Abstract
The MSH3 gene is one of the DNA mismatch repair (MMR) genes that has undergone somatic mutation frequently in MMR-deficient cancers. MSH3, together with MSH2, forms the MutSβ heteroduplex, which interacts with interstrand cross-links (ICLs) induced by drugs such as cisplatin and psoralen. However, the precise role of MSH3 in mediating the cytotoxic effects of ICL-inducing agents remains poorly understood. In this study, we first examined the effects of MSH3 deficiency on cytotoxicity caused by cisplatin and oxaliplatin, another ICL-inducing platinum drug. Using isogenic HCT116-derived clones in which MSH3 expression is controlled by shRNA expression in a Tet-off system, we discovered that MSH3 deficiency sensitized cells to both cisplatin and oxaliplatin at clinically relevant doses. Interestingly, siRNA-induced down-regulation of the MLH1 protein did not affect MSH3-dependent toxicity of these drugs, indicating that this process does not require participation of the canonical MMR pathway. Furthermore, MSH3-deficient cells maintained higher levels of phosphorylated histone H2AX and 53BP1 after oxaliplatin treatment in comparison with MSH3-proficient cells, suggesting that MSH3 plays an important role in repairing DNA double strand breaks (DSBs). This role of MSH3 was further supported by our findings that MSH3-deficient cells were sensitive to olaparib, a poly(ADP-ribose) polymerase inhibitor. Moreover, the combination of oxaliplatin and olaparib exhibited a synergistic effect compared with either treatment individually. Collectively, our results provide novel evidence that MSH3 deficiency contributes to the cytotoxicity of platinum drugs through deficient DSB repair. These data lay the foundation for the development of effective prediction and treatments for cancers with MSH3 deficiency.
Keywords: Cancer Tumor Promoter, Cancer Therapy, DNA Damage, DNA Repair, Tumor, Cisplatin, Colon Cancer, DNA Repair, MSH3, Oxaliplatin
Introduction
The DNA mismatch repair (MMR)6 system, composed of several proteins such as MLH1, MSH2, MSH6, MSH3, and PMS2, eliminates replication errors and maintains genomic stability. MutSα, an MSH2/MSH6 heterodimer, recognizes single base mismatches, whereas MutSβ, an MSH2/MSH3 heterodimer, primarily recognizes 2–4-bp insertion-deletion loops (1, 2). The MutL complex, mainly MutLα, an MLH1/PMS2 heterodimer, forms a ternary complex with a MutS heterodimer that binds to DNA mismatches during replication and leads to recruitment of other proteins to complete the process of DNA MMR. Germ line mutations in MMR genes result in Lynch syndrome, which is characterized by hereditary predisposition to cancers with microsatellite instability (MSI) in the colon, endometrium, ovaries, and urinary tract (3, 4). In contrast, MMR deficiency resulting from MLH1 promoter methylation causes sporadic MSI tumors, including colorectal cancer (CRC) (∼15%), endometrial cancer (20–25%), and ovarian cancer (∼12%) (4–6).
The MMR system also participates in repairing certain DNA adducts generated by DNA damaging agents such as alkylating agents and 6-thioguanine. The primary cytotoxic lesion generated by alkylating agents is O6-methylguanine (MeG), which causes MeG -T mispairs (7). MutSα recognizes these mispairs and recruits MutLα for the subsequent repair reactions (8, 9). Loss of MutSα or MutLα renders a cell tolerant to the cytotoxic effects of these drugs, suggesting that these two complexes are also linked to a signal transduction pathway that leads to cell growth arrest or cell death (10, 11).
On the other hand, MutSβ recognizes interstrand cross-links (ICLs) generated by DNA cross-linkers such as psoralen and cisplatin. MutSβ is involved in the recognition and uncoupling of the psoralen-induced ICLs in mammalian cell extracts (12). Recently, it has been shown that MutSβ interacts with Xeroderma pigmentosum group A-Replication Protein A or Xeroderma pigmentosum group C-RAD23B, both of which are involved in nucleotide excision repair, in the recognition of psoralen ICLs and promotes the nucleotide excision repair process (13, 14). The level of homologous recombination (HR) that repairs ICLs is also dependent on MutSβ but not on MutSα or MLH1. These results suggest that MutSβ may cooperate with the nucleotide excision repair, HR and Fanconi anemia proteins for repairing psoralen-induced ICLs (15). In addition, MutSβ also binds to cisplatin-induced ICLs together with PARP-1, DNA ligase III, XRCC-1, Ku80, and Ku70, suggesting that MutSβ may also cooperate with other repair pathways to recognize and repair platinum drug-induced ICLs (16).
Oxaliplatin, a third generation platinum drug, is one of the key drugs that is currently being used for the treatment of CRC patients. Similar to cisplatin, oxaliplatin also forms intrastrand cross-links and ICLs (17). However, the detailed molecular mechanisms involved in repair and the cytotoxic effects of oxaliplatin-induced adducts, especially ICLs, have not been explored extensively.
Considering that the MutSβ complex plays a role in repairing ICLs, we hypothesized that MSH3 deficiency may halt the repair of ICLs induced by platinum drugs, resulting in enhanced cytotoxicity of these drugs in cancer patients. Additionally, because MSH3 deficiency results in suppressed HR (15), and HR-defective cells are hypersensitive to poly(ADP-ribose) polymerase (PARP) inhibitors (18, 19), we further hypothesized that MSH3 deficiency may also result in sensitization of cells to PARP inhibitors. In MSI CRC, frequent frameshift mutations (20–50%) within the mononucleotide A8 repeats in exon 7 of MSH3 results in loss or reduction of MSH3 (20–22). Recently, we found that MSH3-negative cancer cell population exists within sporadic CRC tissues that exhibit low levels of MSI and/or elevated microsatellite alterations at tetranucleotide repeats (EMAST) (23). Intuitively, if MSH3 deficiency dictates the toxicity of platinum drugs and PARP inhibitors in a clinical setting, MSH3 status can be used as a predictive marker for the chemotherapeutic outcome in patients with MSH3-deficient cancers. To explore this possibility, using isogenic cell lines in which MSH3 protein expression can be regulated thorough shRNA expression in a Tet-off system, we investigated the effect of MSH3 deficiency on the cellular sensitivity to two platinum drugs and a well known PARP inhibitor. Herein, we provide novel molecular evidence that MSH3 deficiency in CRC cell lines contributes toward the cytotoxicity of platinum drugs, especially as a result of compromised double strand break (DSB) repair.
EXPERIMENTAL PROCEDURES
Reagents
Cisplatin, oxaliplatin, N-methyl-N′-nitro-N-nitrosoguanidine, and propidium iodide were purchased from Sigma-Aldrich. Olaparib, a PARP inhibitor, was purchased from Selleck Chemicals (Houston, TX).
Cell Lines and Cell Culture
The human colon cancer cell lines HCT116, HCT116+ch.3 (HCT116+3), HCT116+ch.3+ch.5 (HCT116+3+5) have been described previously (10, 23). HCT116+3+5 cells were stably transfected with a tetracycline-regulated retroviral vector, the TMP (Open Biosystems, Huntsville, AL) that encodes shRNA against MSH3. Stable MSH3-deficient clones G1, G2, and G5 were isolated (see “Results” and Ref. 23). HCT116, HCT116+3, and HCT116+3+5 cells were grown in Iscove's Modified Dulbecco's Medium (Invitrogen) with 10% fetal bovine serum. The G1, G2, and G5 cells were maintained in Iscove's Modified Dulbecco's Medium with 10% fetal bovine serum and 0.6 μg/ml of puromycin. To turn off the expression of MSH3 shRNA, 1 μg/ml of doxycycline was added to the culture medium.
Western Blot Analysis
Proteins from cell lysates were prepared, separated on SDS-PAGE, and transferred to PVDF membranes as described previously (23). Anti-human MSH3 mouse monoclonal antibody (dilution: 1:250, clone 52, BD Pharmingen, San Jose, CA), anti-human MLH1 mouse monoclonal antibody (1:200, G168–728, BD Pharmingen) and anti-β-actin antibody (1:10,000, clone AC-15, Sigma-Aldrich) were used as primary antibodies for the detection of specific proteins. Goat anti-mouse antibody (1:3000, catalog no. sc-2005, Santa Cruz Biotechnology, Santa Cruz, CA) was used as a secondary antibody. The signal amplification and detection was achieved by exposing the membrane to ECL reagent (GE Healthcare), followed by visualization on the Storm imaging system (Amersham Biosciences).
Clonogenic Survival Assay
Two hundred cells were seeded in each well of a six-well plate. For the measurement of the cytotoxicity caused by cisplatin or oxaliplatin, the cells were treated with the drugs for 24 h once the cells were attached to the plate. For the measurement of the cytotoxicity caused by olaparib, cells were treated continuously with the drug during the experiments. After 8–10 days, the number of colonies (colonies with >50 cells) were counted, and the relative change in clonogenic survival of drug-treated versus untreated cells was determined.
Cell Cycle Analysis
One million cells were seeded in 10-cm plates. Once attached, the cells lines were treated with oxaliplatin for 24 h. After an additional 48 h, cells were washed twice with cold PBS and fixed in cold 70% ethanol at −20 °C overnight or for several days. The ethanol-fixed cells (2 × 106) were subsequently washed with PBS twice and incubated with 300 μl of PBS and 0.15% RNase A for 15 min at 37 °C. The cells were stained with 75 μg/ml propidium iodide for 30 min and then analyzed for DNA content using a FACSCantoII flow cytometer (BD Biosciences). Cell cycle data were analyzed by Flowjo software (Tree Star, Ashland, OR).
Proliferation Assay
The proliferation index was measured by BrdU incorporation in HCT116+3+5 and G5 cells, 48 h after the initial 24-h treatment with oxaliplatin (Cell Proliferation ELISA, BrdU, Roche Diagnostics). Experiments were performed in triplicate, and data were obtained from three or four independent experiments.
siRNA Treatment
MLH1 siRNA, MSH3 siRNA, and nontargeted siRNA were purchased from Dharmacon (Lafayette, CO). Two hundred thousand cells were seeded in 24-well plates. After an overnight incubation, the cells were transfected with 83 nm of the targeted siRNAs or nontargeted siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Two days after transfection, the cells were harvested and replated for clonogenic survival assays.
Immunofluorescence Staining
Ten thousand cells were grown on glass coverslips in a 12-well plate. The cells were fixed with 4% paraformaldehyde (pH 7.5) in PBS for 15 min, permeabilized with 0.3% Triton X-100 for 5 min, and then blocked with 10% goat serum (Invitrogen) for 1 h. The cells were subsequently incubated with an anti-active caspase-3 antibody (1:500, G748, Promega, Madison, WI), an anti-phosphorylated histone H2AX (pH2AX) antibody (1:5000, JBW301, Millipore Corp., Billerica, MA), or an anti-53BP1 antibody (1:600, ab21083, Abcam, Cambridge, MA) for 1 h, followed by a secondary antibody (1:800, Alexa Fluor 555 goat anti-mouse or anti-rabbit antibody, Invitrogen) for 40 min. Prolong Gold with DAPI (Invitrogen) was used in the mounting medium. The images were obtained using AxioSkop2 multichannel epifluorescence microscope equipped with AxioVision software (Carl Zeiss, Thornwood, NY).
RESULTS
MSH3 Expression Is Controlled by Doxycycline in MSH3-deficient Clones
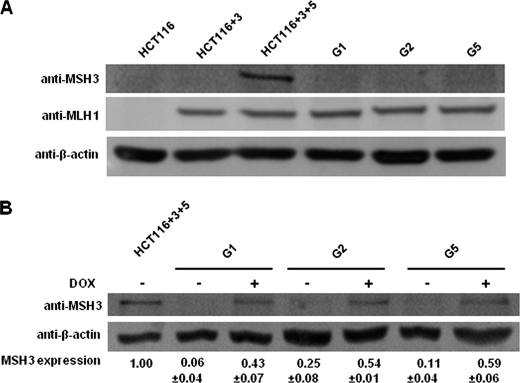
We first determined whether MSH3 expression in G1, G2, and G5 cell clones of HCT116 CRC cells is controlled by doxycycline. We used HCT116 and HCT116+3 as negative controls and HCT116+3+5 as a positive control for MSH3 expression. HCT116 and HCT116+3 cells showed no detectable MSH3 protein expression (Fig. 1A), which is consistent with HCT116 cells harboring homozygous frameshift mutations in a mononucleotide repeat of the MSH3 exon 7 (23). HCT116+3+5, generated from MSH3-deficient HCT116+3 by transfer of a copy of chromosome 5, showed MSH3 expression. Although no MSH3 was detected in G1, G2 and G5 clones in the absence of doxycycline, addition of doxycycline restored MSH3 expression in all of these clones to ∼40–60% of the levels of parental HCT116+3+5 (Fig. 1, A and B). We believe it is technically challenging to expect complete blockade for the production of MSH3 shRNA in these cell lines even in the presence of doxycycline. However, we think this protein level is enough to analyze the effect of MSH3 on drug sensitivity in this study because we have shown previously that this level of MSH3 in G5 cells is enough to recover MSH3 functions regarding the EMAST phenotype in vitro (23).
FIGURE 1.
MSH3 expression of the HCT116+3+5-derived clones stably transfected with MSH3 shRNA is controlled by a Tet-off system. A, Western blot analysis of MSH3, MLH1, and β-actin in HCT116, HCT116+3, HCT116+3+5, and the three HCT116+3+5-derived clones, G1, G2, and G5 cells. B, Western blot analysis of MSH3 and β-actin in HCT116+3+5, G1, G2, and G5 cells cultured in the medium with and without 1 μg/ml doxycycline. Relative MSH3 expression was calculated by densitometry, and the results were obtained from three or more independent experiments.
MSH3-deficient Cells Are More Sensitive to Cisplatin and Oxaliplatin Than MSH3-proficient Cells
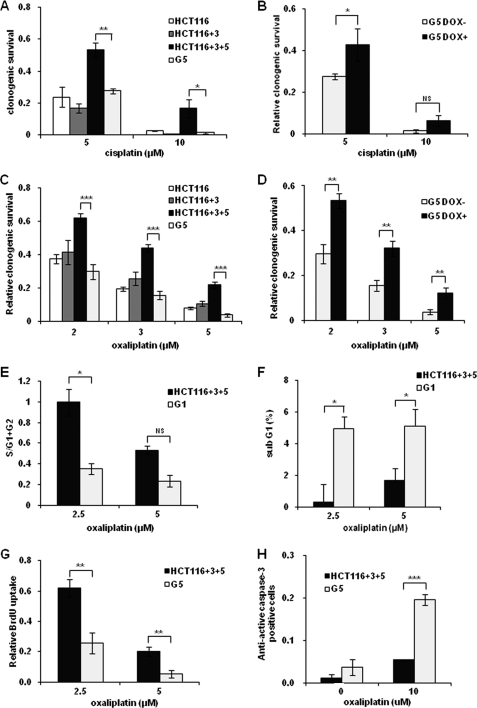
To determine whether MSH3 status affects cellular sensitivity to two platinum drugs, we first examined the clonogenic survival of HCT116 and HCT116-derived cell lines in cisplatin-treated cells. No significant differences in cisplatin sensitivity were observed between MLH1 and MSH3-deficient HCT116 and MSH3-deficient HCT116+3 cell lines, whereas higher resistance was observed in MSH3-proficient HCT116+3+5 cell lines (Fig. 2A). Among various cell lines, the MSH3-deficient G5 clone was more sensitive than its parental HCT116+3+5 (Fig. 2A). To further confirm that MSH3 existence influenced cytotoxicity induced by cisplatin, we compared the clonogenic survival of G5 cells in the presence and absence of doxycycline. We found that restoration of MSH3 expression desensitized the cells to cisplatin (5 μm; Fig. 2B). These results indicate that MSH3 depletion leads to the sensitization of cells to cisplatin. We also analyzed clonogenic survival of the other clones, G1 and G2 and found that these clones behaved similarly to G5 (data not shown). This further strengthened the possible role of MSH3 in the cytotoxicity caused by cisplatin. Next, we determined whether MSH3 deficiency also influenced cellular sensitivity to oxaliplatin. Surprisingly, the MSH3-deficient HCT116, HCT116+3, and G5 clones were significantly more sensitive to oxaliplatin than the parental HCT116+3+5, as was the case for cisplatin (Fig. 2C). Furthermore, we observed that the restoration of MSH3 in the MSH3-deficient cells led to restoration of oxaliplatin insensitivity (Fig. 2D). Next, we compared the rate of growth inhibition and the levels of apoptosis between MSH3-proficient and -deficient cells after oxaliplatin treatment. We discovered that the degree of cell growth inhibition (Fig. 2E) and the levels of apoptosis were significantly higher (Fig. 2F) in MSH3-deficient cells than in MSH3-proficient cells, using flow cytometry. We also confirmed that cell proliferation was decreased, and the apoptotic fraction was increased in MSH3-deficient cells treated with oxaliplatin, using a BrdU assay and an immunofluorescence assay, respectively (Fig. 2, G and H). These results are consistent with our findings on growth inhibition obtained via clonogenic assays.
FIGURE 2.
MSH3-deficient cells are more sensitive to cisplatin and oxaliplatin than MSH3-proficient cells. A, clonogenic survival fraction of HCT116, HCT116+3, HCT116+3+5, and G5 cells treated with cisplatin. B, clonogenic survival fraction of G5 cells cultured with and without 1 μg/ml doxycycline, which were treated with cisplatin. C, clonogenic survival fraction of HCT116, HCT116+3, HCT116+3+5, and G5 cells treated with oxaliplatin. D, clonogenic survival fraction of G5 cells cultured with and without doxycycline (DOX) 1 μg/ml, which were also treated with oxaliplatin. Shown are a decrease in S-phase population (E) and an increase in sub-G1 population (F) of the HCT116+3+5 and G1 cells. G, decrease in relative BrdU incorporation compared with nontreated controls. H, increase in anti-active caspase-3 positive cells in immunofluorescence in the HCT116+3+5 and G5 cells. Data are represented as means ± S.E. from three or more independent experiments. The statistical difference was determined by a two-sided Student's t test. *, **, and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively. NS indicates a p = 0.05 or more. Representative data from one of the three MSH3-deficient clones is shown in this figure.
Depletion of MSH3 by siRNA Transfection also Sensitizes Cells to Cisplatin and Oxaliplatin
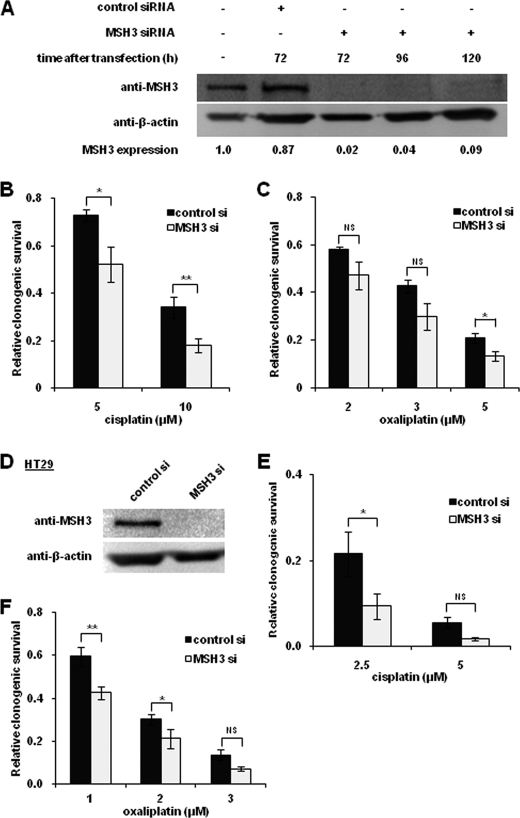
To further confirm the role of MSH3-related sensitization to cisplatin and oxaliplatin, we next determined the clonogenic survival frequencies of cells transiently transfected with MSH3 siRNA and nontargeted siRNA. In these experiments, we confirmed that MSH3 protein expression was significantly diminished 72 h after siRNA transfection (98% MSH3 expression inhibition compared with untreated cells) in HCT116+3+5 cells (Fig. 3A). HCT116+3+5 cells were transfected with MSH3 siRNA, and the cells were exposed to cisplatin (5 and 10 μm) or oxaliplatin (2 and 5 μm) 48 h after transfection. As shown in Fig. 3, B and C, transfection of MSH3 siRNA rendered HCT116+3+5 cells more susceptible to both cisplatin and oxaliplatin in comparison with cell lines transfected with nontargeted siRNA. To further confirm this increased sensitivity to platinum drugs in MSH3-depleted cells, we also transfected MSH3 siRNA or nontargeted siRNA into another colon cancer cell line, HT29. We confirmed that MSH3 was repressed almost completely in HT29 cells (Fig. 3D) and that HT29 cells treated with MSH3 siRNA became more sensitive to both cisplatin and oxaliplatin (Fig. 3, E and F). These results further strengthen our findings that MSH3 deficiency sensitizes cells to both cisplatin and oxaliplatin.
FIGURE 3.
Transient depletion of MSH3 by siRNA also sensitizes HCT116 + 3+5 cells to cisplatin and oxaliplatin. A, Western blot analysis of MSH3 and β-actin in MSH3-depleted HCT116+3+5 cells by transient siRNA (si) transfection. Comparison of the clonogenic survival fraction of HCT116+3+5 cell lines treated with cisplatin (B) and oxaliplatin (C) after transfection with nontargeted (control) siRNA and with MSH3 siRNA. D, Western blot analysis of MSH3 and β-actin in HT29 cells treated with nontargeted siRNA and with MSH3 siRNA. Cells were extracted 72 h after siRNA transfection. Comparison of clonogenic survival fractions of HT29 cells treated with cisplatin (E) and oxaliplatin (F) after transfection with nontargeted siRNA and MSH3 siRNA. Data are represented as mean ± S.E. from five independent experiments. The statistical difference was determined by a two-sided Student's t test. * and ** represent p < 0.05 and p < 0.01, respectively; NS represents p > 0.05.
Cisplatin or Oxaliplatin Sensitivity in MSH3-proficient and MSH3-deficient Cells Occurs Independently of MLH1 Status in Colon Cancer Cells
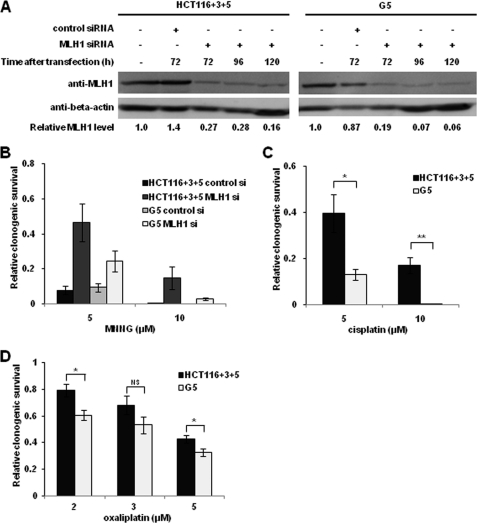
From a clinical viewpoint, it is important to determine whether the MSH3 status influences sensitivity to cisplatin and oxaliplatin in patients with cancers that are also MLH1-deficient. To address this question, we compared the sensitivity of MSH3-deficient G5 cells and MSH3-proficient cells with cisplatin and oxaliplatin by inducing siRNA mediated down-regulation of MLH1 expression (Fig. 4A). When MLH1 was down-regulated in both HCT116+3+5 and G5 cells transfected with MLH1 siRNA, both cell lines became more resistant to N-methyl-N′-nitro-N-nitrosoguanidine in comparison with cell lines treated with nontargeted siRNA (Fig. 4B), validating the functional repression of MLH1 in our experimental conditions (10, 11). Interestingly, in this scenario, we observed that G5 cells were more sensitive to cisplatin and oxaliplatin (2 and 5 μm) than HCT116+3+5 (Fig. 4, C and D). These results further support the notion that MSH3-dependent sensitivity to cisplatin and oxaliplatin occurs independently of MLH1 status.
FIGURE 4.
Transient depletion of MLH1 by siRNA does not affect the resistance to cisplatin and oxaliplatin in MSH3-proficient and -deficient cells. A, Western blot analysis of MLH1 and β-actin in MLH1-depleted HCT116+3+5 and G5 cells following transient siRNA transfection. B, Clonogenic survival fraction of HCT116+3+5 and G5 cells treated with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) after transfection with control siRNA or MLH1 siRNA. Comparison of the clonogenic survival fraction between MLH1-depleted HCT116+3+5 and G5 cells treated with cisplatin (C) and oxaliplatin (D). Data are represented as mean ± S.E. from four or more independent experiments. The statistical difference was determined by a two-sided Student's t test. * and ** represent p < 0.05 and p < 0.01, respectively; NS represents p > 0.05.
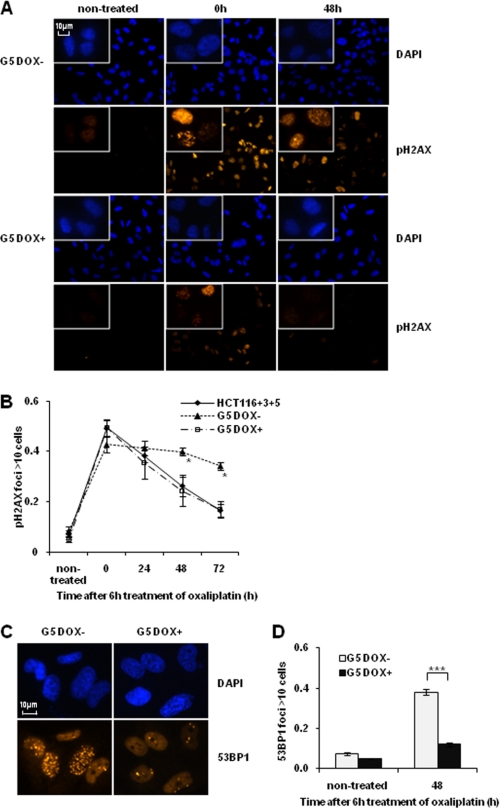
MSH3-deficient Cells Demonstrate Sustained Levels of pH2AX and 53BP1 after Oxaliplatin Treatment
Platinum drugs induce DNA intrastrand cross-links and ICLs, and some of the lesions eventually lead to secondary DNA DSBs, presumably as a result of a collapsed replication fork (24). To determine whether MSH3 is involved in the repair of DSBs, we next analyzed time-dependent changes in the levels of nuclear pH2AX, a surrogate marker for DNA DSBs (25) using immunofluorescence staining. We found that there were no differences in the number of pH2AX foci-positive cells before and after oxaliplatin treatment in the MSH3-proficient and -deficient cell lines. In contrast, we observed a lower rate of reduction in the number of pH2AX foci-positive cells in the MSH3-deficient G5 cells compared with both MSH3-restored G5 cells and the HCT116+3+5 cell lines during a 48- and 72-h treatment with oxaliplatin (Fig. 5, A and B), indicating that DSB repair is compromised only in MSH3-deficient cell lines. To further confirm this DSB repair inefficiency, we also performed immunofluorescence assays using an anti-53BP1 antibody, another marker for detecting DNA DSB. We confirmed sustained levels of 53BP1 in MSH3-deficient G5 cells after oxaliplatin treatment (Fig. 5, C and D). These results suggest that the higher sensitivity of MSH3-deficient cells to oxaliplatin may in part be due to reduced DNA DSB repair efficiency, rather than a quantitative difference in the burden of DNA damage induced by treatment.
FIGURE 5.
MSH3-deficient cells show a decrease in DNA double strand break repair efficiency. A, immunofluorescence staining for pH2AX foci formation in the HCT116+3+5, G5 with doxycycline (DOX) and G5 cells without doxycycline. The cells were treated with 5 μm oxaliplatin for 6 h and were analyzed by immunofluorescence after the indicated hours. B, inefficient decline of pH2AX-positive cells in the MSH3-deficient cells. C, immunofluorescence staining for 53BP1 foci formation in G5 with doxycycline and G5 cells without doxycycline. The cells were treated with 5 μm oxaliplatin for 6 h and were analyzed by immunofluorescence after 48 h. At least 100 cells were counted in each slide. Data are represented as mean ± S.E. from three or four independent experiments. The statistical difference between MSH3-deficient and -proficient G5 was determined by a two-sided Student's t test. * and *** represent p < 0.05 and p < 0.001, respectively.
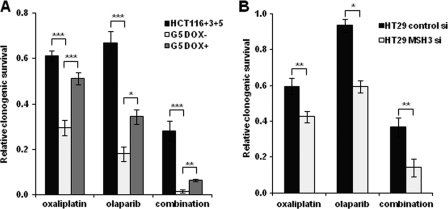
MSH3-deficient Cells Are also Sensitive to Olaparib, a PARP Inhibitor
PARP inhibitors increase the number of single strand breaks, which eventually leads to DNA DSBs that are repaired by the HR system. HR-defective cells are hypersensitive to PARP inhibitors because of their inability to repair these DSBs (18, 19). The possible role of MSH3 in DSB repair evidenced from our results (Fig. 5) prompted us to further examine whether MSH3-deficient cells are also sensitive to PARP inhibitors. As shown in Fig. 6A, MSH3-deficient G5 cells were more sensitive to olaparib than the MSH3-restored G5 cell line. These data clearly support the role of MSH3 in DSB repairs in CRC cells. Moreover, the combination of oxaliplatin and olaparib exhibited a synergistic effect in cytotoxicity in the MSH3-deficient G5 cells compared with the parental HCT116+3+5 cells. We also confirmed this effect in two other colon cancer cell lines HT29 (Fig. 6B) and SW480 (data not shown) in a transient knockdown system using MSH3 siRNA. These results suggest the promising potential for a combination therapy of platinum drugs and PARP inhibitors in MSH3-deficient cancers.
FIGURE 6.
MSH3-deficient cells are sensitive to olaparib, a PARP inhibitor, and the combination with oxaliplatin. A, clonogenic survival of HCT116+3+5, G5 without doxycycline (DOX), and G5 cells with doxycycline, which were treated with 2 μm of oxaliplatin, 2 μm of olaparib, and the combination of these two drugs. B, clonogenic survival of HT29 cells, which were treated with 1 μm oxaliplatin, 2 μm olaparib, and the combination of these two drugs. Data are represented as mean ± S.E. from three or more independent experiments. The statistical difference was determined by a two-sided Student's t test. *, **, and *** represent p < 0.05, p < 0.01 and p < 0.001, respectively.
DISCUSSION
The aim of the present study was to elucidate whether loss of MSH3 affects cellular sensitivity to platinum drugs. It was anticipated that such data could lead to establishment of the diagnostic and therapeutic strategy that MSH3 status may be used as a predictive marker for chemotherapeutic outcome in patients with MSH3-deficient tumors. Using the isogenic colon cancer cell lines in which MSH3 expression is regulated by a Tet-off system, we demonstrate that the depletion of MSH3 expression in colon cancer cells sensitized them to cisplatin, and also to oxaliplatin and a PARP inhibitor. Our data suggest that these effects can be best explained by the reduced ability of MSH3-deficient cells to repair DSBs that are incurred following treatment with these drugs. To the best of our knowledge, this is the first report demonstrating that selective inhibition of MSH3 increases cellular sensitivity to platinum drugs and a PARP inhibitor. Moreover, we demonstrate that the MSH3-dependent increase in sensitivity to cisplatin and oxaliplatin is not influenced by down-regulation of MLH1 and is probably independent of the canonical MMR system.
The role of MutS and MutL homologues in repair of ICLs has been well studied using psoralen ICLs (12–15, 26). These data suggest that MutSβ is involved in both recognition and processing of certain types of ICLs in cooperation with other proteins such as nucleotide excision repair and HR proteins and the fact that MutSβ also functions in ICL repair independent of its primary role in MMR. Our finding that MSH3-depleted cells are sensitive to cisplatin and oxaliplatin, and that this occurs independent of MLH1 function, is consistent with these findings with psoralen ICLs.
Our results together with previous studies support the idea that MutSβ is involved in the repair of toxic DSBs induced by ICL adducts. First, there is existing evidence that MSH3 is co-localized to DSB lesions induced by laser (27) and by carcinogens such as chromium(VI) (28). Second, we have observed sustained levels of pH2AX and 53BP1 that co-localize with DSBs in MSH3-deficient cells after oxaliplatin treatment compared with MSH3-proficient cells. Third, MSH3-deficient cells are sensitive to a PARP inhibitor that induces DSBs. Thus, our results suggest that unrepaired DSBs due to MSH3 deficiency are the direct cause of cell death. However, a recent study has shown that tumors occurring in MSH2-null mice are more resistant to cisplatin and the combination of 5-Fluorouracil plus oxaliplatin than tumors in mice that have MSH2 G674D mutations (29). Interestingly, although this missense mutation results in loss of MMR activity, it still retains sensitivity to DNA damage. These results suggest that MSH2 has distinctive functions in MMR activity and chemosensitivity (29). MSH2 and MLH1 have been shown to be required for the activation of various proteins involved in apoptotic pathways such as JNK and c-Abl after cisplatin treatment (30); however, it is not clear whether MutSα or MutSβ or both are involved in the signaling pathways induced by cisplatin or oxaliplatin. However, because our results indicate that loss of MSH3 increases the sensitivity to cisplatin and oxaliplatin, it is likely that MutSβ is mainly involved in the repair of DNA damage, and MutSα is involved in both the repair and signaling pathways that lead to cell death. Further studies are needed to elucidate the exact role of MutSα or MutSβ in repair for DNA damage and in damage signaling caused by these drugs.
Our results regarding the sensitivity of MSH3-deficient cells to cisplatin and oxaliplatin are inconsistent with a previous report by Vaisman et al. (31). That study reported that the sensitivity to these drugs did not differ between the MSH3-deficient HHUA cells and the MSH3-proficient HHUA complemented with chromosome 5 (31). In their study, the influence by hundreds of other genes of chromosome 5 could not be excluded; therefore, we believe that our present data are more robust as we used isogenic clones of HCT116 colon cancer cells, in which MSH3 expression was regulated selectively as needed.
From a clinical standpoint, our results suggest that a considerable population of patients with MSI CRC might benefit from oxaliplatin-based treatment regimens, PARP inhibitors, or in particular, a combination of the two. In CRC, many recent studies have shown that patients with stage III MSI cancer do not benefit from 5-Fluorouracil adjuvant chemotherapy (32–34). Moreover, Bertagnolli et al. (35) reported that patients with stage III MSI-CRC benefit from adjuvant chemotherapy containing 5-Fluorouracil and irinotecan, whereas another study has reported that these patients received no benefit from this adjuvant treatment (36). These inconsistent results raise the possibility that there may be subgroups of patients that have different chemosensitivities among MSI CRC. For instance, our results suggest that there are at least two subpopulations of MLH1-deficient CRC, MSH3-proficient and MSH3-deficient CRC, and these may respond differentially to oxaliplatin, a PARP inhibitor, and their combination depending on the MSH3 status.
Although our data indicate that MSH3-deficient cells are hypersensitive to combination therapy, a synergistic effect was even noted in the MSH3-proficient cells (Fig. 6). We speculate that MSH3 is just one of the several factors that determine cellular sensitivity to these drugs. The precise molecular details on the specific steps and how MSH3 or the MutSβ complex is involved in repair of platinum-induced DNA damage or PARP inhibitor-induced damage still remain elusive. Nonetheless, we believe that our data indicating this hypersensitivity of MSH3-deficient cancer cells especially to the combination therapy in comparison with MSH3-proficient cells at clinically relevant doses could have important clinical implications in terms of managing MSH3-deficient cancers in the future.
In addition to MSH3, several other DNA repair genes are mutated in MSI cancers. MRE11A and RAD50, whose products are in the DSB repair complex MRE11A-hRAD50-NBS1, are among the most frequently mutated genes in MSI cancers (22). Mutations in MRE11A and RAD50 have been shown to increase sensitivity to irinotecan, which induces secondary DSBs in cultured cells (37, 38). We have also confirmed that MSH3 deficiency sensitized cells to SN-38, an active metabolite of irinotecan (data not shown). Moreover, loss of the phosphatase and tensin homologue, another gene frequently mutated in MSI cancer, has been shown to sensitize cells to PARP inhibitors through inefficiency of HR repair (39, 40). Thus, analyzing the genes or proteins that are involved in DSB repair could be helpful for predicting the therapeutic response in patients with MSI cancer. Clinical studies to validate predictive markers for drug therapy in MSI cancer are warranted.
Previously, we demonstrated that loss of MSH3 expression caused the EMAST and MSI-low phenotypes and that focal loss of MSH3 expression was associated with EMAST in sporadic CRC tissues (23). Moreover, most MSI-low CRCs and some proportion of MSS tumors exhibited EMAST, suggesting that these tissues might have experienced MSH3 deficiency (23). MSH3 deficiency is possibly related to disease progression in MLH1-deficient CRC (20), and MSI-low CRCs have poor prognosis (41, 42), raising the possibility that loss of MSH3 may be related to the promotion of metastasis or recurrence of CRC. In this scenario, treatment of sporadic CRC containing MSH3-negative cancer cell populations with platinum drugs or PARP inhibitors, or both, may inhibit disease progression.
In conclusion, we show that MSH3-deficient cells are sensitive to cisplatin, oxaliplatin, and a PARP inhibitor, possibly resulting from reduced repair of DNA DSBs. Our findings could contribute to a better understanding of the role of MSH3 in DNA repair and drug sensitivity and to better predicting and improving the therapeutic outcome of patients with MSH3-deficient cancers.
Acknowledgments
We thank Thomas Jascur and Yan Shen for technical assistance.
This work was supported, in whole or in part, by NCI, National Institutes of Health Grants R01 CA72851 and CA129286. This work was also supported by funds from the Baylor Research Institute (to C. R. B. and A. G.).
- MMR
- mismatch repair
- MSI
- microsatellite instability
- CRC
- colorectal cancer
- ICL
- interstrand cross-link
- HR
- homologous recombination
- PARP
- poly(ADP-ribose) polymerase
- EMAST
- elevated microsatellite alterations at tetranucleotide repeats
- DSB
- double strand break
- pH2AX
- phosphorylated histone H2AX.
REFERENCES
- 1. Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 2. Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 3. Hendriks Y. M., de Jong A. E., Morreau H., Tops C. M., Vasen H. F., Wijnen J. T., Breuning M. H., Bröcker-Vriends A. H. (2006) CA Cancer J. Clin. 56, 213–225 [DOI] [PubMed] [Google Scholar]
- 4. Boland C. R., Goel A. (2010) Gastroenterology 138, 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Black D., Soslow R. A., Levine D. A., Tornos C., Chen S. C., Hummer A. J., Bogomolniy F., Olvera N., Barakat R. R., Boyd J. (2006) J. Clin. Oncol. 24, 1745–1753 [DOI] [PubMed] [Google Scholar]
- 6. Pal T., Permuth-Wey J., Kumar A., Sellers T. A. (2008) Clin. Cancer Res. 14, 6847–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duckett D. R., Drummond J. T., Murchie A. I., Reardon J. T., Sancar A., Lilley D. M., Modrich P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6443–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van't Wout K., te Riele H. (1999) Nat. Genet. 23, 359–362 [DOI] [PubMed] [Google Scholar]
- 9. Abuin A., Zhang H., Bradley A. (2000) Mol. Cell. Biol. 20, 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koi M., Umar A., Chauhan D. P., Cherian S. P., Carethers J. M., Kunkel T. A., Boland C. R. (1994) Cancer Res. 54, 4308–4312 [PubMed] [Google Scholar]
- 11. Hawn M. T., Umar A., Carethers J. M., Marra G., Kunkel T. A., Boland C. R., Koi M. (1995) Cancer Res. 55, 3721–3725 [PubMed] [Google Scholar]
- 12. Zhang N., Lu X., Zhang X., Peterson C. A., Legerski R. J. (2002) Mol. Cell. Biol. 22, 2388–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao J., Jain A., Iyer R. R., Modrich P. L., Vasquez K. M. (2009) Nucleic Acids Res. 37, 4420–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vasquez K. M. (2010) Environ. Mol. Mutagen 51, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang N., Liu X., Li L., Legerski R. (2007) DNA Repair 6, 1670–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu G., Lippard S. J. (2009) Biochemistry 48, 4916–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaney S. G., Campbell S. L., Bassett E., Wu Y. (2005) Crit. Rev. Oncol. Hematol. 53, 3–11 [DOI] [PubMed] [Google Scholar]
- 18. Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T. (2005) Nature 434, 913–917 [DOI] [PubMed] [Google Scholar]
- 19. Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., Ashworth A. (2005) Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 20. Plaschke J., Krüger S., Jeske B., Theissig F., Kreuz F. R., Pistorius S., Saeger H. D., Iaccarino I., Marra G., Schackert H. K. (2004) Cancer Res. 64, 864–870 [DOI] [PubMed] [Google Scholar]
- 21. Miquel C., Jacob S., Grandjouan S., Aimé A., Viguier J., Sabourin J. C., Sarasin A., Duval A., Praz F. (2007) Oncogene 26, 5919–5926 [DOI] [PubMed] [Google Scholar]
- 22. Hewish M., Lord C. J., Martin S. A., Cunningham D., Ashworth A. (2010) Nat. Rev. Clin. Oncol. 7, 197–208 [DOI] [PubMed] [Google Scholar]
- 23. Haugen A. C., Goel A., Yamada K., Marra G., Nguyen T. P., Nagasaka T., Kanazawa S., Koike J., Kikuchi Y., Zhong X., Arita M., Shibuya K., Oshimura M., Hemmi H., Boland C. R., Koi M. (2008) Cancer Res. 68, 8465–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinz J. M. (2010) Environ. Mol. Mutagen 51, 582–603 [DOI] [PubMed] [Google Scholar]
- 25. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. (2008) Nat. Rev. Cancer 8, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Q., Vasquez K. M. (2008) PLoS Genet. 4, e1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong Z., Jiang J., Hashiguchi K., Hoshi M., Lan L., Yasui A. (2008) J. Cell Sci. 121, 3146–3154 [DOI] [PubMed] [Google Scholar]
- 28. Reynolds M. F., Peterson-Roth E. C., Bespalov I. A., Johnston T., Gurel V. M., Menard H. L., Zhitkovich A. (2009) Cancer Res. 69, 1071–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kucherlapati M. H., Lee K., Nguyen A. A., Clark A. B., Hou H., Jr., Rosulek A., Li H., Yang K., Fan K., Lipkin M., Bronson R. T., Jelicks L., Kunkel T. A., Kucherlapati R., Edelmann W. (2010) Gastroenterology 138, 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nehmé A., Baskaran R., Nebel S., Fink D., Howell S. B., Wang J. Y., Christen R. D. (1999) Br. J. Cancer 79, 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaisman A., Varchenko M., Umar A., Kunkel T. A., Risinger J. I., Barrett J. C., Hamilton T. C., Chaney S. G. (1998) Cancer Res. 58, 3579–3585 [PubMed] [Google Scholar]
- 32. Ribic C. M., Sargent D. J., Moore M. J., Thibodeau S. N., French A. J., Goldberg R. M., Hamilton S. R., Laurent-Puig P., Gryfe R., Shepherd L. E., Tu D., Redston M., Gallinger S. (2003) N. Engl. J. Med. 349, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Popat S., Hubner R., Houlston R. S. (2005) J. Clin. Oncol. 23, 609–618 [DOI] [PubMed] [Google Scholar]
- 34. Sargent D. J., Marsoni S., Monges G., Thibodeau S. N., Labianca R., Hamilton S. R., French A. J., Kabat B., Foster N. R., Torri V., Ribic C., Grothey A., Moore M., Zaniboni A., Seitz J. F., Sinicrope F., Gallinger S. (2010) J. Clin. Oncol. 28, 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bertagnolli M. M., Niedzwiecki D., Compton C. C., Hahn H. P., Hall M., Damas B., Jewell S. D., Mayer R. J., Goldberg R. M., Saltz L. B., Warren R. S., Redston M. (2009) J. Clin. Oncol. 27, 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tejpar S., Bosman F., Delorenzi M., Fiocca R., Yan P., Klingbiel D., Dietrich D., Van Cutsem E., Labianca R., Roth A. (2009) J. Clin. Oncol. 27, 4001 [Google Scholar]
- 37. Vilar E., Scaltriti M., Balmaña J., Saura C., Guzman M., Arribas J., Baselga J., Tabernero J. (2008) Br. J. Cancer 99, 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez R., Hansen L. T., Phear G., Scorah J., Spang-Thomsen M., Cox A., Helleday T., Meuth M. (2008) Clin. Cancer Res. 14, 5476–5483 [DOI] [PubMed] [Google Scholar]
- 39. Mendes-Pereira A. M., Martin S. A., Brough R., McCarthy A., Taylor J. R., Kim J. S., Waldman T., Lord C. J., Ashworth A. (2009) EMBO Mol. Med. 1, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McEllin B., Camacho C. V., Mukherjee B., Hahm B., Tomimatsu N., Bachoo R. M., Burma S. (2010) Cancer Res. 70, 5457–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohonen-Corish M. R., Daniel J. J., Chan C., Lin B. P., Kwun S. Y., Dent O. F., Dhillon V. S., Trent R. J., Chapuis P. H., Bokey E. L. (2005) J. Clin. Oncol. 23, 2318–2324 [DOI] [PubMed] [Google Scholar]
- 42. Wright C. M., Dent O. F., Newland R. C., Barker M., Chapuis P. H., Bokey E. L., Young J. P., Leggett B. A., Jass J. R., Macdonald G. A. (2005) Gut. 54, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]