safety point of view. The successful formation of a disulfide bond in this mutant was verified by DSC (supplemental Table S1) and free thiol-scavenging experiments using Ellman's reagent (data not shown). We attribute its lack of activity under reducing conditions to the effect of the G293C mutation alone (Fig. 4) because T65C by itself does not affect PLY activity (data not shown). Also noteworthy is PLY(G293C,I329C,C428A), whose hemolytic activity was 3 orders of magnitude lower than that of wild-type PLY under nonreducing conditions. The partial inactivation of this mutant indicates that the Cys-293–Cys-329 disulfide bond impedes but does not completely prevent conformational changes required for membrane insertion. In addition, a disulfide bond between residues 293 and 329 is predicted to pull the side chain of residue 293 away from two of its nearest and largest neighbors, Thr-65 and Asn-66, thus reducing steric hindrance to pore-forming conformational change. Upon reduction of the disulfide bond, restoration of steric interference between the side chains of Cys-293, Thr-65, and Asn-66 results in complete inactivation. We therefore conclude that steric effects resulting from the G293C modification are a more effective inactivation mechanism than the linking of D3 domain residue 293 to D1 domain residue 329 via a disulfide bond.
The final class of mutants in this study had rigidity-conferring Pro substitutions introduced at hinge residue sites as a strategy for detoxification. Although computational tools for dynamic domain and hinge prediction have been in existence for over a decade, examples of the practical application of these tools in protein design are rare (27). All PLY mutants designed based on this approach showed reduced hemolytic activity, with PLY(E29P,E352P) being completely attenuated. Despite the success of this strategy in attenuating toxin activity, the concomitant decrease in neutralizing antibodies elicited by PLY(E352P) and PLY(E29P,E352P) makes these mutations less interesting from a vaccine development perspective.
Achievement of safety and efficacy is the ultimate challenge in vaccinology. Antigen modifications intended to eliminate toxicity may have unexpected effects on the correct presentation of protective epitopes. Therefore, our engineered PLY mutants were prioritized not only by loss of hemolytic activity but also by their ability to elicit neutralizing antibodies. PLY(T65C,G293C,C428A), which is attenuated by a dual mechanism and elicits the desired antibody response, is currently in Phase I clinical trials.
Although genetic detoxification of bacterial toxins has been reported (43, 44), the application of rigorous structure-driven methods to identify suitable mutation sites is yet to be fully exploited. As demonstrated herein, structure-based antigen design is a viable approach for eliminating toxin activity while retaining the features necessary for inducing neutralizing antibodies. Historically, chemical methods like formalin inactivation have been used to detoxify virulence factors (45). However, these methods are less desirable from a regulatory and production standpoint due to their nonspecific nature. A systematic structure-guided approach has the potential to save time and resources in vaccine development and holds promise for making progress on difficult but critical antigen targets that have thus far defied traditional approaches (46).
Acknowledgments
J. A. Y. thanks Jan Switzer for excellent technical assistance. B. C. thanks Elena Newman for excellent technical help with the DSC and CD experiments. We are grateful to Dr. Robert Charlebois for critical reading of the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Figs. S1 and S2, Tables S1 and S2, and additional references.
- PLY
- pneumolysin
- PFO
- perfringolysin O
- DSC
- differential scanning calorimetry
- MACPF
- membrane attack complex/perforin.
REFERENCES
- 1. World Health Organization (2003) Wkly. Epidemiol. Rec. 78, 110–119 [Google Scholar]
- 2. Paton J. C., Boslego J. W. (2008) in Pneumococcal Vaccines: The Impact of Conjugate Vaccine (Siber G. R., Klugman K. P., Makela P. H. eds) pp. 421–436, ASM Press, Washington, D. C [Google Scholar]
- 3. Dormitzer P. R., Ulmer J. B., Rappuoli R. (2008) Trends Biotechnol. 26, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berman H. M., Westbrook J. D., Gabanyi M. J., Tao W., Shah R., Kouranov A., Schwede T., Arnold K., Kiefer F., Bordoli L., Kopp J., Podvinec M., Adams P. D., Carter L. G., Minor W., Nair R., La Baer J. (2009) Nucleic Acids Res. 37, D365–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dill K. A., Ozkan S. B., Shell M. S., Weikl T. R. (2008) Annu. Rev. Biophys. 37, 289–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schueler-Furman O., Wang C., Bradley P., Misura K., Baker D. (2005) Science 310, 638–642 [DOI] [PubMed] [Google Scholar]
- 7. Ash W. L., Zlomislic M. R., Oloo E. O., Tieleman D. P. (2004) Biochim. Biophys. Acta 1666, 158–189 [DOI] [PubMed] [Google Scholar]
- 8. Lindahl E., Sansom M. S. (2008) Curr. Opin. Struct. Biol. 18, 425–431 [DOI] [PubMed] [Google Scholar]
- 9. Pletz M. W., Maus U., Krug N., Welte T., Lode H. (2008) Int. J. Antimicrob. Agents 32, 199–206 [DOI] [PubMed] [Google Scholar]
- 10. Black S., Eskola J., Whitney C., Shinefiled H. (2008) in Vaccines (Plotkin S. A., Orenstein W., Offit P. eds) pp. 531–567, W. B. Saunders Co., Philadelphia [Google Scholar]
- 11. Paton J. C. (1996) Trends Microbiol. 4, 103–106 [DOI] [PubMed] [Google Scholar]
- 12. Feldman C., Munro N. C., Jeffery P. K., Mitchell T. J., Andrew P. W., Boulnois G. J., Guerreiro D., Rohde J. A., Todd H. C., Cole P. J. (1991) Am. J. Respir. Cell Mol. Biol. 5, 416–423 [DOI] [PubMed] [Google Scholar]
- 13. Mitchell T. J., Mendez F., Paton J. C., Andrew P. W., Boulnois G. J. (1990) Nucleic Acids Res. 18, 4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrante A., Rowan-Kelly B., Paton J. C. (1984) Infect. Immun. 46, 585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paton J. C., Ferrante A. (1983) Infect. Immun. 41, 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexander J. E., Lock R. A., Peeters C. C., Poolman J. T., Andrew P. W., Mitchell T. J., Hansman D., Paton J. C. (1994) Infect. Immun. 62, 5683–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lock R. A., Paton J. C., Hansman D. (1988) Microb. Pathog. 5, 461–467 [DOI] [PubMed] [Google Scholar]
- 18. Paton J. C., Lock R. A., Hansman D. J. (1983) Infect. Immun. 40, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert R. J. (2002) Cell. Mol. Life Sci. 59, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berry A. M., Alexander J. E., Mitchell T. J., Andrew P. W., Hansman D., Paton J. C. (1995) Infect. Immun. 63, 1969–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korchev Y. E., Bashford C. L., Pederzolli C., Pasternak C. A., Morgan P. J., Andrew P. W., Mitchell T. J. (1998) Biochem. J. 329, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirkham L. A., Kerr A. R., Douce G. R., Paterson G. K., Dilts D. A., Liu D. F., Mitchell T. J. (2006) Infect. Immun. 74, 586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossjohn J., Feil S. C., McKinstry W. J., Tweten R. K., Parker M. W. (1997) Cell 89, 685–692 [DOI] [PubMed] [Google Scholar]
- 24. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tilley S. J., Orlova E. V., Gilbert R. J., Andrew P. W., Saibil H. R. (2005) Cell 121, 247–256 [DOI] [PubMed] [Google Scholar]
- 26. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]
- 27. Wriggers W., Schulten K. (1997) Proteins 29, 1–14 [PubMed] [Google Scholar]
- 28. Nöllmann M., Gilbert R., Mitchell T., Sferrazza M., Byron O. (2004) Biophys. J. 86, 3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walz T. (2005) Mol. Cell 18, 393–394 [DOI] [PubMed] [Google Scholar]
- 30. Rossjohn J., Polekhina G., Feil S. C., Morton C. J., Tweten R. K., Parker M. W. (2007) J. Mol. Biol. 367, 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polekhina G., Giddings K. S., Tweten R. K., Parker M. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hotze E. M., Wilson-Kubalek E. M., Rossjohn J., Parker M. W., Johnson A. E., Tweten R. K. (2001) J. Biol. Chem. 276, 8261–8268 [DOI] [PubMed] [Google Scholar]
- 33. Ramachandran R., Tweten R. K., Johnson A. E. (2004) Nat. Struct. Mol. Biol. 11, 697–705 [DOI] [PubMed] [Google Scholar]
- 34. Reiter Y., Brinkmann U., Jung S. H., Pastan I., Lee B. (1995) Protein Eng. 8, 1323–1331 [DOI] [PubMed] [Google Scholar]
- 35. Sowdhamini R., Srinivasan N., Shoichet B., Santi D. V., Ramakrishnan C., Balaram P. (1989) Protein Eng. 3, 95–103 [DOI] [PubMed] [Google Scholar]
- 36. Saunders F. K., Mitchell T. J., Walker J. A., Andrew P. W., Boulnois G. J. (1989) Infect. Immun. 57, 2547–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. MacArthur M. W., Thornton J. M. (1991) J. Mol. Biol. 218, 397–412 [DOI] [PubMed] [Google Scholar]
- 38. Hadders M. A., Beringer D. X., Gros P. (2007) Science 317, 1552–1554 [DOI] [PubMed] [Google Scholar]
- 39. Rosado C. J., Buckle A. M., Law R. H., Butcher R. E., Kan W. T., Bird C. H., Ung K., Browne K. A., Baran K., Bashtannyk-Puhalovich T. A., Faux N. G., Wong W., Porter C. J., Pike R. N., Ellisdon A. M., Pearce M. C., Bottomley S. P., Emsley J., Smith A. I., Rossjohn J., Hartland E. L., Voskoboinik I., Trapani J. A., Bird P. I., Dunstone M. A., Whisstock J. C. (2007) Science 317, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 40. Chou P. Y., Fasman G. D. (1974) Biochemistry 13, 222–245 [DOI] [PubMed] [Google Scholar]
- 41. Choi E. J., Mayo S. L. (2006) Protein Eng. Des. Sel. 19, 285–289 [DOI] [PubMed] [Google Scholar]
- 42. Vieille C., Burdette D. S., Zeikus J. G. (1996) Biotechnol. Annu. Rev. 2, 1–83 [DOI] [PubMed] [Google Scholar]
- 43. Giannini G., Rappuoli R., Ratti G. (1984) Nucleic Acids Res. 12, 4063–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Del Giudice G., Rappuoli R. (1999) Schweiz. Med. Wochenschr. 129, 1744–1748 [PubMed] [Google Scholar]
- 45. Rappuoli R. (1994) Vaccine 12, 579–581 [DOI] [PubMed] [Google Scholar]
- 46. Douek D. C., Kwong P. D., Nabel G. J. (2006) Cell 124, 677–681 [DOI] [PubMed] [Google Scholar]
- 47. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]