Abstract
SmgGDS is an atypical guanine nucleotide exchange factor (GEF) that promotes both cell proliferation and migration and is up-regulated in several types of cancer. SmgGDS has been previously shown to activate a wide variety of small GTPases, including the Ras family members Rap1a, Rap1b, and K-Ras, as well as the Rho family members Cdc42, Rac1, Rac2, RhoA, and RhoB. In contrast, here we show that SmgGDS exclusively activates RhoA and RhoC among a large panel of purified GTPases. Consistent with the well known properties of GEFs, this activation is catalytic, and SmgGDS preferentially binds to nucleotide-depleted RhoA relative to either GDP- or GTPγS-bound forms. However, mutational analyses indicate that SmgGDS utilizes a distinct exchange mechanism compared with canonical GEFs and in contrast to known GEFs requires RhoA to retain a polybasic region for activation. A homology model of SmgGDS highlights an electronegative surface patch and a highly conserved binding groove. Mutation of either area ablates the ability of SmgGDS to activate RhoA. Finally, the in vitro specificity of SmgGDS for RhoA and RhoC is retained in cells. Together, these results indicate that SmgGDS is a bona fide GEF that specifically activates RhoA and RhoC through a unique mechanism not used by other Rho family exchange factors.
Keywords: Exchange, Exchangers, Ras, Rho, Signal Transduction, GEF, Guanine Nucleotide Exchange Factor, RhoA, RhoC
Introduction
Rho family GTPases play essential signaling roles in a wide variety of cellular processes ranging from migration (1), to cell cycle regulation (2), neurite outgrowth (1, 3), exocytosis (4), and mitosis (5). As nucleotide-dependent switches, Rho GTPases exist in two states: an inactive GDP-bound state and an active GTP-bound state in which they signal to downstream partners. Rho family guanine nucleotide exchange factors (RhoGEFs)2 stimulate the activation of Rho GTPases by promoting the dissociation of GDP from the inactive GTPase and the subsequent binding of GTP. High resolution structures have allowed elucidation of the mechanism of nucleotide exchange for eukaryotic exchange factors of the Dbl (6, 7) and DOCK (8) families as well as bacterial exchange factors such as SopE (9) and IpgB2 (10). SmgGDS is a eukaryotic GEF composed of entirely armadillo (ARM) repeats that does not belong to either the Dbl or DOCK family (11). Despite its early identification as a GEF (12, 13), details of the mechanism by which SmgGDS stimulates nucleotide exchange remain sparse.
SmgGDS was originally characterized as activating multiple GTPases, including Ras family members Rap1A (14), Rap1B (13), and K-ras (14), as well as the Rho family members Rac1 (15), Rac2 (16), Cdc42 (17), RhoA (12), and RhoB (12). However, early experiments using SmgGDS were performed with crudely purified protein samples (12, 13) and yielded inconsistent reports of GTPase specificity. In addition, some reports claimed that SmgGDS only activated prenylated small GTPases (14, 18) whereas others reported activity independent of prenylation state (16, 19). SmgGDS has only been well characterized for its ability to activate RhoA (19), but nothing is known about the regions of SmgGDS necessary to catalyze exchange.
The biological function of SmgGDS has also been questioned. SmgGDS has not yet been shown to activate any small GTPases in vivo. An alternative function of controlling the nucleocytoplasmic shuttling of small GTPases has been proposed (20). Despite the unknowns surrounding the function of SmgGDS, it has been implicated in a number of disease states. For example, SmgGDS has been found to be up-regulated in both prostate cancer (21) and non-small cell lung carcinoma (22) where it promotes both proliferation and migration.
This study revisits the GTPase specificity of SmgGDS using highly purified proteins and for the first time implicates specific residues of SmgGDS as being necessary for exchange upon small GTPases. We show that SmgGDS is solely able to activate RhoA and RhoC using in vitro nucleotide exchange assays. The activation depends on the presence of an intact polybasic region on the C terminus of the RhoA and does not utilize the canonical mechanism of nucleotide exchange seen with Dbl family exchange factors. SmgGDS also requires an electronegative patch and a highly conserved binding pocket on its surface to activate RhoA. Finally, we show that transfection of cells with SmgGDS leads to specific increases in the levels of active RhoA and RhoC, but not other GTPases, including RhoB.
EXPERIMENTAL PROCEDURES
Molecular Constructs
SmgGDS-558 (GenBank accession: NM_174666, bovine) was kindly provided by L. Quilliam. SmgGDS (GenBank accession: NM_001100426) was obtained from the IMAGE Consortium. PCR amplification was used to subclone each isoform into a modified pET-21a vector (Novagen) using a ligation-independent cloning strategy (LIC) (23). The pLIC-His vector expresses an N-terminal His6 tag, a tobacco etch virus cleavage site, and the inserted protein construct. Rho and Ras family GTPases were cloned into bacterial expression vectors as described previously (6, 24, 25). The mammalian expression constructs of human SmgGDS-558 (GenBank accession: NP_001093899) and SmgGDS (GenBank accession: NP_001093897) in pcDNA3.1+ (Invitrogen) were kind gifts from C. Williams. Point mutations in constructs were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and were verified by DNA sequencing of the entire open reading frame.
Protein Expression and Purification
SmgGDS and GTPase constructs were transformed into BL21(DE3) Escherichia coli, grown at 37 °C in LB medium supplemented with 0.1 mg/ml ampicillin to an A600 of 0.6–0.8, and then induced for 15–18 h with 200 μm IPTG. Cells were harvested and lysed, and His6-tagged proteins were purified via Ni2+ affinity chromatography. The His6 tag was cleaved from SmgGDS and RhoA with tobacco etch virus protease. Proteins were further purified with size-exclusion chromatography. Proteins were then concentrated, and the final concentration was determined using A280 and the extinction coefficient as calculated by ProtParam (ExPASy server) (26) prior to storage at −80 °C.
Gel Filtration Binding Assay
SmgGDS was incubated on ice for 30 min with a 2-fold molar excess of RhoA in buffer containing 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 5% glycerol, 1 mm DTT, and either 25 mm EDTA (nucleotide-free buffer), 2 mm MgCl2, and 30 μm GDP (GDP buffer), or 2 mm MgCl2 and 30 μm GTPγS (GTPγS buffer). Protein was then separated using a Superdex-75 gel filtration column (GE Healthcare), and fractions were analyzed via SDS-PAGE.
Guanine Nucleotide Exchange Assays
The ability of SmgGDS to catalyze guanine nucleotide exchange was determined with a MANT-GDP loading assay as described previously (27). Exchange assays were performed with a LS-55 fluorescence spectrometer (PerkinElmer Life Sciences) with λex = 360 nm and λem = 430 nm and slits of 5 nm. The exchange assay buffer was 50 mm NaCl, 20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 5% glycerol, 1 mm DTT, and 400 nm MANT-GDP. Dissociation of nucleotide from RhoA preloaded with MANT-GDP was measured in a buffer containing 50 mm NaCl, 20 mm Tris-HCl, pH 8.0, 100 μm MgCl2, and 100 μm free GDP.
Homology Modeling
Because there is not a template structure in the Protein Data Bank with enough ARM repeats to model all of SmgGDS, the SmgGDS sequence was broken into four sections of overlapping ARM repeats (1–184, 118–362, 254–528, and 472–608), and a homology model of each section was created using β-catenin (Protein Data Bank ID code 2Z6G) as a template (28). Alignment of SmgGDS repeats to β-catenin was computed with the HHpred server, and homology models were created using the InsightII molecular modeling package (Accelrys, San Diego, CA). Overlapping repeats were superimposed to yield a single model of SmgGDS that was analyzed with Profiles3D, yielding a score of 0.87, indicating the model characteristics are similar to known protein structures (29). The electrostatic surface potential of the homology model was calculated using PBEQ Solver (30) and displayed using PyMOL. A multiple sequence alignment of SmgGDS sequences from 23 different species was aligned using ClustalX (31). The clustal consensus scores from the multiple sequence alignment were mapped onto the surface of SmgGDS and colored according to degree of sequence conservation.
Rho and Rac Family Activity Assays
Rho family activity assays were performed as described previously (32). SmgGDS constructs were transfected into HEK293 cells using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Cells were grown for 24 h and lysed in 50 mm Tris-HCl, pH 7.6, 500 mm NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 10 mm MgCl2, 200 μm orthovanadate, and protease inhibitors to assess Rho activity and 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Triton X-100, 10 mm MgCl2, 200 μm orthovanadate, and protease inhibitors to assess Rac activity. Lysates were clarified by centrifugation, equalized for total volume and protein concentration, and rotated for 30 min with 30 μg of purified GST-RBD (Rho binding domain of either Rhotekin for Rho or PAK for Rac bound to glutathione-Sepharose beads). The bead pellets were washed in 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Triton X-100, 10 mm MgCl2, 200 μm orthovanadate, with protease inhibitors, and subsequently processed for SDS-PAGE.
Western Blotting
Cell lysates were subjected to SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). For Western blotting, membranes were incubated with primary antibody overnight at 4 °C and secondary antibodies for 1 h at room temperature. Pan-Rho and Rac1 antibodies were obtained from BD Transduction. RhoA, B, and C specific antibodies were obtained from Santa Cruz Biotechnology. Blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and visualized using Kodak BioMax film (Kodak).
Circular Dichroism
Purified SmgGDS proteins at a concentration of 0.15 mg/ml in 20 mm sodium phosphate, pH 7.5, were analyzed by circular dichroism spectroscopy at 25 °C using a Pistar-180 spectrometer with a 0.5 mm path length. Data were plotted as mean residue ellipticity as a function of wavelength and were analyzed for α-helical content using CDPro software.
RESULTS
SmgGDS Specifically Activates RhoA and RhoC
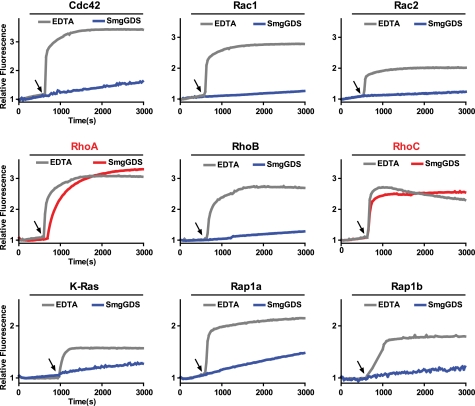
Previous studies of the specificity of SmgGDS are contradictory. Furthermore, these studies indicate that SmgGDS can activate GTPases from both the Rho and Ras families, a highly unusual property for a GEF. Consequently, a panel of small GTPases previously reported to be SmgGDS substrates was purified to homogeneity and tested for its capacity to be activated by purified SmgGDS (Fig. 1). In this assay, SmgGDS increased the nucleotide exchange rate for RhoA and RhoC but not for other Rho or Ras family GTPases. All tested GTPases retained the capacity to exchange nucleotide upon the addition of EDTA, a small molecule chelator of Mg2+, which is an essential co-factor for nucleotide binding and hydrolysis. These results strongly indicate that SmgGDS specifically activates RhoA and RhoC relative to other GTPase.
FIGURE 1.
Purified SmgGDS specifically activates RhoA and RhoC in vitro. The intrinsic exchange of the indicated GTPase (1 μm) was measured for 600 s in exchange buffer. At the indicated time (arrow), SmgGDS (50 μm) or EDTA (25 μm) was added to stimulate nucleotide exchange.
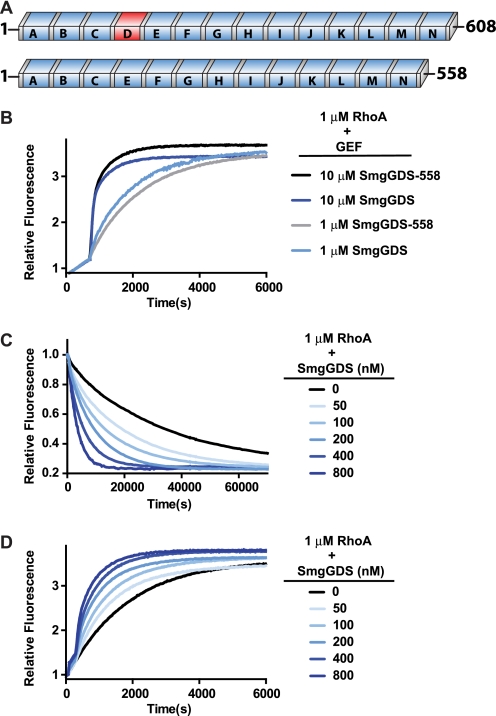
Activation of RhoA by SmgGDS Is Catalytic and Independent of SmgGDS Isoform
Two major splice variants of SmgGDS are expressed in human tissues, a 608-amino acid protein (SmgGDS) containing 14 ARM repeats and a 558-amino acid protein (SmgGDS-558) containing 13 ARM repeats (Fig. 2A). The fourth ARM repeat is not present in the shorter isoform. There is no significant difference in the activation of RhoA by SmgGDS-558 and SmgGDS, indicating that the isoforms have equivalent nucleotide exchange abilities in vitro (Fig. 2B). Also, no difference was detected in GTPase specificity between SmgGDS and SmgGDS-558 (data not shown). Thus, the remainder of the in vitro exchange assays in this paper utilize the longer SmgGDS isoform.
FIGURE 2.
Activation of RhoA by SmgGDS is catalytic and independent of SmgGDS isoform. A, domain architecture of SmgGDS isoforms is shown with conserved ARM repeats (blue boxes) and inserted repeat (red). B, intrinsic exchange of RhoA (1 μm) was measured for 600 s in exchange buffer before stimulation of nucleotide exchange with the indicated concentration of SmgGDS or SmgGDS-558. C, MANT-GDP-loaded RhoA (1 μm) was incubated in unloading exchange buffer for 600 s before addition of SmgGDS at the indicated concentrations to stimulate nucleotide exchange. D, RhoA (1 μm) was incubated in unloading exchange buffer with 400 nm MANT-GDP in place of the free GDP for 600 s before addition of SmgGDS at the indicated concentrations to stimulate nucleotide exchange.
Consistent with the ability of GEFs to stimulate both the loading and unloading of guanine nucleotides at catalytic concentrations, SmgGDS catalyzes the unloading of MANT-GDP from RhoA as indicated by a decrease in fluorescence, as well as the loading of MANT-GDP in a concentration-dependent fashion (Fig. 2, C and D). The slower unloading rates likely reflect: (i) the slower off-rate of MANT-labeled nucleotides from RhoA and (ii) the potential of the fluorophore to interfere with exchange factor binding (33). Due to these potential caveats, loading of labeled nucleotide is the preferred method to examine SmgGDS function and is used in all subsequent studies.
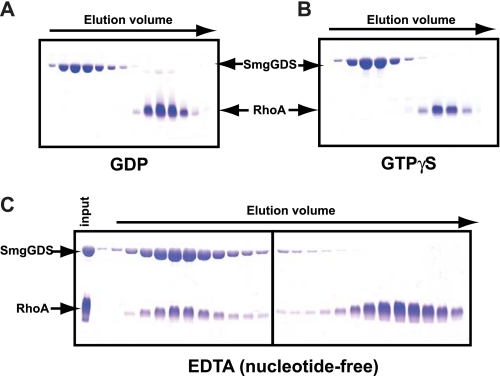
SmgGDS Preferentially Forms a High Affinity Complex with Nucleotide-free RhoA
GEFs directly and preferentially stabilize nucleotide-free GTPases relative to either GDP- or GTP-bound forms. Consequently, size-exclusion chromatography was used to test the capacity of SmgGDS to interact differentially with RhoA bound to either GDP or GTPγS or stripped of nucleotide with EDTA (Fig. 3). Under these conditions, SmgGDS forms a stable complex with only nucleotide-depleted RhoA consistent with a bona fide GEF.
FIGURE 3.
SmgGDS preferentially forms a high affinity complex with nucleotide-depleted RhoA possessing a polybasic region. SmgGDS was incubated on ice for 30 min with a 2-fold molar excess of RhoA loaded with GDP (A), GTPγS (B), or in the presence of EDTA (C), separated over size-exclusion chromatography, and fractions were analyzed via SDS-PAGE.
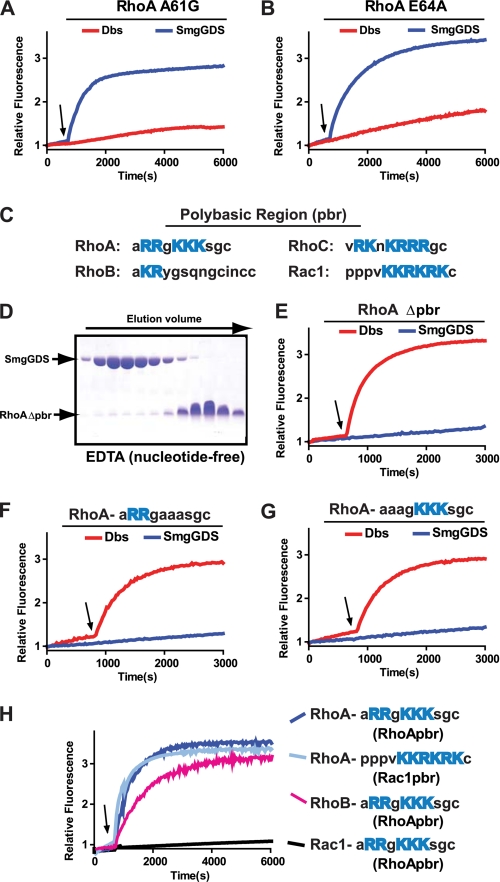
SmgGDS Has a Unique Exchange Mechanism Compared with Dbl Family RhoGEFs and Bacterial Exchange Factors
The exchange mechanism has been elucidated for canonical Dbl family GEFs and bacterial exchange factors such as SopE and IpgB2. Despite having distinct structures, both types of GEF stabilize nearly identical conformations of the GTPase switch regions. In particular, both GEFs stabilize Rho family GTPases such that Ala-61 (RhoA numbering) in switch 2 occludes the binding site for Mg2+, and this occlusion is stabilized by a salt bridge between Lys-18 of the phosphate binding loop and Glu-64 of switch 2. Mutations of either Ala-61 or Glu-64 of RhoA ablated the ability of Dbs (a Dbl family GEF) to catalyze exchange. In contrast, these mutations had no effect on exchange catalyzed by SmgGDS (Fig. 4, A and B). The fact that RhoA residues Ala-61 and Glu-64 are not essential for SmgGDS-catalyzed nucleotide exchange supports the conclusion that SmgGDS activates RhoA by a distinct mechanism relative to other GEFs.
FIGURE 4.
SmgGDS utilizes a novel exchange mechanism to activate RhoA. A, B, E–G, intrinsic exchange of 1 μm GTPase was measured for 600 s in exchange buffer, and at the indicated time (arrow) SmgGDS (50 μm) or Dbs (200 nm) was added to stimulate nucleotide exchange. A, RhoA A61G. B, RhoA E64A. C, sequence comparison of Rho family member polybasic regions with basic residues highlighted in blue. D, SmgGDS incubated on ice for 30 min with a 2-fold molar excess of RhoA Δpbr in the presence of EDTA, separated over size-exclusion chromatography, and fractions analyzed via SDS-PAGE. E, RhoA Δpbr. F, RhoA R182A+R183A. G, RhoA K185A+K186A+K187A. H, intrinsic exchange of the indicated GTPase construct (1 μm) measured for 600 s in exchange buffer. At the indicated time (arrow), SmgGDS (50 μm) was added to stimulate nucleotide exchange.
SmgGDS Requires a C-terminal Polybasic Region to Interact with RhoA
SmgGDS activated RhoA and RhoC, yet was inert toward RhoB (Fig. 1). Sequence alignment of RhoA, B, and C reveals a high degree of similarity with the exception of the C-terminal polybasic regions (Fig. 4C). Indeed, RhoB lacks a strong polybasic sequence, implicating this motif as critical for SmgGDS binding. Consistent with this assumption, SmgGDS and a truncated form of RhoA lacking its C-terminal polybasic region, RhoA Δpbr, failed to form a high affinity complex upon gel-exclusion chromatography under conditions designed to favor nucleotide depletion (Fig. 4D). In addition, SmgGDS was unable to activate either RhoA Δpbr or RhoA containing mutations in the polybasic region, whereas Dbs retained activity on all proteins (Fig. 4, E–G). These data are consistent with previous reports that SmgGDS requires the polybasic region of RhoA for binding (20, 34, 35). To define structural aspects of the polybasic region required for SmgGDS activity, several chimeric GTPase having altered polybasic tails were tested for activation (Fig. 4H). In particular, RhoA having the polybasic tail of Rac1 retained its capacity to be activated by SmgGDS. Furthermore, RhoB, which is normally not a substrate for SmgGDS, was efficiently activated when possessing the polybasic tail of RhoA. In contrast, Rac1 possessing the polybasic region of RhoA failed to be activated by SmgGDS. These data illustrate that the presence of a strong polybasic region is necessary for the interaction of SmgGDS and Rho isoforms, but the specific sequence of the polybasic tail is not essential. Importantly, these results also indicate that there are differences outside of the polybasic region that account for the ability of SmgGDS to exchange upon RhoA or RhoB, but not Rac1.
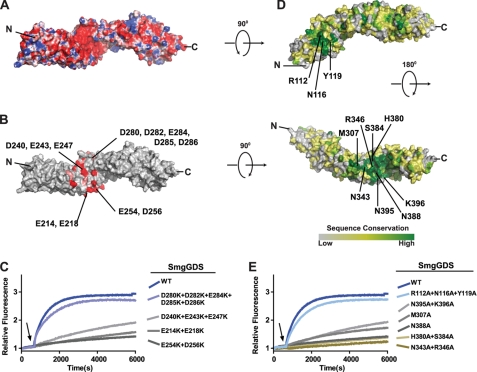
Surface Electrostatics and Conservation Highlight Portions of SmgGDS Required for Activation of RhoA
The necessity of a polybasic region on RhoA suggests that it interacts with a complementary acidic region on the surface of SmgGDS. Lacking a high resolution structure of SmgGDS, we created a homology model using the ARM repeats of β-catenin as a template structure. The electrostatic potential surface was calculated, and a region of strong electronegative charge was observed near the center of the protein (Fig. 5A). To identify which acidic residues were likely to be critical for interaction with RhoA, a multiple sequence alignment was created of SmgGDS sequences from 23 different species. The alignment was analyzed to determine acidic residues that were highly or completely conserved (Fig. 5B). Charge reversal mutations were made in clusters of the conserved acidic residues, and the mutant SmgGDS proteins were tested for their ability to activate RhoA (Fig. 5C). As predicted, mutating any of the conserved acidic clusters that comprise the electronegative patch on SmgGDS ablated the ability of SmgGDS to activate RhoA. However, mutating a modestly conserved acidic cluster on the opposite surface of SmgGDS had no effect on the ability of SmgGDS to activate RhoA. Circular dichroism spectroscopy was used to confirm that mutant SmgGDS proteins had the same secondary structure characteristics as the wild-type protein and were not misfolded (supplemental Fig. S1).
FIGURE 5.
Electronegative patch and highly conserved binding groove on SmgGDS facilitate activation of RhoA. A, electrostatic surface potential of SmgGDS homology model with acidic regions in red and basic regions in blue. B, homology model of SmgGDS with completely (red) and highly (pink) conserved acidic residues. C, intrinsic exchange of RhoA (1 μm) measured for 600 s in exchange buffer. At the indicated time (arrow), wild-type or mutant SmgGS (50 μm) was added to stimulate nucleotide exchange. D, homology model of SmgGDS colored according to ClustalX consensus score from a multiple sequence alignment of full-length SmgGDS isoforms. E, intrinsic exchange of RhoA (1 μm) measured for 600 s in exchange buffer. At the indicated time (arrow), wild-type or mutant SmgGDS (50 μm) was added to stimulate nucleotide exchange.
Having observed that the conserved acidic residues were essential for the ability of SmgGDS to activate RhoA, we investigated whether there were additional conserved regions on the surface of SmgGDS that could be implicated in the interaction with RhoA. Using the multiple sequence alignment of SmgGDS, we mapped the sequence conservation of SmgGDS onto the surface of the homology model (Fig. 5D). Two regions of high sequence conservation were observed. The smaller region of sequence conservation was between the N terminus and the electronegative patch whereas the larger region was in a superhelical groove formed by the curvature of the ARM repeats and was C-terminal to the electronegative patch. Mutations of conserved residues were generated, and mutant proteins were tested for their ability to activate RhoA (Fig. 5E). Whereas mutating the N-terminal conserved region (R112A+N116A+Y119A) did not affect the ability of SmgGDS to exchange upon RhoA, mutations in the C-terminal binding groove either impaired (M307A; N395A+K396A) or abolished (N388A; H380A+S384A; N343A+K396A) activity on RhoA. None of the SmgGDS mutations had altered α-helical content as measured by circular dichroism spectroscopy, indicating that they retained their proper fold (supplemental Fig. S1). These results suggest that the conserved binding groove of SmgGDS might bind the body of the GTPase with the polybasic tail extending over the electronegative surface patch.
SmgGDS Specifically Activates RhoA and RhoC in Cells
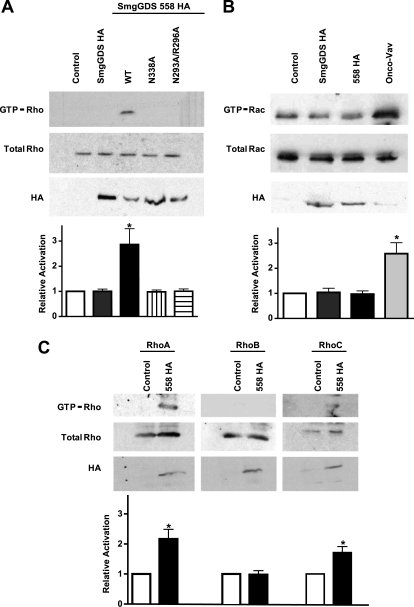
SmgGDS isoforms were transiently transfected into HEK293 cells, and lysates were probed for active Rho proteins using an effector pulldown assay (Fig. 6A). Interestingly, and in contrast to the equivalent activities of the two isoforms defined in vitro, only the SmgGDS-558 isoform activated Rho GTPases in cells. This activation was abolished by substitutions in the conserved binding groove of SmgGDS-558 equivalent to mutations in SmgGDS shown earlier to prevent the in vitro ability of SmgGDS to activate RhoA. Similar assays conducted with Rac1 showed no activation by either SmgGDS isoform (Fig. 6B). Using antibodies specific to the various Rho isoforms, SmgGDS-558 was observed to activate only RhoA and RhoC, but not RhoB (Fig. 6C). Hence, the specificity profiles of SmgGDS in vitro and SmgGDS-558 in cells are identical.
FIGURE 6.
SmgGDS specifically activates RhoA in cells. HEK293 cells were transfected with the indicated GEF expression vectors and then lysed. Active GTPase was precipitated from lysate with GST-RBD (Rho) or GST-PBD (Rac) beads. Pulldown (GTP-bound GTPase) and lysates (total GTPase) were detected by Western blotting. Expression of GEF was confirmed with immunoblotting for HA antibody. Experiments were performed in triplicate, and mean band intensity relative to the control is graphed with S.E. (error bars). A, active Rho detected with a pan-Rho antibody. B, active Rac detected with a pan-Rac antibody. C, active RhoA, RhoB, and RhoC detected with RhoA-, RhoB-, and RhoC-specific antibodies.
DISCUSSION
Distinct types of GEF domains invariably activate specific families of GTPases. For example, DH domains activate Rho family GTPases whereas Cdc25 domains activate Ras family GTPases. However, SmgGDS, which is predicted to be a single domain, has been reported to activate both Rho and Ras family GTPases. In addition, the literature describing GTPase activation by SmgGDS is contradictory. For example, SmgGDS has been reported to activate Rap1b (36), RhoB (14), as well as neither of these GTPases (12, 13). Early work was performed with SmgGDS purified from brain extracts and contamination by other GEFs could account for the conflicting results. In addition, it has been shown that SmgGDS can associate with other exchange factors such as β-Pix, lending credence to this possibility (37). Using highly purified recombinant proteins, we have demonstrated that SmgGDS is only capable of stimulating nucleotide exchange on RhoA and RhoC in vitro. This narrow substrate specificity more closely aligns with the typical restriction of distinct GEFs to specific families of GTPases.
SmgGDS has also been proposed to have an alternative function in regulating the nucleocytoplasmic transport of small GTPases (20). Our present studies do not examine this role of SmgGDS, and it is possible that the protein may have multiple biological roles. However, we show convincing evidence that SmgGDS possesses the characteristics expected of a true GEF. First, it is able to catalyze both the loading and the unloading of MANT-GDP on RhoA in a catalytic and concentration-dependent fashion. In addition, it specifically forms a high affinity complex only with nucleotide-depleted RhoA and not with GDP- or GTPγS-bound RhoA, displaying a key characteristic of GEFs to stabilize the nucleotide-free state.
Divergent GEFs, such as SopE and Dbs, despite having unique tertiary structures, stabilize essentially identical conformations of nucleotide-free GTPases. These results lead to the suggestion that all GEFs for Rho family GTPases would promote similar nucleotide-free states (38). Residues Ala-61 and Glu-64 of RhoA are essential for exchange catalyzed by Dbl family GEFs. Our data show that these residues are not essential for the ability of SmgGDS to activate RhoA, indicating that SmgGDS has a unique exchange mechanism that stabilizes a distinct nucleotide-free state of the GTPase. DOCK9 has also been shown to stabilize a nucleotide-free conformation of Cdc42 which does not rely on residues analogous to Ala-61 or Glu-64 of RhoA (8). DOCK9 catalyzes nucleotide exchange by the direct insertion of a valine side chain into the nucleotide binding pocket. Although there is no significant homology between the GEF region of DOCK9 and SmgGDS, the possibility that the two classes of GEF work by similar mechanisms cannot be ruled out.
This work is the first identification of regions of SmgGDS that are essential for its ability to activate RhoA. The electrostatic surface potential generated from the SmgGDS homology model highlights a conserved electronegative patch that when mutated ablates the ability of SmgGDS to activate RhoA. This functionally important electronegative patch suggests a complementary electropositive binding surface on RhoA. It seems likely that this surface on RhoA corresponds to its polybasic tail, which we have shown is essential for SmgGDS-catalyzed exchange.
A second, highly conserved binding surface was also identified in the groove of the ARM repeat superhelical structure. Mutation of residues in this conserved binding surface also renders SmgGDS unable to activate RhoA. Given the relative positions of the two regions, we postulate that the body of nucleotide-depleted RhoA will lie in the conserved groove and that the polybasic tail of RhoA will extend over the surface of the electronegative region. Without any information on the orientation of RhoA with respect to the surface of SmgGDS it is not possible to model complex formation accurately, but given the distance between the conserved binding groove and the electronegative region (∼25 Å) and the length of the polybasic region (∼32 Å if fully extended) such a conformation is physically reasonable.
Given that both SmgGDS and SmgGDS-558 showed equivalent capacity to activate RhoA in vitro, it is surprising that only SmgGDS-558 activated RhoA in cells. Interestingly, recent results from the Williams laboratory suggest that SmgGDS-558 specifically binds prenylated GTPases in cells, whereas the larger SmgGDS isoform specifically interacts with nonprenylated GTPases (39). As the majority of Rho in cells is expected to be prenylated, this may explain our observation of preferential activation of Rho in cells by SmgGDS-558. It may be that the two isoforms differentially interact with cellular factors that affect their ability to catalyze nucleotide exchange. Indeed, it is tempting to speculate that the highly conserved patch containing Arg-112, Asn-116, and Tyr-119, found directly N-terminal to the extra ARM repeat of SmgGDS, may be an important binding motif. The three-dimensional context of this motif relative to the GTPase binding surface will vary between the two SmgGDS isoforms. This variation may differentially affect interactions with cellular factors to modify exchange capacity.
A unique property of SmgGDS is that it requires the RhoA polybasic region to interact with and promote nucleotide exchange upon RhoA. To date, no other nucleotide exchange factor clearly requires the polybasic region of the GTPase for exchange. Indeed, the polybasic region of GTPases is typically used for proper membrane localization and is dispensable for direct protein/protein interactions. The polybasic region of the GTPase ends at the CAAX box, with the cysteine residue being the site of prenylation for the GTPase. Early reports on the function of SmgGDS suggested that SmgGDS required prenylation of the GTPase to stimulate exchange and was capable of causing extraction of GTPase from cell membranes (14, 40). Given the close proximity of the prenylation site and the polybasic region and recent observations that prenylation status influences the SmgGDS/GTPase interaction in cells, it would be interesting to investigate how prenylation of GTPase alters affinity for either SmgGDS isoform.
This study is the first identification of SmgGDS residues essential for nucleotide exchange. SmgGDS is observed to specifically activate only RhoA and RhoC in vitro, and this specificity is confirmed in cells. The mechanism of nucleotide exchange is shown to be distinct from Dbl family GEFs and to require the presence of a C-terminal polybasic region in the GTPase. Future structural characterization of a SmgGDS·RhoA complex will be necessary for a full understanding of the SmgGDS nucleotide exchange mechanism.
Acknowledgments
We thank Carol Williams for helpful discussions and Ashutosh Tripathy for assistance collecting circular dichroism spectroscopy data.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM062299 (to J. S.) and R01-GM029860 (to K. B.). This work was also supported by American Cancer Society Grant PF-09-119-01-CSM (to E. M.-B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- GEF
- guanine nucleotide exchange factor
- ARM
- armadillo
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- MANT
- N-methylanthraniloyl
- RBD
- Rho-binding domain.
REFERENCES
- 1. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 2. Coleman M. L., Marshall C. J., Olson M. F. (2004) Nat. Rev. Mol. Cell Biol. 5, 355–366 [DOI] [PubMed] [Google Scholar]
- 3. Nikolic M. (2002) Int. J. Biochem. Cell Biol. 34, 731–745 [DOI] [PubMed] [Google Scholar]
- 4. Wu H., Rossi G., Brennwald P. (2008) Trends Cell Biol. 18, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narumiya S., Yasuda S. (2006) Curr. Opin. Cell Biol. 18, 199–205 [DOI] [PubMed] [Google Scholar]
- 6. Worthylake D. K., Rossman K. L., Sondek J. (2000) Nature 408, 682–688 [DOI] [PubMed] [Google Scholar]
- 7. Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 8. Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D. (2009) Science 325, 1398–1402 [DOI] [PubMed] [Google Scholar]
- 9. Buchwald G., Friebel A., Galán J. E., Hardt W. D., Wittinghofer A., Scheffzek K. (2002) EMBO J. 21, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klink B. U., Barden S., Heidler T. V., Borchers C., Ladwein M., Stradal T. E., Rottner K., Heinz D. W. (2010) J. Biol. Chem. 285, 17197–17208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peifer M., Berg S., Reynolds A. B. (1994) Cell 76, 789–791 [DOI] [PubMed] [Google Scholar]
- 12. Isomura M., Kaibuchi K., Yamamoto T., Kawamura S., Katayama M., Takai Y. (1990) Biochem. Biophys. Res. Commun. 169, 652–659 [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto T., Kaibuchi K., Mizuno T., Hiroyoshi M., Shirataki H., Takai Y. (1990) J. Biol. Chem. 265, 16626–16634 [PubMed] [Google Scholar]
- 14. Mizuno T., Kaibuchi K., Yamamoto T., Kawamura M., Sakoda T., Fujioka H., Matsuura Y., Takai Y. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiraoka K., Kaibuchi K., Ando S., Musha T., Takaishi K., Mizuno T., Asada M., Ménard L., Tomhave E., Didsbury J., Snyderman R., Takai Y. (1992) Biochem. Biophys. Res. Commun. 182, 921–930 [DOI] [PubMed] [Google Scholar]
- 16. Chuang T. H., Xu X., Quilliam L. A., Bokoch G. M. (1994) Biochem. J. 303, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yaku H., Sasaki T., Takai Y. (1994) Biochem. Biophys. Res. Commun. 198, 811–817 [DOI] [PubMed] [Google Scholar]
- 18. Hiroyoshi M., Kaibuchi K., Kawamura S., Hata Y., Takai Y. (1991) J. Biol. Chem. 266, 2962–2969 [PubMed] [Google Scholar]
- 19. Hutchinson J. P., Eccleston J. F. (2000) Biochemistry 39, 11348–11359 [DOI] [PubMed] [Google Scholar]
- 20. Lanning C. C., Ruiz-Velasco R., Williams C. L. (2003) J. Biol. Chem. 278, 12495–12506 [DOI] [PubMed] [Google Scholar]
- 21. Zhi H., Yang X. J., Kuhnmuench J., Berg T., Thill R., Yang H., See W. A., Becker C. G., Williams C. L., Li R. (2009) J. Pathol. 217, 389–397 [DOI] [PubMed] [Google Scholar]
- 22. Tew G. W., Lorimer E. L., Berg T. J., Zhi H., Li R., Williams C. L. (2008) J. Biol. Chem. 283, 963–976 [DOI] [PubMed] [Google Scholar]
- 23. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 24. Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., Sondek J. (2002) Nat. Struct. Biol. 9, 468–475 [DOI] [PubMed] [Google Scholar]
- 25. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rojas R. J., Kimple R. J., Rossman K. L., Siderovski D. P., Sondek J. (2003) Comb. Chem. High Throughput Screen. 6, 409–418 [DOI] [PubMed] [Google Scholar]
- 28. Xing Y., Takemaru K., Liu J., Berndt J. D., Zheng J. J., Moon R. T., Xu W. (2008) Structure 16, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eisenberg D., Lüthy R., Bowie J. U. (1997) Methods Enzymol. 277, 396–404 [DOI] [PubMed] [Google Scholar]
- 30. Soundararajan M., Turnbull A., Fedorov O., Johansson C., Doyle D. A. (2008) Proteins 72, 498–505 [DOI] [PubMed] [Google Scholar]
- 31. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Arthur W. T., Ellerbroek S. M., Der C. J., Burridge K., Wennerberg K. (2002) J. Biol. Chem. 277, 42964–42972 [DOI] [PubMed] [Google Scholar]
- 33. Mazhab-Jafari M. T., Marshall C. B., Smith M., Gasmi-Seabrook G. M., Stambolic V., Rottapel R., Neel B. G., Ikura M. (2010) J. Biol. Chem. 285, 5132–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vikis H. G., Stewart S., Guan K. L. (2002) Oncogene 21, 2425–2432 [DOI] [PubMed] [Google Scholar]
- 35. Williams C. L. (2003) Cell. Signal. 15, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 36. Kaibuchi K., Mizuno T., Fujioka H., Yamamoto T., Kishi K., Fukumoto Y., Hori Y., Takai Y. (1991) Mol. Cell. Biol. 11, 2873–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin E. Y., Lee C. S., Cho T. G., Kim Y. G., Song S., Juhnn Y. S., Park S. C., Manser E., Kim E. G. (2006) J. Biol. Chem. 281, 35954–35964 [DOI] [PubMed] [Google Scholar]
- 38. Erickson J. W., Cerione R. A. (2004) Biochemistry 43, 837–842 [DOI] [PubMed] [Google Scholar]
- 39. Berg T. J., Gastonguay A. J., Lorimer E. L., Kuhnmuench J. R., Li R., Fields A. P., Williams C. L. (2010) J. Biol. Chem. 285, 35255–35266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hori Y., Kikuchi A., Isomura M., Katayama M., Miura Y., Fujioka H., Kaibuchi K., Takai Y. (1991) Oncogene 6, 515–522 [PubMed] [Google Scholar]