Abstract
TRPM1 is the founding member of the melastatin subgroup of transient receptor potential (TRP) proteins, but it has not yet been firmly established that TRPM1 proteins form ion channels. Consequently, the biophysical and pharmacological properties of these proteins are largely unknown. Here we show that heterologous expression of TRPM1 proteins induces ionic conductances that can be activated by extracellular steroid application. However the current amplitudes observed were too small to enable a reliable biophysical characterization. We overcame this limitation by modifying TRPM1 channels in several independent ways that increased the similarity to the closely related TRPM3 channels. The resulting constructs produced considerably larger currents after overexpression. We also demonstrate that unmodified TRPM1 and TRPM3 proteins form functional heteromultimeric channels. With these approaches, we measured the divalent permeability profile and found that channels containing the pore of TRPM1 are inhibited by extracellular zinc ions at physiological concentrations, in contrast to channels containing only the pore of TRPM3. Applying these findings to pancreatic β cells, we found that TRPM1 proteins do not play a major role in steroid-activated currents of these cells. The inhibition of TRPM1 by zinc ions is primarily due to a short stretch of seven amino acids present only in the pore region of TRPM1 but not of TRPM3. Combined, our data demonstrate that TRPM1 proteins are bona fide ion-conducting plasma membrane channels. Their distinct biophysical properties allow a reliable identification of endogenous TRPM1-mediated currents.
Keywords: Ion Channels, Pancreatic Islet, Retina, TRP Channels, Zinc, Ion Channel Pharmacology, Ion Channel Selectivity, Retinal Bipolar Cell
Introduction
TRPM1 (Transient receptor potential melastatin 1) was identified in 1998 as a protein down-regulated in highly metastatic melanoma cells (1). Since then, it has been established that the expression level of TRPM1 is a clinically useful prognostic marker after diagnosis of melanoma (2–4). In melanocytes, TRPM1 expression is necessary for the function of endogenous nonselective cation channels with constitutive activity, because experimentally down-regulating TRPM1 expression diminished these currents (5).
Recently, TRPM1 has also been implicated in an entirely different disease, congenital stationary night blindness. Initially, it was found that TRPM1 mRNA levels are severely down-regulated in horses with this condition (6). This has since been extended by showing that TRPM1-deficient mice also have severe visual impairments (7, 8) and that in some cases of human congenital stationary night blindness, patients bear mutations in TRPM1 (9–12). TRPM1 mRNA is highly expressed in a subpopulation of bipolar cells (13–15), at least some of which are ON-bipolar cells. In ON-bipolar cells, glutamate released from rod photoreceptor cells activates mGluR6 metabotropic glutamate receptors (16, 17). Those receptors in turn activate a poorly understood signaling cascade that involves Gαo proteins (18, 19) and leads to closure of a constitutively active, nonselective cationic conductance (20, 21). TRPM1 proteins are necessary for this transduction pathway to work. In TRPM1-deficient mice, the constitutively active currents are not observed, and no response can be obtained from rod ON-bipolar cells with the appropriate glutamatergic stimulation (7, 8, 22). Based on these results and because of the vast amount of accumulated knowledge that shows that the majority of TRP proteins forms nonselective cation channels (23), it was suggested that TRPM1 proteins actually form the pore of the transduction channel in rod ON-bipolar cells.
In agreement with this, Koike et al. (8) found that overexpression of TRPM1 proteins in CHO cells induces a constitutively active conductance. Others have not found any evidence for spontaneous currents after heterologous overexpression of TRPM1 but found increased nonselective currents when overexpressing TRPM1 in melanocytes that, however, also express these proteins endogenously (5). It should be noted that the currents observed after overexpression were very small and sometimes were only measured under highly artificial ionic conditions (8). This raises the question of whether overexpressed TRPM1 proteins do really form plasma membrane channels. An alternative explanation for the reported data is that overexpression of TRPM1 proteins might, in some cases, lead to the up-regulation of another, unrelated channel protein. Furthermore, in melanocytes and bipolar cells, it might be possible that TRPM1 does not participate in forming an ion-conducting pore, but that it is simply necessary for the function of the observed nonselective ionic conductances, for example by being a compulsory accessory subunit.
To answer this important question, a detailed description of the functional characteristics, including the biophysical and pharmacological properties of heterologously expressed TRPM1 channels, would be desirable. However, because of the aforementioned difficulties in studying overexpressed TRPM1 channels, this information has so far not been forthcoming. In this study, we provide very strong evidence that TRPM1 proteins are capable of forming ion-conducting channels in the plasma membrane. By utilizing several strategies to increase the current amplitude through the pore of TRPM1, we were able to characterize this pore, revealing differences to the pore of the most closely related channel TRPM3 (24), especially with respect to the action of Zn2+ ions.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293 and a HEK293-derived cell line stably transfected with Myc-TRPM3α2-EYFP were cultured as described (25). We also followed the previously published procedures to isolate and culture mouse (C57BL/6N) pancreatic islet cells (26). We identified pancreatic β cells by their response to pregnenolone sulfate (26). COS cells were cultured in the same conditions as HEK293 cells but using DMEM (Invitrogen) supplemented with 10% FCS as medium.
Mutagenesis and Construction of Chimeric Constructs
Point mutations were generated in a similar way as described previously (27) using circular, double-stranded plasmid DNA templates, appropriate primers, and the Phusion polymerase (New England Biolabs).
All of the mutants and chimeras were based on the N-terminally Myc-tagged cDNA of TRPM1Δ1066–1071 (accession number AJ867483) encoding 1622 amino acid residues (28). To obtain TRPM1Δex11 (see Fig. 1B), two PmeI restriction sites were introduced into the TRPM1 cDNA, and a PmeI fragment (nucleotides 1319–1508) was replaced with the phosphorylated oligonucleotides 5′-TTG GCC CAC ACT GGC CGG T-3′ and 5′-ACC GGC CAG TGTG GGC CAA-3′, which were hybridized with each other, leading to a TRPM1 construct missing exactly exon 11 (Pro-446 to Trp-502).
FIGURE 1.
Alignment of TRPM1 and TRPM3 sequences representing some mutations used in this study. A, alignment of the primary sequence of the pore region between transmembrane domains 5 and 6 of TRPM3 and TRPM1. Restriction sites for the enzyme HpaI (arrows) were introduced by silent mutations. The region swapped in the chimeric proteins is located between the HpaI restriction sites. Also indicated is histidine 1034 (H1034), the place for the three point mutations (H1034F, E1036N, and K1038E; triple mutation), and the LYAMEIN motif studied in Fig. 10. B, amino acid sequence alignment of TRPM1 (AJ867483), TRPM3 (AJ544535), TRPM2 (BAD83707), TRPM4 (AJ575814), TRPM5 (AF177473), TRPM6 (AY135644), TRPM7 (AY032951), and TRPM8 (AY095352) showing that the sequence of TRPM1 exon 11 is lacking in TRPM3 and the other TRPM channels. Accordingly, TRPM1Δex11 was constructed by deleting the 57 amino acids encoded by exon 11.
To delete the motif comprising seven amino acids from the TRPM1 pore (LYAMEIN motif; see Fig. 1A), single PmeI and SmaI restriction sites were introduced into the TRPM1 cDNA. The PmeI/SmaI fragment (nucleotides 3179–3225) was subsequently exchanged by the phosphorylated oligonucleotides 5′-TTG CAG ACC AGA TAG ACC CTC CTT GT-3′ and 5′-ACA AGG AGG GTC TAT CTG GTC TGC AA-3′ that were hybridized with each other. Similarly, the introduction of the LYAMEIN motif into the TRPM3 pore (variant α2; Ref. 25; accession number AJ544535) was achieved by introducing single ClaI and SmaI restrictions sites into the TRPM3 cDNA and replacing a ClaI/SmaI fragment (nucleotides 3189–3217) by the phosphorylated oligonucleotides 5′-CGA TCT CTA TGC CAT GGA AAT CAA CCC TCC CTG TGG ACA GAA TGA GAC CC-3′ and 5′-GGG TCT CATT CTG TCC ACA GGG AGG GTT GAT TTC CAT GGC ATA GAG AT-3′, again hybridized before ligation.
Chimeras of TRPM1 and TRPM3 were generated by introducing silent HpaI restriction sites (Fig. 1A), which were identified with the aid of the “silent” program included in the European Molecular Biology software suite (29) and subsequent swapping of fragments.
All of the modified constructs were verified by sequencing of both strands. For heterologous overexpression, the wild type and mutant cDNAs were introduced into the bicistronic vector pCAGGSM2-IRES-GFP (30) that carried an adapted multiple cloning site and had its HpaI restriction sites removed.
For FRET measurements, DNA fragments encoding TRPM1 (AJ867482) and TRPM3 (AJ544532), each carrying a Myc epitope at their N terminus, were introduced into the SmaI sites of the vectors pECFP-N1 and pEYFP-N1. The resulting constructs allowed expression of the following fusion proteins: Myc-TRPM1-ECFP, Myc-TRPM1-EYFP, Myc-TRPM3-ECFP, and Myc-TRPM3-EYFP.
Electrophysiology and Ca2+ Imaging
We used the standard whole cell patch-clamp technique (31) and applied voltage ramps (from −115 to +85 mV) at a rate of ∼1/s from a holding potential of −15 mV on stably or transiently transfected HEK293 cells. Patch pipettes were pulled from borosilicate glass tubing (Science Products, Hofheim, Germany) and had resistances of 2.5–5 mΩ in standard bath solution. Polyfect (Qiagen) was used according to the instructions of the manufacturer for transient transfections. When co-transfecting TRPM1 and TRPM3 for electrophysiological experiments, we used a plasmid ratio of 3:1 to reduce the formation of TRPM3 homomultimeric channels except where explicitly stated otherwise. Typically, 24 h after transfection, the cells were split and reseeded in culture dishes to reduce cell density. Single cells not in contact with neighboring cells were used 10–96 h after transfection.
The composition of the intracellular (pipette) solution was 80–90 mm CsAsp, 45 mm CsCl, 4 mm Na2ATP, 10 mm BAPTA, 5 mm EDTA, and 10 mm HEPES. The pH was adjusted to 7.2 with CsOH (adding ∼60 mm Cs+ to the solution), and the osmolality was adjusted to values within the range of 305–320 mosm. The standard bath solution contained 145 mm NaCl, 3 mm KCl, 10 mm CsCl, 2 mm MgCl2, 2 mm CaCl2, and 10 mm HEPES. A pH of 7.2 was obtained with NaOH (2–5 mm). The liquid junction potential between the pipette solution and the standard bath solution (15 mV) was corrected for. Pregnenolone sulfate (from 50 mm stock in Me2SO) and ZnCl2 (0.001–1 mm) were added to yield the final concentrations as indicated. The solution containing 120 mm Ca2+ was composed of 120 mm CaCl2, 10 mm HEPES, and 16 mm d-glucose. Approximately 3 mm N-methyl-d-glucamine was used to adjust the pH to 7.2. The solutions containing 10 mm of a divalent cation was composed of 10 mm XCl2 (where X represents the divalent cation in question), 10 mm HEPES, and 265–290 mm d-glucose or mannitol. N-Methyl-d-glucamine (1–10 mm) was used to adjust the pH to 7.2 or to 7.0–7.1 in case of Zn2+ (to avoid precipitation). All of the extracellular solutions had an osmolality of 320–335 mosm, adjusted with d-glucose. Solution exchange was performed with a custom-build gravity-driven system with a small dead space. The data were analyzed offline by extracting the current amplitudes at +80 and −80 mV. Statistical tests were performed with Excel or GraphPad prism (version 3.0).
For Ca2+ imaging, the cells were loaded for 30 min with 5 μm of the AM-ester of Fura-2 (Sigma). The procedures for measuring the ratio of fluorescence intensities at 340 nm and 380 nm were standard and described in detail previously (26).
We used Student's t test, when appropriate with Bonferroni correction, or analysis of variance with Newman-Keul's post test. Values of p < 0.05 were considered significant and are denoted with a single asterisk (*). p < 0.01 is shown with two asterisks (**), and triple asterisks (***) indicate p < 0.001.
Co-immunoprecipitation
Co-immunoprecipitation was performed essentially as described (32). Four dishes (diameter, 3 cm) of HEK293 cells stably transfected with TRPM3 and grown to 70–80% confluence were each transfected with 3 μg of pCAGGS-IRES-GFP constructs encoding mouse TRPM1. The cells were washed twice 48 h after transfection with ice-cold phosphate-buffered saline (136 mm NaCl, 2.7 mm KCl, 1.47 mm KH2PO4, 8 mm Na2HPO4, pH 7.4), lysed in 1.0 ml of ice-cold radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-Cl, pH 8.0, 0.5% sodium deoxycholate, 1% Igepal (Sigma), 0.1% SDS, 5 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 1 μm benzamidine, 0.3 μm aprotinin, 1 μg/ml leupeptin, 1 μm pepstatin, 1 μg/ml antipain, 1 mm phenanthroline, and 0.9 μm iodacetamide), and homogenized at 4 °C by forcing through a 27-gauge cannula. After centrifugation for 15 min at 12,000 × g, the supernatant was transferred to a fresh tube. As a measure for the amount of proteins introduced into immunoprecipitation (input), of the solubilized proteins was directly denaturated by adding loading buffer (120 mm Tris-Cl, pH 6.8, 8% sodium dodecyl sulfate, 20% glycerine, 0.01% bromphenol blue, 10% 2-mercaptoethanol). 100 μl of protein G-Sepharose (GE Healthcare) equilibrated three times with 1 ml of radioimmune precipitation assay buffer were incubated with solubilized proteins for 2 h and centrifuged for 3 min at 1000 × g to remove proteins that interact nonspecifically with the Sepharose.
The remaining supernatant was incubated with 5 μg of affinity-purified polyclonal anti-TRPM3 antibodies (25) or anti-TRPM1 antibodies (directed against the C terminus of TRPM1, accession numbers NP_001034193, residues 1602–1622) for 4 h at 4 °C under continuous shaking. Immunocomplexes were precipitated by incubation with 100 μl of protein G-Sepharose for 2 h, centrifuged, and washed four times with 1 ml of radioimmune precipitation assay buffer. Complexes bound to protein G-Sepharose were dissolved by incubation in 50 μl of loading buffer at 95 °C for 5 min. The samples were centrifuged briefly at 12,000 × g, and each sample was subjected to SDS-PAGE. Immunoblots were performed as described (25) using anti-TRPM1- and anti-TRPM3 antibodies and corresponding horseradish peroxidase-labeled secondary antibodies. The labeled proteins were visualized using the Renaissance Western lightning chemiluminescence reagent plus (PerkinElmer).
FRET Measurements
Static FRET was measured as donor recovery during acceptor bleach. As donor we used eCFP5 and as acceptor we used eYFP, each fused to the C terminus of the channel protein of interest. In co-transfections for FRET measurements, we transfected three times more DNA of the CFP-containing vector than of the YFP containing vector to minimize the fraction of multimeric channels containing only YFP and no CFP. Fluorescence of transfected live cells (in standard bath solution) was excited with light from a xenon light bulb that passed through a monochromator (Polychrome IV; TILL Photonics, Gräfelfing, Germany), a dual pass excitation filter, and a dichroic mirror with two reflective wavelength ranges (both optical components from the “51017” CFP/YFP set, Chroma). We used a 40× (NA = 1.3) oil immersion objective on a Nikon TE2000 inverted microscope. The recorded area was limited with the viewfinder device (TILL Photonics) to a rectangle that contained a single cell. The emitted light was split by a second dichroic mirror (50% at 505 nm) in two channels that were each equipped with a emission filter (470/30 nm and 535/30 nm). The light intensity was measured by two photodiodes (TILL Photonics), and the analog signals were digitized with an EPC 10 (HEKA, Lambrecht, Germany). The software Patchmaster controlled the monochromator, the shutter, and the data acquisition. The stimulus protocol consisted of repeated pairs of light flashes (50 ms) at 435 and 500 nm. After measuring 10 pairs (to determine the baseline), another 50 pairs of light flashes were applied, with the shutter opened for 5 s after each pair of light flashes to bleach the acceptor (enhanced YFP) with light of 480 nm. After finishing this protocol, the microscope stage was moved slightly to move the cell out of the field of view, and the background was measured for both excitation wavelengths.
The data were analyzed with the aid of custom software written in Matlab. After background subtraction, the stability of the baseline recordings (first 10 measured values) was assessed, and only cells with good baseline stability were analyzed further. The FRET efficiency was calculated individually for each cell according to the formula given in Ref. 33.
RESULTS
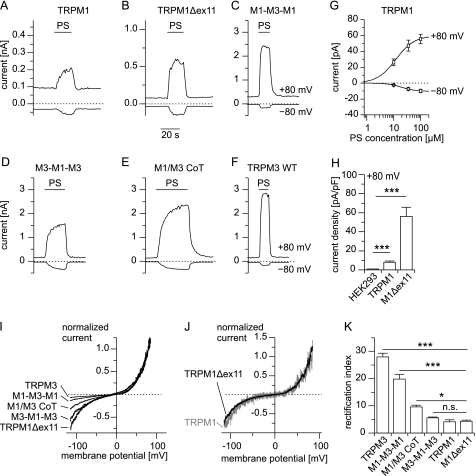
To test whether TRPM1 proteins form functional ion channels, we overexpressed TRPM1 proteins in HEK293 cells and measured the membrane currents with the whole cell patch-clamp technique. Under our standard recording conditions, we did not observe constitutively active conductances in transfected cells, in agreement with a previous report (5). We recently described that pregnenolone sulfate (PS) strongly activates heterologously and endogenously expressed TRPM3 channels (26). Because TRPM3 is the closest relative to TRPM1 (34), we tested whether PS was also capable of activating TRPM1 proteins. Indeed, we observed a reversible increase in inward and outward current upon application of PS in cells transfected with TRPM1 but not in control cells (Fig. 2, A, G, and H). However, the observed current densities were very small and were only marginally larger when we applied an increased PS concentration (100 μm; Fig. 2G). Furthermore, the current amplitudes were variable across different transfections. We considered these current amplitudes too small for a meaningful biophysical analysis. Additionally, these very small amplitudes raise doubts regarding whether the observed currents indeed flow through pores formed by TRPM1 proteins, because it is conceivable that overexpressing TRPM1 proteins might lead to altered expression of other channel-forming proteins.
FIGURE 2.
HEK293 cells transfected with TRPM1 display steroid-evoked currents with rectification behavior distinct from TRPM3 transfected cells. A–F, typical current traces (at −80 and +80 mV) of HEK293 cells transfected with the indicated constructs during application of PS (35 μm). A graphical representation of the chimeric constructs used is given in supplemental Fig. S1. G, dose-response curve for currents induced by PS in cells transiently overexpressing TRPM1 (n = 6–7). H, statistical analysis of the current density (+80 mV) of the currents induced by 35 μm PS in cells overexpressing TRPM1 or TRPM1Δex11 compared with untransfected control cells (n = 17–19). I, normalized (to +80 mV) current-voltage relationship of the PS-induced currents for the indicated constructs. J, direct comparison of background-subtracted and normalized current-voltage relationship after overexpression of TRPM1 or TRPM1Δex11. K, statistical analysis of the rectification index (RI = I+80 mV/I−80 mV * (−1)) calculated from recordings similar to those shown in I and J (n = 11–44).
Three Strategies to Increase Current Amplitudes in TRPM1-expressing Cells
We devised three independent experimental strategies to augment the current amplitudes through putative TRPM1 channels. First we reasoned that it may be possible, given the close sequence homology between TRPM1 and TRPM3 in the pore region (Fig. 1A), to transplant the pore region of TRPM1 into TRPM3 proteins and vice versa. We were surprised to find that both chimeric proteins constructed accordingly (M1-M3-M1 and M3-M1-M3; supplemental Fig. S1) reliably supported large PS-induced currents after overexpression in HEK293 cells (Fig. 2, C and D). This finding opened the possibility of studying the properties of the TRPM1 pore. However, the approach taken suffers from the drawback that the backbone of TRPM3 possibly influences the properties of the TRPM1 pore. However, the use of the construct M1-M3-M1 (TRPM3 pore inserted into TRPM1) as control allowed estimation of the impact of the pore transplantation on the biophysical properties of the already well characterized TRPM3 pore.
In the second approach, we identified an exon (exon 11) that is present in all TRPM1 cDNA clones but has no corresponding homologous region neither in TRPM3 nor in any other TRPM channel (Fig. 1B). It encodes 57 amino acids in the N-terminal cytosolic region of TRPM1. Reasoning that the region encoded by exon 11 may reduce the current amplitude, we constructed TRPM1Δex11 that lacked exon 11 (Fig. 1B). When overexpressing TRPM1Δex11, we observed PS-induced current amplitudes that were on average 6.9 times larger than those obtained by overexpressing wild type TRPM1 (Fig. 2, B and H). The advantage of this approach, therefore, appears to be that the TRPM1 protein was much less altered compared with our first approach and not changed at all in the transmembrane region. The disadvantage of this approach was that the current amplitudes were still relatively small.
In our final approach, we considered the propensity of TRP channels to form heteromultimers (35) that has also been observed in TRPM6 and TRPM7 (36, 37), close relatives of TRPM1 and TRPM3. We therefore co-expressed TRPM1 and TRPM3 with a cDNA ratio of 3:1, to avoid the formation of significant amounts of TRPM3 homomultimeric channels.
TRPM1 and TRPM3 Form Heteromultimeric Channels
In cells co-expressing TRPM1 and TRPM3, we recorded large PS-induced currents that had conspicuously larger inward currents than those obtained from cells expressing TRPM3 alone (Fig. 2, E and F). Indeed, quantitative inspection of the rectification properties of the I/V curves from all constructs revealed that channels with the pore region of TRPM1 had significantly larger relative inward currents (normalized to outward currents at +80 mV) compared with channels with a pore of TRPM3 (Fig. 2, I–K). Importantly, the rectification properties of TRPM1Δex11 and wild type TRPM1 were indistinguishable from each other (Fig. 2J). This systematic variation of the rectification properties clearly indicates different properties of TRPM1 and TRPM3 channels that can be studied with the approaches outlined above.
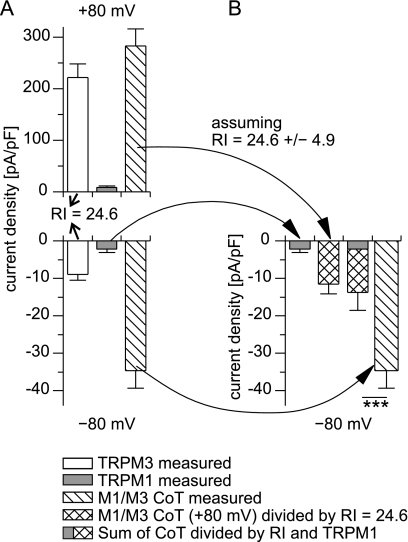
Also, cells co-expressing TRPM1 and TRPM3 had altered rectification properties that were quantitatively in between those of cells expressing only TRPM3 or only TRPM1. Initially, two interpretations of this result are possible. First, it is conceivable that cells co-expressing TRPM1 and TRPM3 form two independent types of channels, each only comprising one type of protein (TRPM1 or TRPM3). The altered rectification observed in those cells would then reflect the average of those two channel types. Alternatively, cells co-expressing TRPM1 and TRPM3 could form heteromultimeric channels that each contain both types of proteins. The altered rectification properties in this scenario then reflect the properties of the heteromultimeric channels. As detailed below, a quantitative analysis of the inward currents observed in cells co-expressing TRPM1 and TRPM3 (Fig. 3) strongly supports the second option (that TRPM1 and TRPM3 proteins together form functional heteromultimeric channels).
FIGURE 3.
The inward current amplitude observed in cells co-transfected with TRPM1 and TRPM3 cannot be explained by assuming the independent formation of homomultimeric TRPM1 and TRPM3 channels. A, average current density values at −80 and +80 mV measured in cells expressing either TRPM3 (n = 38) or TRPM1 (n = 12) alone or both together (ratio TRPM1:TRPM3 = 3:1, n = 37). From the averaged values of the PS-induced currents of TRPM3, a RI of 24.6 ± 4.9 (S.E.) was calculated (arrows). B, inward current amplitudes are given, assuming the hypothesis that all PS-induced channels are either TRPM3 channels (cross-hatched bar, calculated with the RI obtained in A), TRPM1 channels (gray bar, as measured in A with cells transfected only with TRPM1), or the sum of both (composite bar). Even the sum of both values (composite bar) is still significantly smaller than the inward current amplitude actually measured from co-transfected cells (hatched bar). This demonstrates that the hypothesis of independent formation of homomultimeric TRPM1 and TRPM3 channels is incompatible with the experimental data. The errors of the experimental values were propagated appropriately in the calculations.
We tried to estimate the maximal amplitude of the inward current density, assuming that cells co-expressing TRPM1 and TRPM3 proteins form only one type of channel, either from TRPM1 or from TRPM3 proteins. Making the assumption that all PS-induced channels are TRPM1 channels, the expected current density was simply taken to be the current density in cells that have been transfected with TRPM1 only (Fig. 3, A and B, gray bars). Note that this is a generous estimate of TRPM1 current amplitude, because in the co-transfected cells, only 75% of the transfected cDNA encodes for TRPM1 proteins.
Because the current amplitudes in cells transfected with TRPM3 are large but variable (depending on time after transfection, age of the cells, and transfection method, among others), we did not use absolute current amplitudes but calculated the rectification index from the averaged values of the PS-induced currents of TRPM3 (Fig. 3A; rectification index (RI) = 24.6 ± 4.9). (Note that this value is slightly different from the values obtained in Fig. 2K, because here we divided the averaged current densities, whereas in Fig. 2K this quotient was calculated for each cell individually and then averaged. This slight quantitative difference is not important to the argument presented here.) Now, making the assumption that in cells co-transfected with TRPM1 and TRPM3, all PS-induced current is flowing through TRPM3 channels, the expected inward current was calculated by taking the RI value of TRPM3 and the outward current density measured in the cells co-transfected with TRPM1 and TRPM3. The resulting value (Fig. 3B, cross-hatched bar) represents the maximal inward current density that can be expected from homomultimeric TRPM3 channels (given the amplitude of the outward current density).
Because the ratio of putative TRPM1 and TRPM3 homomultimeric channels is not known, we took the sum of both previously calculated values (Fig. 3B, composite bar). This represents the unfavorable (and physically impossible) assumption that all of the inward current results at the same time from both homomultimeric TRPM1 and homomultimeric TRPM3 channels. However, even with this disadvantageous calculation, the resulting inward current density is significantly smaller than the PS-induced current density actually measured in cells co-transfected with TRPM1 and TRPM3 (Fig. 3B, hatched bar). This demonstrates that the amplitude of the inward current density in cells co-expressing TRPM1 and TRPM3 cannot be explained by assuming independent formation of homomultimeric channels. Hence, we have to assume that TRPM1 and TRPM3 proteins interact with each other in the co-transfected cells.
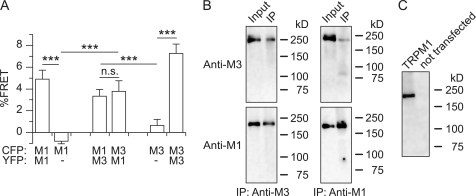
To add further evidence for the interaction of TRPM1 and TRPM3 proteins, we performed FRET experiments with TRPM1 and TRPM3 proteins tagged with GFP variants at their C termini. FRET signals were obtained from single cells, not differentiating between intracellular proteins and proteins at the plasma membrane. We found that TRPM1 and TRPM3 co-expressed in a single cell produced a positive FRET signal, whereas we did not observe such a signal in cells expressing only CFP-tagged proteins (Fig. 4A). Additionally, TRPM3 immunoprecipitated with an antibody that recognized TRPM1 (as demonstrated in Fig. 4C) and vice versa from cells overexpressing both proteins (Fig. 4B). Together, these data constitute decisive evidence that TRPM1 and TRPM3 proteins interact with each other and thereby form functional ion channels that have altered biophysical properties. These data also provide evidence that in these heteromultimeric channels, wild type TRPM1 proteins might indeed be pore-forming subunits of ion channels.
FIGURE 4.
Co-expressed TRPM1 and TRPM3 proteins interact with each other biochemically. A, significant FRET signals were obtained from cells co-expressing constructs encoding for TRPM1 and TRPM3 proteins fused C-terminally to either CFP or YFP. This shows that the distance between TRPM1 and TRPM3 proteins in the intact cells was very small (n = 13–25). B, HEK293 cells stably transfected with TRPM3 and transiently transfected with TRPM1 were used to immunoprecipitate (IP) proteins with anti-TRPM3 (left-hand panels) or anti-TRPM1 (right-hand panels) antibodies. After blotting, TRPM3 proteins (upper panels) and TRPM1 (lower panels) were detected with the antibodies indicated. The results indicate that TRPM1 and TRPM3 proteins interact with each other and co-immunoprecipitate. C, Western blot showing that the anti-TRPM1 antibody recognized TRPM1 proteins in transfected COS cells but not in untransfected control cells.
The Pore of TRPM1 Channels Is Permeable for Ca2+ but Not for Zn2+ Ions
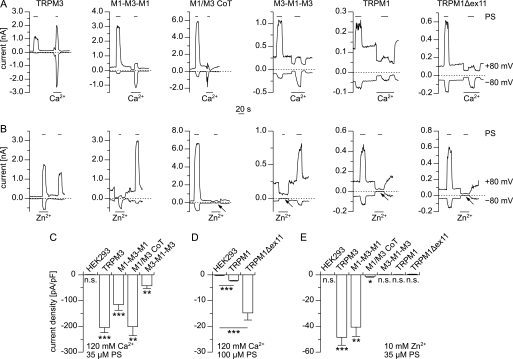
Having demonstrated that TRPM1 proteins can participate in the pore of ion channels, we aimed at characterizing the permeation properties of the TRPM1 pore. First, we tested for Ca2+ permeability by using bi-ionic conditions with 120 mm Ca2+ at the extracellular side (Fig. 5A). Upon application of PS, we observed increased inward currents with all constructs (Fig. 5, C and D). Even when we overexpressed wild type TRPM1 proteins, the resulting PS-induced Ca2+ current was significantly larger than in control cells (Fig. 5D). These data show that the TRPM1 pore is permeable for Ca2+ ions. This finding was corroborated by experiments on Fura-2-loaded cells transfected with either TRPM1Δex11 or TRPM1 (supplemental Fig. S2). Application of PS clearly induced an increase in the Fura-2 ratio in the transfected cells but not in untransfected control cells, consistent with a PS-induced opening of a Ca2+-permeable pore.
FIGURE 5.
Channels containing the pore region of TRPM1 are permeable to Ca2+ but not to Zn2+. A, typical current traces (at −80 and +80 mV) measured from cells transfected with the indicated construct. PS was applied (as indicated by the bars) in standard bath solution and in a solution containing only Ca2+ ions (120 mm) as cationic charge carriers. B, similar experiments as shown in A, but using a solution containing 10 mm Zn2+ (instead of Ca2+). In cells expressing constructs with the pore of TRPM1, inward currents carried by Zn2+ were very small or undetectable (arrows). C and D, statistical analysis of inward current densities of PS-induced currents with Ca2+ as the only extracellular cation (n = 7–10 in C, and n = 18–20 in D). Significant inward currents could be observed with all constructs, showing that the pores of all constructs were permeable to Ca2+. E, inward current density of currents carried only by Zn2+ (n = 5–18). The absence of significant inward currents in constructs containing the pore of TRPM1 shows that this pore is impermeable for Zn2+ ions. The small currents after co-transfecting TRPM1 and TRPM3 may possibly be accounted for by a small number of homomultimeric TRPM3 channels. The statistical tests in C and E were performed to evaluate the difference from 0.
Because we have recently shown that the pore of TRPM3 is highly permeable for Zn2+ ions (38), we next repeated the electrophysiological experiments with 10 mm Zn2+ on the extracellular side to test for Zn2+ permeability (Fig. 5B). Although channels containing the TRPM3 pore but not the TRPM1 pore (wild type TRPM3 and M1-M3-M1) showed large Zn2+-carried inward currents, those currents were hardly visible in cells co-transfected with TRPM1 and TRPM3 and entirely absent in cells transfected with constructs containing only the pore region of TRPM1 (Fig. 5E). These data establish that the pore of TRPM1 is essentially impermeable for Zn2+ ions.
Divalent Permeability Profile of the TRPM1 Pore
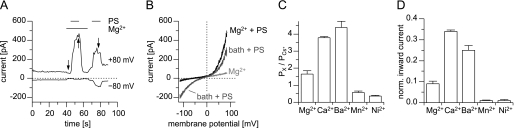
We extended these studies with Ca2+ and Zn2+ to other divalent cations (Fig. 6) by studying the PS-evoked currents in cells expressing TRPM1Δex11. We found that all divalent cations tested (Mg2+, Mn2+, Ba2+, and Ni2+) permeated through channels formed by TRPM1Δex11. Analysis of the reversal potentials revealed permeability ratios for the divalent cations (relative to Cs+) of 0.36 ± 0.04 (Ni2+) to 4.39 ± 0.37 (Ba2+). Thus, the pore of TRPM1 is permeable to these cations but to a lesser extent than the pore of TRPM3 (38). A direct comparison of the relative divalent permeabilities of these two channels is given in supplemental Fig. S3.
FIGURE 6.
Relative permeability ratios of the TRPM1 pore. A, exemplary recording of currents (+80 and −80 mV) from a HEK293 cell transfected with TRPM1Δex11. Bi-ionic conditions with 10 mm Mg2+ and 50 μm PS were applied as indicated. B, corresponding current voltage relationships, obtained at time points indicated by arrows in A. C, statistical analysis of relative permeability ratios obtained with different divalent cations from cells expressing TRPM1Δex11 (n = 6–9). The pore of TRPM1 is permeable to all divalent cations tested with the exception of Zn2+. For a direct comparison with TRPM3 channels, see supplemental Fig. S3. D, inward current amplitudes (normalized to the current amplitude measured with standard bath solution) for the different divalent cations measured under the same bi-ionic conditions (n = 6).
From these data, we also analyzed the conductivity of the TRPM1 pore for the different divalent cations by measuring the inward currents at −80 mV and normalizing them to the current amplitude obtained in standard bath solution (Fig. 6D). Contrary to the situation in TRPM3 (38), we found that for the pore of TRPM1, the rank order of relative permeabilities matched the rank order of conductivities rather closely (Fig. 6, C and D). However, the divalent conductivity of the TRPM1 pore was much less than the divalent conductivity of TRPM3, because the divalent cations at a concentration of 10 mm all produced less inward current than the standard bath solution.
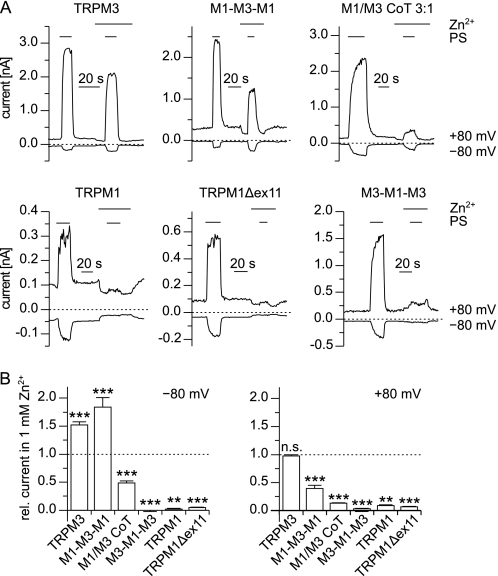
TRPM1 Channels Are Inhibited by Extracellular Zn2+ Ions
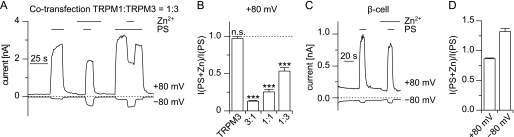
Close examination of the data presented in Fig. 5B shows that during application of the Zn2+ solution not only inward currents but also outward currents were inhibited in channels harboring the pore of TRPM1. This may indicate that extracellular Zn2+ inhibits the TRPM1 pore. To test this possibility, we performed experiments in which we added 1 mm Zn2+ to the standard extracellular solution (Fig. 7). The currents evoked by application of PS in the presence of 1 mm Zn2+ were unaltered (+80 mV) or increased (−80 mV) in cells expressing TRPM3 (Fig. 7B). Also, the inward currents through M1-M3-M1 were increased by the presence of 1 mm Zn2+ (Fig. 7, A and B). This may be due to the increased number of charge carriers, because the TRPM3 pore is highly permeable for Zn2+ ions (Ref. 38 and Fig. 5, B and E). In contrast, 1 mm Zn2+ strongly inhibited the channels that contained the pore of TRPM1 (Fig. 7, A and B). Importantly, also the channels obtained after co-transfecting TRPM1 and TRPM3 were strongly inhibited by the addition of 1 mm Zn2+ to the extracellular solution, indicating that the TRPM1 subunits in the pore of these channels exert a dominant effect over the TRPM3 subunits (Fig. 7, A and B). Because we used a co-transfection ratio of 3:1 (TRPM1:TRPM3), we wondered whether this dominant effect only occurs at higher concentrations of the TRPM1 subunit. Altering the transfection ratio to 1:1 and then to 1:3 (Fig. 8, A and B), we observed that the inhibition caused by zinc was reduced dose-dependently depending on the fraction of TRPM1 subunits. Quantitatively, these experiments are difficult to interpret because of the increased formation of TRPM3 homomultimeric channels. However, they clearly show that even a minor contribution of TRPM1 proteins to PS-sensitive channels can be detected by investigating the zinc sensitivity of these channels. Together, these data show that zinc ions inhibit the channel pores containing TRPM1 proteins but not those consisting of TRPM3 proteins.
FIGURE 7.
Channels containing the pore region of TRPM1 are inhibited by extracellular Zn2+. A, example traces of recordings (−80 and +80 mV) from cells transfected with the indicated constructs. PS (100 μm for TRPM1 and TRPM1Δex11, 35 μm for other constructs) was applied in standard bath solution and in standard bath solution supplemented with 1 mm Zn2+. In channels containing the pore region of TRPM1, the addition of 1 mm Zn2+ caused a severe suppression of the PS-induced currents. B, statistical analysis of recordings similar to those shown in A for the constructs indicated (n = 5–18). The amplitude of the PS-induced current (same concentrations of PS as in A) in solution containing 1 mm Zn2+ was normalized to the amplitude of the current in standard bath solution. The statistical tests evaluated the differences from 1 (dashed line). The addition of 1 mm Zn2+ increased the inward currents (−80 mV, left hand panel) through TRPM3 and M1-M3-M1 but reduced these currents in the other constructs. Outward currents (+80 mV, right-hand panel) through all constructs but TRPM3 were inhibited by 1 mm Zn2+.
FIGURE 8.
The extent of inhibition of PS-induced currents by Zn2+ depends on the ratio of TRPM1:TRPM3 expression. A, example trace of a cell co-transfected with TRPM1 and TRPM3 in a ratio of 1:3. PS (35 μm) and Zn2+ ions (1 mm) were applied as indicated. Zn2+ still inhibited the PS-induced outward currents, although the transfection regime allowed for substantial amounts of TRPM3 homomultimeric channels to form. B, statistical analysis of the inhibition of the PS-induced outward current (+80 mV) by 1 mm Zn2+. The PS-induced outward current (+80 mV) in the presence of 1 mm Zn2+ was divided by the amplitude of the PS-induced outward current in the absence of Zn2+. The cells were transfected either with TRPM3 or co-transfected with TRPM1 and TRPM3 in the ratio indicated (n = 18–19). The data for TRPM3 and the 3:1 transfection are the same as in Fig. 7B and shown for comparison. Statistical tests were performed to evaluate the difference from 1 (dashed line). At all co-transfection ratios, the inhibition by Zn2+ was highly significant. C, similar recording as shown in A but from a primary pancreatic β cell. D, statistical analysis of the inhibition caused by application of 1 mm Zn2+ in β cells at −80 and +80 mV (n = 61). Although there is significant reduction of the outward current in the presence of 1 mm Zn2+, the amount of inhibition is small compared with the inhibition observed with the unfavorable transfection ratio of 1:3 (B).
To apply these findings to endogenous PS-sensitive channels, we turned to primary mouse pancreatic β cells, because we previously reported that these cells endogenously express functional PS-activated channels, for which TRPM3 proteins play an indispensable role (26). However, the precise molecular composition of these channels is still unclear. We tested whether TRPM1 proteins participate in the formation of PS-activated channels in pancreatic β cells by testing whether 1 mm Zn2+ was capable of reducing the PS-induced currents in these cells (Fig. 8, C and D, and supplemental Fig. S4). Although we tested a large number of cells (n = 61) from both female and male animals with a diverse range of age, we did not find an indication that the addition of 1 mm Zn2+ reduced the PS-induced current amplitude in primary mouse pancreatic β cells consistently. The reduction that we observed at +80 mV (to 86 ± 1.3% of control values) is less than those observed even for the unfavorable transfection ratio 1:3 (Fig. 8B). At −80 mV, we did not observe any reduction of the current amplitude caused by application of Zn2+. These data indicate that TRPM1 proteins do not play a major role in PS-activated channels in primary pancreatic β cells.
Dose Dependence of Zn2+ Inhibition of the TRPM1 Pore
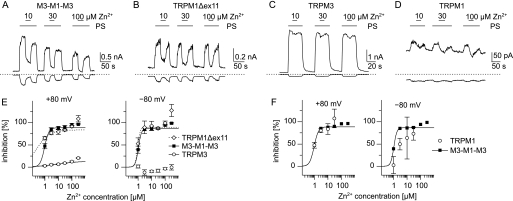
Having determined that Zn2+ ions inhibit the TRPM1 pore, we next aimed to determine the dose dependence of this effect. We found that applying 1 μm Zn2+ already reduced the response to 100 μm PS to roughly 50% in inward and outward currents in cells expressing M3-M1-M3 or TRPM1Δex11 (Fig. 9, A, B, and E), but not in TRPM3-expressing cells (Fig. 9, C and E). In TRPM1-expressing cells, the zinc ions appear to exert an inhibitory effect as potently as in M3-M1-M3 and TRPM1Δex11-expressing cells (Fig. 9, D and F); however, because of the small amplitude of the currents, the data are much less reliable, especially for inward currents. These data establish that the pore of TRPM1 channels is inhibited with an IC50 value of ∼1 μm.
FIGURE 9.
Dose dependence of the inhibition of PS-induced currents by zinc. A–D, typical current recordings (−80 and +80 mV) from cells expressing the indicated constructs. PS (100 μm) and Zn2+ (at the given concentrations) were applied as indicated. E, dose-response curves of the inhibitory effect of Zn2+ on cells expressing the constructs indicated. Measured data points between 1 and 30 μm were fit to Hill functions (lines). F, direct comparison of the dose-response curves for M3-M1-M3 (■) and wild type TRPM1 (○) constructed as in E, showing that wild type TRPM1 channels are inhibited by Zn2+ in a quantitatively similar way as the artificial chimera that only contains the pore of TRPM1 (n = 4–18 per data point in E and F).
Molecular Determinants of Zn2+ Inhibition of TRPM1
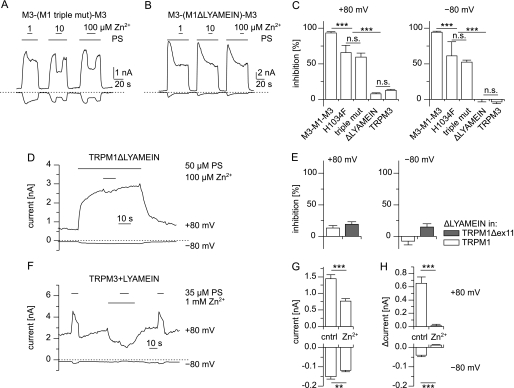
The pore region between TRPM1 and TRPM3 is highly homologous, and the differences can be interpreted (as aligned in Fig. 1A) as 15 point mutations and one deletion from TRPM1 (LYAMEIN) and one insertion into TRPM1 (TI). This high degree of homology allows testing in a straightforward manner which of those alterations is responsible for the high sensitivity to Zn2+ present in TRPM1 but absent in TRPM3. We first mutated a histidine residue (His-1034) in the pore region of TRPM1 (Fig. 1A) to phenylalanine present at the homologous position in TRPM3 (H1034F). Because two other amino acids in close proximity differ in TRPM1 with respect to TRPM3, we also created the triple mutation that changes the entire locus from the one found in TRPM1 to the corresponding sequence in TRPM3 (Fig. 1A). Finally, we constructed the pore region of TRPM1 that lacks the seven-amino acid motif that is also absent in TRPM3 (ΔLYAMEIN). The three modified TRPM1 pore regions (H1034F, triple mutation, and ΔLYAMEIN) were first placed in the TRPM3 backbone, so that they could directly be compared with M3-M1-M3 and TRPM3. We assayed the PS-induced currents in these constructs with and without Zn2+ ions added. At 100 μm Zn2+, in H1034F and in the triple mutant, a modest but significant reduction in the inhibitory capacity of Zn2+ was noted (Fig. 10, A and C). However, in the construct lacking the LYAMEIN motif, no inhibition induced by the Zn2+ ions was seen (Fig. 10, B and C). This strongly indicates that the LYAMEIN motif is the essential difference between the pore region of TRPM3 and TRPM1 responsible for the Zn2+ inhibition. We then repeated these experiments with constructs in which LYAMEIN motif was deleted from wild type TRPM1 and TRPM1Δex11 and obtained essentially the same results, because no inhibitory effect of 100 μm Zn2+ could be detected in cells transfected with these constructs (Fig. 10, D and E).
FIGURE 10.
Identification of the molecular determinants that cause the inhibition of TRPM1 by Zn2+. A and B, exemplary current traces obtained from cells expressing constructs with mutated pores of TRPM1 placed in TRPM3 channels. In the construct used in A, three point mutations were introduced in the pore region, whereas in B seven amino acids (LYAMEIN motif) were deleted (see Fig. 1A). C, statistical analysis of the inhibition caused by application of 100 μm Zn2+ from constructs used in A and B (n = 4–18). The values for M3-M1-M3 and TRPM3 (taken from Fig. 9E) are presented for comparison. Although mutating a single amino acid (H1034F) and mutating three amino acids (H1034F, E1036N, and K1038E; triple mut) reduced the inhibitory effect of Zn2+, Zn2+ inhibition was completely abolished by the removal of the LYAMEIN sequence from the pore of TRPM1. D, recording from a cell expressing a construct in which the LYAMEIN sequence was deleted from wild type TRPM1 proteins. E, statistical analysis of similar recordings as shown in D. The LYAMEIN sequence was removed from wild type TRPM1 and TRPM1Δex11 proteins. In both constructs, 100 μm Zn2+ did not cause a strong inhibition of the PS-induced currents (n = 6–9). F, typical recording (+80 and −80 mV) of a HEK293 cell transfected with TRPM3+LYAMEIN. PS (35 μm) and Zn2+ (1 mm) were added to the standard bath solution as indicated. G and H, statistical analysis of the magnitude of the constitutive current (G) and the PS-induced current (H) without (control) and during application of 1 mm Zn2+. The upper panel shows outward (+80 mV) currents, and the lower panel shows inward (−80 mV) currents (n = 17). The addition of zinc significantly reduced the current amplitudes of the constitutive and the PS-evoked currents.
To further establish the importance of the LYAMEIN motif, we constructed the inverse mutant and introduced this motif into the pore region of TRPM3 proteins. Testing this construct, we found that the addition of the LYAMEIN motif to the pore of TRPM3 caused a substantially increased spontaneous activity that was strongly reduced by the addition of 1 mm Zn2+ (Fig. 10, F and G). Furthermore, the PS-induced increase in current amplitude that could be measured on top of the spontaneous activity was also reduced by the addition of 1 mm Zn2+ to the extracellular medium (Fig. 10, F and H). Together these data show that the introduction of the LYAMEIN motif into the pore of TRPM3 renders this pore sensitive to inhibition by Zn2+ ions and thus provides strong independent evidence for the importance of this motif.
DISCUSSION
In the past, it has been difficult to determine whether TRPM1 proteins, like most other members of the TRP family, participate in forming ion-conducting channels in the plasma membrane (23). After overexpression of TRPM1 channels, an early report found elevated Ca2+ permeability of the plasma membrane (39), and recently, an increased membrane conductance was reported (8). Additionally, TRPM1 proteins have been shown to be necessary for endogenous, constitutively active nonselective cation conductances in melanocytes (5) and ON-bipolar cells (7, 8, 22). However, these data do not allow the conclusion that TRPM1 proteins themselves participate in an ion-conducting pathway. To prove this, it is necessary to demonstrate that the biophysical properties of the resulting channels can predictably be altered by experimental manipulations (40). Here we show that co-expression of TRPM1 and TRPM3 alters several biophysical properties compared with wild type TRPM3 channels, i.e. rectification (Fig. 2), permeability to Zn2+ (Fig. 5), and inhibition by Zn2+ ions (Fig. 7). Importantly, we exclude the possibility that the currents observed in co-transfected cells are mediated by a mixture of separate TRPM1 and TRPM3 channels (Fig. 3). We thus establish that TRPM1 and TRPM3 proteins form functional heteromultimeric channels. A parsimonious interpretation of this finding is that TRPM1 participates in the formation of the pore and thereby alters the biophysical properties of these channels.
This interpretation received strong support from additional, independent experiments, because our data also provide decisive evidence that the pore of TRPM1 proteins is capable of forming functional membrane channels on its own. Two independent lines of evidence led us to this conclusion. The first line came from chimeric channels that contain the TRPM1 pore in the TRPM3 backbone (M3-M1-M3), as well as in TRPM1 channels that lack exon 11 (TRPM1Δex11). Both constructs display much larger currents after overexpression and have very similar biophysical properties (Figs. 2, 5, and 7), making it unlikely that the alterations of the primary sequence have distorted the biophysical properties of the ion-conducting pathway. In support of this, we also found that a control construct (M1-M3-M1) retained much of the biophysical properties of TRPM3. More importantly, however, wild type TRPM1 channels, when activated with PS, also share all of the biophysical properties displayed by the artificial constructs containing the TRPM1 pore (e.g. inhibition by Zn2+), reinforcing our conclusion that TRPM1 proteins are bona fide ion channels with biophysical properties distinct from TRPM3 channels.
The second line of evidence came from experiments in which we removed the LYAMEIN motif from TRPM1 and added these seven amino acids to TRPM3. These manipulations removed Zn2+ inhibition from TRPM1 and transferred it to TRPM3. Therefore, these experiments show that the pore properties of TRPM1 proteins can be altered predictably and thus allow identifying TRPM1 as a true pore forming protein.
Channels containing the pore of TRPM1 are potently inhibited by Zn2+ ions, whereas TRPM3 channels conduct these ions well (Figs. 7–9 and Ref. 38). Interestingly, it is not uncommon that closely related ion channels are differentially regulated by Zn2+ ions. Well established examples are NMDA receptors (41, 42), T-type voltage-gated Ca2+ channels (43), and acid-sensing ion channels (44), where zinc sensitivity is differentially present in closely related isoforms or even in splice variants.
The closest relatives to TRPM3 and TRPM1 channels, TRPM6 and TRPM7, also form heteromultimeric channels (36, 37). Both are also highly permeable to divalent cations, including Zn2+ ions (36, 37, 45–47). This is also true for dTRPM, the only TRPM channel present in Drosophila melanogaster (48). The most striking difference in primary sequence between TRPM1 on one hand and TRPM3, TRPM6, TRPM7, and dTRPM on the other hand is the addition of seven amino acids in the pore region (the LYAMEIN motif), making it a prime candidate for mediating the zinc sensitivity of TRPM1. Indeed, as already discussed above, removal of this motif from the pore of TRPM1 abolished the inhibition by Zn2+, whereas the addition of this motif to the pore of TRPM3 introduces inhibition by Zn2+ ions (Fig. 10). Interestingly, introducing another motif of 12 amino acids in the same position of TRPM3 channels yields the naturally occurring splice variant TRPM3α1 that has very poor or no permeability for divalent cations (25). Therefore, it appears that the properties of the pore of TRPM1 and TRPM3 channels can be fine-tuned to different permeation profiles for divalent cations by subtle alterations of the length and amino acid composition of this short stretch. Likely, it will be possible to obtain significant knowledge about the structural correlate of divalent permeation profiles by systematically studying this protein region. Although our data clearly show that the LYAMEIN motif is the most important determinant of Zn2+ sensitivity, other parts of the pore also appear to play a role. Mutating a single histidine residue of the TRPM1 pore (His-1034) significantly attenuated the inhibition by 100 μm Zn2+. This residue is located at a distance of 32 amino acids to the LYAMEIN motif, showing that the pore properties of TRPM1 and TRPM3 channels are not determined by this short, localized region alone but rather are encoded by an extended set of residues.
TRPM1 channels are inhibited by Zn2+ ions with an IC50 of approximately 1 μm (Fig. 9). This value is much higher than the resting free concentration of zinc that in the brain has been estimated to be 19 nm (49). However, zinc is often co-released with glutamate, and in the tiny synaptic cleft, free zinc concentrations reaching or exceeding 1 μm may very well be obtained (50–54). It is significant that in the retina, release of zinc from the terminals of rod photoreceptors has been demonstrated (55, 56), because TRPM1 has been shown to be located at the postsynaptic membrane of bipolar dendrites at the photoreceptor-to-bipolar cell synapse (7, 8), where it will be exposed to zinc released from photoreceptor cells during darkness. If TRPM1 proteins form, as proposed, the transduction channel in ON-bipolar cells, the rapid release of zinc at the onset of darkness might quickly inhibit TRPM1 channels, possibly faster than the transduction cascade initiated by glutamate binding to mGluR6 receptors could turn off these channels. Such a mechanism could therefore significantly improve the temporal resolution of the information transmitted by bipolar cells. However, the extent of inhibition of rod bipolar cell TRPM1 channels by Zn2+ during darkness is unclear. It appears not to be complete because application of mGluR6 antagonists in retina slices that retained light sensitivity increased the channel activity even in darkness (57).
In conclusion, our results provide compelling evidence that TRPM1 proteins are functional ion-conducting plasma membrane channels. Our approach opens the way for a detailed biophysical and pharmacological characterization of TRPM1 channels, and we thus demonstrate that TRPM1 proteins are inhibited by low micromolar concentrations of Zn2+. Our findings allow distinguishing TRPM1 channels from their close homologs TRPM3, TRPM6, and TRPM7 and enable the elucidation in vivo of their physiological and pathophysiological function.
Acknowledgments
We thank Dr. Veit Flockerzi for support and help with anti-TRPM1 antibodies and co-immunoprecipitation. We also thank Dr. Roger Hardie for helpful discussions and him and Dr. Andreas Beck for critically reading the manuscript. The excellent technical assistance of Heidi Löhr, Ute Soltek, and Martin Simon-Thomas is gratefully acknowledged.
This work was supported by Deutsche Forschungsgemeinschaft (GK 1326, SFB 530, SFB 894 and Emmy Noether-program), HOMFOR, and Forschungskommission der Universität des Saarlandes.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- eCFP
- enhanced cyan fluorescent protein
- YFP
- yellow fluorescent protein
- PS
- pregnenolone sulfate
- RI
- rectification index.
REFERENCES
- 1. Duncan L. M., Deeds J., Hunter J., Shao J., Holmgren L. M., Woolf E. A., Tepper R. I., Shyjan A. W. (1998) Cancer Res. 58, 1515–1520 [PubMed] [Google Scholar]
- 2. Duncan L. M., Deeds J., Cronin F. E., Donovan M., Sober A. J., Kauffman M., McCarthy J. J. (2001) J. Clin. Oncol. 19, 568–576 [DOI] [PubMed] [Google Scholar]
- 3. Carlson J. A., Ross J. S., Slominski A., Linette G., Mysliborski J., Hill J., Mihm M., Jr. (2005) J. Am. Acad. Dermatol. 52, 743–778 [DOI] [PubMed] [Google Scholar]
- 4. Erickson L. A., Letts G. A., Shah S. M., Shackelton J. B., Duncan L. M. (2009) Mod. Pathol. 22, 969–976 [DOI] [PubMed] [Google Scholar]
- 5. Oancea E., Vriens J., Brauchi S., Jun J., Splawski I., Clapham D. E. (2009) Science Signal. 2, ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellone R. R., Brooks S. A., Sandmeyer L., Murphy B. A., Forsyth G., Archer S., Bailey E., Grahn B. (2008) Genetics 179, 1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morgans C. W., Zhang J., Jeffrey B. G., Nelson S. M., Burke N. S., Duvoisin R. M., Brown R. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19174–19178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koike C., Obara T., Uriu Y., Numata T., Sanuki R., Miyata K., Koyasu T., Ueno S., Funabiki K., Tani A., Ueda H., Kondo M., Mori Y., Tachibana M., Furukawa T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Audo I., Kohl S., Leroy B. P., Munier F. L., Guillonneau X., Mohand-Saïd S., Bujakowska K., Nandrot E. F., Lorenz B., Preising M., Kellner U., Renner A. B., Bernd A., Antonio A., Moskova-Doumanova V., Lancelot M. E., Poloschek C. M., Drumare I., Defoort-Dhellemmes S., Wissinger B., Léveillard T., Hamel C. P., Schorderet D. F., De Baere E., Berger W., Jacobson S. G., Zrenner E., Sahel J. A., Bhattacharya S. S., Zeitz C. (2009) Am. J. Hum. Genet. 85, 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Genderen M. M., Bijveld M. M., Claassen Y. B., Florijn R. J., Pearring J. N., Meire F. M., McCall M. A., Riemslag F. C., Gregg R. G., Bergen A. A., Kamermans M. (2009) Am. J. Hum. Genet. 85, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z., Sergouniotis P. I., Michaelides M., Mackay D. S., Wright G. A., Devery S., Moore A. T., Holder G. E., Robson A. G., Webster A. R. (2009) Am. J. Hum. Genet. 85, 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura M., Sanuki R., Yasuma T. R., Onishi A., Nishiguchi K. M., Koike C., Kadowaki M., Kondo M., Miyake Y., Furukawa T. (2010) Mol. Vis. 16, 425–437 [PMC free article] [PubMed] [Google Scholar]
- 13. Kim D. S., Matsuda T., Cepko C. L. (2008) J. Neurosci. 28, 7748–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim D. S., Ross S. E., Trimarchi J. M., Aach J., Greenberg M. E., Cepko C. L. (2008) J. Comp. Neurol. 507, 1795–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakajima Y., Moriyama M., Hattori M., Minato N., Nakanishi S. (2009) J. Biochem. 145, 811–818 [DOI] [PubMed] [Google Scholar]
- 16. Nakajima Y., Iwakabe H., Akazawa C., Nawa H., Shigemoto R., Mizuno N., Nakanishi S. (1993) J. Biol. Chem. 268, 11868–11873 [PubMed] [Google Scholar]
- 17. Masu M., Iwakabe H., Tagawa Y., Miyoshi T., Yamashita M., Fukuda Y., Sasaki H., Hiroi K., Nakamura Y., Shigemoto R. (1995) Cell 80, 757–765 [DOI] [PubMed] [Google Scholar]
- 18. Snellman J., Kaur T., Shen Y., Nawy S. (2008) Prog. Retin. Eye Res. 27, 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okawa H., Pahlberg J., Rieke F., Birnbaumer L., Sampath A. P. (2010) J. Gen. Physiol. 136, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nawy S., Jahr C. E. (1990) Nature 346, 269–271 [DOI] [PubMed] [Google Scholar]
- 21. Nawy S., Jahr C. E. (1991) Neuron 7, 677–683 [DOI] [PubMed] [Google Scholar]
- 22. Shen Y., Heimel J. A., Kamermans M., Peachey N. S., Gregg R. G., Nawy S. (2009) J. Neurosci. 29, 6088–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Owsianik G., Talavera K., Voets T., Nilius B. (2006) Annu. Rev. Physiol. 68, 685–717 [DOI] [PubMed] [Google Scholar]
- 24. Oberwinkler J., Phillipp S. E. (2007) Handb. Exp. Pharmacol. 179, 253–267 [DOI] [PubMed] [Google Scholar]
- 25. Oberwinkler J., Lis A., Giehl K. M., Flockerzi V., Philipp S. E. (2005) J. Biol. Chem. 280, 22540–22548 [DOI] [PubMed] [Google Scholar]
- 26. Wagner T. F., Loch S., Lambert S., Straub I., Mannebach S., Mathar I., Düfer M., Lis A., Flockerzi V., Philipp S. E., Oberwinkler J. (2008) Nat. Cell Biol. 10, 1421–1430 [DOI] [PubMed] [Google Scholar]
- 27. Braman J., Papworth C., Greener A. (1996) Methods Mol. Biol. 57, 31–44 [DOI] [PubMed] [Google Scholar]
- 28. Lis A., Wissenbach U., Philipp S. E. (2005) Naunyn Schmiedebergs Arch. Pharmacol. 371, 315–324 [DOI] [PubMed] [Google Scholar]
- 29. Rice P., Longden I., Bleasby A. (2000) Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 30. Warnat J., Philipp S., Zimmer S., Flockerzi V., Cavalié A. (1999) J. Physiol. 518, 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Pflügers Arch. 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 32. Philipp S., Strauss B., Hirnet D., Wissenbach U., Mery L., Flockerzi V., Hoth M. (2003) J. Biol. Chem. 278, 26629–26638 [DOI] [PubMed] [Google Scholar]
- 33. Amiri H., Schultz G., Schaefer M. (2003) Cell Calcium 33, 463–470 [DOI] [PubMed] [Google Scholar]
- 34. Lee N., Chen J., Sun L., Wu S., Gray K. R., Rich A., Huang M., Lin J. H., Feder J. N., Janovitz E. B., Levesque P. C., Blanar M. A. (2003) J. Biol. Chem. 278, 20890–20897 [DOI] [PubMed] [Google Scholar]
- 35. Schaefer M. (2005) Pflügers Arch. 451, 35–42 [DOI] [PubMed] [Google Scholar]
- 36. Chubanov V., Waldegger S., Mederos y Schnitzler M., Vitzthum H., Sassen M. C., Seyberth H. W., Konrad M., Gudermann T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2894–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M., Jiang J., Yue L. (2006) J. Gen. Physiol. 127, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagner T. F., Drews A., Loch S., Mohr F., Philipp S. E., Lambert S., Oberwinkler J. (2010) Pflügers Arch. 460, 755–765 [DOI] [PubMed] [Google Scholar]
- 39. Xu X. Z., Moebius F., Gill D. L., Montell C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10692–10697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Voets T., Nilius B. (2003) Cell Calcium 33, 299–302 [DOI] [PubMed] [Google Scholar]
- 41. Paoletti P., Ascher P., Neyton J. (1997) J. Neurosci. 17, 5711–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mony L., Kew J. N., Gunthorpe M. J., Paoletti P. (2009) Br. J. Pharmacol. 157, 1301–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jeong S. W., Park B. G., Park J. Y., Lee J. W., Lee J. H. (2003) Neuroreport 14, 1537–1540 [DOI] [PubMed] [Google Scholar]
- 44. Chu X. P., Wemmie J. A., Wang W. Z., Zhu X. M., Saugstad J. A., Price M. P., Simon R. P., Xiong Z. G. (2004) J. Neurosci. 24, 8678–8689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nadler M. J., Hermosura M. C., Inabe K., Perraud A. L., Zhu Q., Stokes A. J., Kurosaki T., Kinet J. P., Penner R., Scharenberg A. M., Fleig A. (2001) Nature 411, 590–595 [DOI] [PubMed] [Google Scholar]
- 46. Monteilh-Zoller M. K., Hermosura M. C., Nadler M. J., Scharenberg A. M., Penner R., Fleig A. (2003) J. Gen. Physiol. 121, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voets T., Nilius B., Hoefs S., van der Kemp A. W., Droogmans G., Bindels R. J., Hoenderop J. G. (2004) J. Biol. Chem. 279, 19–25 [DOI] [PubMed] [Google Scholar]
- 48. Georgiev P., Okkenhaug H., Drews A., Wright D., Lambert S., Flick M., Carta V., Martel C., Oberwinkler J., Raghu P. (2010) Cell Metab. 12, 386–397 [DOI] [PubMed] [Google Scholar]
- 49. Frederickson C. J., Giblin L. J., Krezel A., McAdoo D. J., Mueller R. N., Zeng Y., Balaji R. V., Masalha R., Thompson R. B., Fierke C. A., Sarvey J. M., de Valdenebro M., Prough D. S., Zornow M. H. (2006) Exp. Neurol. 198, 285–293 [DOI] [PubMed] [Google Scholar]
- 50. Vogt K., Mellor J., Tong G., Nicoll R. (2000) Neuron 26, 187–196 [DOI] [PubMed] [Google Scholar]
- 51. Li Y., Hough C. J., Suh S. W., Sarvey J. M., Frederickson C. J. (2001) J. Neurophysiol. 86, 2597–2604 [DOI] [PubMed] [Google Scholar]
- 52. Komatsu K., Kikuchi K., Kojima H., Urano Y., Nagano T. (2005) J. Am. Chem. Soc. 127, 10197–10204 [DOI] [PubMed] [Google Scholar]
- 53. Frederickson C. J., Giblin L. J., 3rd, Balaji R. V., Rengarajan B., Masalha R., Frederickson C. J., Zeng Y., Lopez E. V., Koh J. Y., Chorin U., Besser L., Hershfinkel M., Li Y., Thompson R. B., Krezel A. (2006) J. Neurosci. Methods 154, 19–29 [DOI] [PubMed] [Google Scholar]
- 54. Paoletti P., Vergnano A. M., Barbour B., Casado M. (2009) Neuroscience 158, 126–136 [DOI] [PubMed] [Google Scholar]
- 55. Redenti S., Chappell R. L. (2005) Vision Res. 45, 3520–3525 [DOI] [PubMed] [Google Scholar]
- 56. Redenti S., Ripps H., Chappell R. L. (2007) Exp. Eye Res. 85, 580–584 [DOI] [PubMed] [Google Scholar]
- 57. Sampath A. P., Rieke F. (2004) Neuron 41, 431–443 [DOI] [PubMed] [Google Scholar]