Abstract
Grass is a clip domain serine protease (SP) involved in a proteolytic cascade triggering the Toll pathway activation of Drosophila during an immune response. Epistasic studies position it downstream of the apical protease ModSP and upstream of the terminal protease Spaetzle-processing enzyme. Here, we report the crystal structure of Grass zymogen. We found that Grass displays a rather deep active site cleft comparable with that of proteases of coagulation and complement cascades. A key distinctive feature is the presence of an additional loop (75-loop) in the proximity of the activation site localized on a protruding loop. All biochemical attempts to hydrolyze the activation site of Grass failed, strongly suggesting restricted access to this region. The 75-loop is thus proposed to constitute an original mechanism to prevent spontaneous activation. A comparison of Grass with clip serine proteases of known function involved in analogous proteolytic cascades allowed us to define two groups, according to the presence of the 75-loop and the conformation of the clip domain. One group (devoid of the 75-loop) contains penultimate proteases whereas the other contains terminal proteases. Using this classification, Grass appears to be a terminal protease. This result is evaluated according to the genetic data documenting Grass function.
Keywords: Crystal Structure, Drosophila, Innate Immunity, Insect, Proteolytic Enzymes, Serine Protease, Toll Receptors, X-ray Crystallography, Proteolytic Cascade
Introduction
Some biological processes such as blood coagulation in mammals or development and immune responses in invertebrates occur after the amplification of a recognition signal by serine proteases (SP)2 that are organized in cascades (1, 2). These SPs are characterized by a modular organization comprising a C-terminal catalytic domain and one or several N-terminal domains (CUB, EGF-like, LDL, CCP, or clip) and are activated in a very specific order. The SPs and SP homologs (SPHs are SPs where the catalytic triad is mutated) that contain one or more clip domains are called clip-SPs and clip-SPHs. The clip domain, which is found in the N-terminal position, consists of 35–55 residues including six strictly conserved cysteines arranged in three disulfide bonds. The clip and the catalytic domains are connected by a linker containing at least one cysteine, which is involved in an interdomain disulfide bond with a cysteine of the SP domain (1). The activation site of clip-SPs is located between the linker and the catalytic domain. After activation of the zymogen, the clip and SP domains remain linked by the interdomain disulfide bond (1).
Clip-SPs were first described for their role during Drosophila embryonic development, where they control the initiation of the dorso-ventral polarity (3). In early embryonic development, a ventral signal triggers the activation of a proteolytic cascade comprising a multidomain protease (gastrulation-defective, GD) and two clip-SPs, Snake and Easter. GD (4) converts Snake into its active form, which then activates Easter, the ultimate protease in the cascade. Easter then processes Spaetzle into the active ligand of the Toll receptor (5) (Fig. 1).
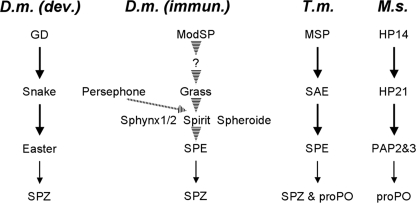
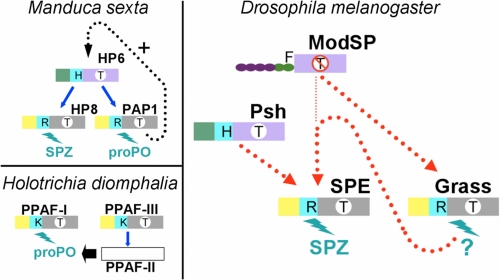
FIGURE 1.
Current knowledge of the proteolytic cascades involving clip-SPs, as described in the literature. The figure depicts the proteolytic cascades of the development and immunity. Left, development in D. melanogaster (D.m. dev.) (3). Center, immunity in D. melanogaster (D.m. immun.) (8, 9). Right, immunity in T. molitor (T.m.) (12) and in M. sexta (M.s.) (15). Arrows indicate experimentally verified direct links. Dashed arrows indicate steps that have not been experimentally verified or in which components of the pathway have not been identified.
To date, clip-SPs have only been identified in invertebrates. In addition to their role in controlling development, several members of the family have been shown to be involved in the activation of immune processes, including melanization via the phenoloxidase system and the production of antimicrobial peptides.
Drosophila has proved to be a powerful genetic model system for unraveling the role of the Toll receptor in the control of antimicrobial peptide synthesis following fungal and Gram-positive bacterial infection. However, none of the SPs described earlier for their role in the embryonic development appears to be required for the accomplishment of the immune function of Toll. The current model proposes that the detection of microbial motifs by appropriate pattern recognition receptors (PRRs) such as PGRP-SA, PGRP-SD, GNBP1, and GNBP3 triggers proteolytic cascades ending with the cleavage of Spaetzle and the subsequent activation of the Toll pathway.
At present, two clip-SPs, namely Spaetzle-processing enzyme (SPE) and Grass, have been demonstrated to participate in this cascade (6, 7). SPE was characterized as the functional equivalent of Dm-Easter, in vivo and in vitro as it processes Spaetzle (6). Grass was identified in the course of an exploratory in vivo RNAi study for the Toll pathway activation and was shown to be specifically associated with signaling in response to Gram-positive infection (8). However, a genetic study using a null mutant of Grass demonstrated that it defines a common protease cascade downstream of the fungal and bacterial pattern recognition receptors (7). More precisely, Grass mutant flies were susceptible to infection by the Gram-positive bacteria Enterococcus faecalis and by the fungus Beauveria bassiana. Grass overexpression in transgenic flies induces a constitutive activation of the Toll pathway that is abolished in SPE mutant flies. Overexpression of both GNBP1 and PGRP-SA also induces the activation of the Toll pathway, which is blocked in Grass mutant flies. A similar result was obtained when overexpressing GNBP3 alone. Taken together, these data suggest that Grass functions downstream of PRRs and upstream of SPE.
In addition to the clip-SPs, a multidomain serine protease, ModSP, was recently identified to be involved in Toll pathway activation (9). Based on epistasic analyses, ModSP was proposed to be the apical SP functioning downstream of the PRRs and upstream of Grass. Additionally, Kambris et al. reported the importance of one clip-SP named spirit and of the SPHs spheroide and sphinx1/2 for the activation of Toll pathway after a fungal or a bacterial infection (8).
Finally, a clip-SP activated independently of PRR named Persephone was identified by an ethylmethanesulfonate-induced mutation screen. Persephone was shown to be involved in response to a fungal infection (10). However, Persephone does not sense fungal molecular patterns downstream of GNBP3; rather, it was more recently proposed that Persephone senses the proteolytic activities elicited by both fungi and Gram-positive bacteria (7, 11).
Despite the accumulating genetic data, the current model of Drosophila Toll activation is still fragmentary (Fig. 1). In particular, no direct link could be established between any of the proteases. Complementary approaches at molecular and biochemical levels are necessary. However, the isolation of proteases for further ex vivo molecular studies is hampered by the small size of Drosophila adults. Insects of larger size and of easily extractable hemolymph, such as Bombyx mori, Manduca sexta, and Tenebrio molitor, are better choices for biochemical characterization (12–17). In particular, proteases and PRRs have been isolated from T. molitor, and a signaling cascade triggering the Toll activation has been reconstituted in vitro. It is composed of an apical modular SP named Tm-MSP and two clip-SPs, Tm-SAE and Tm-SPE. Tm-SPE cleaves Spaetzle in vitro (12, 13) (Fig. 1). In M. sexta, a three-step cascade has also been described for the prophenoloxidase (proPO) activation: HP14, a modular SP similar to Tm-MSP and Dm-ModSP, activates the clip-SP HP21, which in turn activates two clip-SPs, PAP2 and PAP3 (Fig. 1).
To date, the detailed molecular mechanisms underlying such cascades have yet to be elucidated. Only two crystal structures have been published. They document the proPO activation in the insect Holotrichia diomphalia. The first structure is that of an inactive SPH named PPAF-II, acting as a cofactor of proPO (18). The second structure is that of the catalytic domain of a proPO-activating enzyme, PPAF-I, that cleaves the proPO into smaller inactive forms (19).
We recently undertook a systematic structural characterization of the extracellular components identified in the activation of the Drosophila Toll receptor (20, 21). In this context, we determined the crystal structure of the clip-SP Grass, in its zymogen form, which represents the first structure of a full-length clip-SP. A detailed analysis was achieved to provide functional significance, and we propose a structure-based classification of the clip-SPs. This enables us to predict the position, penultimate or terminal, of any clip-SP within a cascade. Using this approach, we propose a new model for the role of Grass in Drosophila Toll pathway activation.
EXPERIMENTAL PROCEDURES
Cloning, Overexpression, and Purification of the Drosophila Proteases
Full-length Grass was cloned into the pMT-V5 His vector and co-transfected into S2 cells with pCoblast vector. Polyclonal and pseudoclonal stable cell lines were selected using blasticidin. After selection, cells were grown in suspension at 24 °C and kept under selection in 1 liter of Schneider's medium (Sigma) containing 5 μg/ml blasticidin (Invivogen), 50 μg/ml streptomycin (Invitrogen), 50 units/ml penicillin (Invitrogen), 2 mm Glutamax (Invitrogen), and 10% heat-inactivated fetal bovine serum. Expression of the secreted protein was induced by the addition of 0.5 mm CuSO4. Six days following induction, cells were aseptically centrifuged, resuspended in 1 liter of fresh medium, and induced again for 6 additional days. The cell culture supernatant was harvested, and after clarification, the recombinant protein was recovered by affinity chromatography (Chelating Sepharose Fast Flow; Amersham Biosciences) by elution with the loading buffer supplemented with 250 mm imidazole. Further purification was performed by size exclusion (Superdex 75 HiLoad 16/60 Prep grade; Amersham Biosciences) in 20 mm Hepes, pH 7.4, 150 mm NaCl.
A specific activation site (DDDDK) was introduced in a two-step strategy using the QuikChange Mutagenesis kit. The modified protein was expressed and purified using the same procedure as for the wild-type protease.
Protease Activation Assays
Grass zymogen (5–10 μg) was incubated with bovine and porcine trypsin (Sigma) (ratio 1/1000) for 20–75 min at room temperature in 10 mm Tris, pH 8, 0–10 mm CaCl2. Grass mutant zymogen (5 μg) was incubated with enterokinase (Invitrogen) (0.01–0.0002 units) for 1 h to overnight, at room temperature and 37 °C. A positive control (with a DDDDK sequence) was included in our experiments for enterokinase digestion. All of the reactions were stopped by the addition of Laemmli buffer and incubation at 95 °C for 5 min and analyzed by SDS-PAGE.
Edman Sequencing
After SDS-PAGE and Coomassie Blue staining, protein bands were excised. Proteins were extracted from the gel and blotted onto polyvinylidene difluoride membranes with the ProSobTM system (Applied Biosystems). The N-terminal sequences of proteins were determined by automated Edman degradation by introducing the blots into a Procise P494 automated protein sequencer (Applied Biosystems). The sequences obtained were compared with sequences in public protein sequence data bases.
Crystallization
Grass zymogen was concentrated up to 4.5 mg/ml. Initial crystallization trials were performed with Crystal Screens 1 and 2 (Hampton Research) using the hanging-drop vapor-diffusion method at 293 K. The drops were composed of equal volumes (1 μl) of protein solution (concentration of 4.5 mg/ml, in 20 mm Hepes, pH 7.4, and 150 mm NaCl buffer) and precipitant solution and were equilibrated against 0.3-ml reservoir volume. Although no crystal was obtained, condition 15, which gave crystalline precipitates, was selected for optimization. Conditions were varied for PEG8000 (22–32%) and for ammonium sulfate (0.16–0.20 m) in 100 mm sodium cacodylate buffer, pH 6.5.
Data Collection
For cryo-cooling, the crystals were soaked for a short time in reservoir solution supplemented with 20% ethylene glycol before being flash-frozen in a nitrogen gas stream at 100 K. X-ray diffraction data were collected to 1.8 Å resolution on beamline ID14-2 of the European Synchrotron Radiation Facility, Grenoble. The diffraction images were processed using MOSFLM (22) and scaled with the program SCALA (23) of the CCP4 suite (Collaborative Computational Project 4, 1994). The crystals belong to space group P212121, with unit-cell parameters a = 78.26 Å, b = 92.04 Å, and c = 113.34 Å. A Matthews coefficient Vm of 2.62 Å3 · Da−1 was calculated assuming two molecules in the asymmetric unit, which corresponds to 53% solvent content by volume.
Structure Resolution
The full-length protease was produced as a zymogen, and its structure was determined at 1.8 Å resolution by molecular replacement using the AMoRe program (24). The crystal structure of trypsin from Fusarium oxysporum (25) deleted of several loops served as the search model. Using data between 8.0 and 3.5 Å, the rotation function yielded one solution, and the translation function gave the position of the two molecules in the asymmetric unit, with a correlation coefficient and an R factor of 43 and 51.7%, respectively. The clip domain was then built in the resulting Fo − Fc electron density map using the Turbo-Frodo program (26). CNS (27), REFMAC (28), and BUSTER (29) refinements were carried out between 20 and 1.8 Å. After performing several cycles of refinement and manual replacement and building on the graphic display with the Turbo-Frodo program (26), the R factor decreased to 17.6% (R-free 20.3%). Strong 2Fo − Fc densities were observed close to the side chain of Asn230 and Asn270 and were assigned to sugars that were built in the densities. Crystallographic and refinement statistics are detailed in supplemental Table 1. Structural figures were generated with PyMOL.
Sequence Alignment of Catalytic Domains
The sequences of clip-SPs of known function, from various insect models (Drosophila melanogaster, B. mori, M. sexta, H. diomphalia, and T. molitor) were retrieved from the NCBI data base. The sequences were aligned using ClustalW2 (30), and the alignment was adjusted manually using superimposed crystal structures of Grass, PPAF-I, PPAF-II, and bovine trypsin. The alignment was displayed using ESPript program.
Accession Code
Atomic coordinates and structure factors have been deposited into the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) under the accession code 2XXL.
RESULTS
Structure of the Catalytic Domain of Grass
The structure of Grass consists of two domains, the clip and the catalytic domains connected by a linker comprising residues 91–118. The SP domain of Grass (Val119–Leu377) exhibits the characteristic polypeptide fold of trypsin-like SPs consisting of two β-barrels made of six β-strands stacked onto one another (Fig. 2A). The superimposition of Grass onto chymotrypsin (PDB code 1GL0) shows that 160 among 240 Cα (66%) of the model display equivalent positions in both molecules with distance between the superimposed residues Cα atoms <1.5 Å. The superimposition onto trypsin (PDB code 3BTE) results in 58% topologically equivalent positions (135 Cα of 230). Most of the inserted residues constitute surface loops named 30, 60, 75, 125, and 201, etc., according to chymotrypsin numbering. The numbering of Grass is that of the precursor, and that of chymotrypsinogen is sometimes indicated in parentheses and denoted with “c” for clarity. The catalytic triad, composed of the three conserved residues His163, Asp223, and Ser318 (corresponding to His57c, Asp102c, and Ser195c) (supplemental Fig. S1), stands in a prearranged conformation superimposable with that of active serine proteases such as trypsin and chymotrypsin. The transition from a zymogen to an active protease is associated with the formation, by proteolytic cleavage, of a new N terminus (Ile16c) which becomes buried within the molecule and after a conformational change in the so-called “activation domain” (residues 16c–19c, 142c–152c, 184c–193c, and 216c–223c) (31). In the recombinant Grass, the activation site (Arg118–Val119) is not cleaved, and several loops (140-loop, 180-loop, and 220-loop) do not stand in the canonical conformation of active SPs. This clearly indicates that the structure of Grass is that of the zymogen. Using the same superimposition strategy, the structure of Grass was also compared with that of Hd-PPAF-I and Hd-PPAF-II (PDB codes 2OLG and 2B9L, respectively).
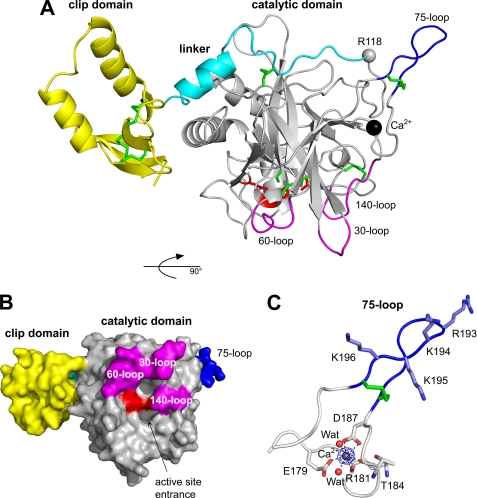
FIGURE 2.
Structure of clip-SP Grass. A, overall structure in ribbon representation, with the three domains, clip, linker, and catalytic, colored in yellow, cyan, and gray, respectively. The disulfide bridges and the catalytic triad are depicted as sticks and colored in green and red, respectively. The calcium ion is represented as a dark sphere and Arg118 (P1 of the activation site) as a gray sphere. The three loops shaping the active site cleft (30, 60, and 140) and the 75-loop are colored in magenta and blue, respectively. B, representation as molecular surface of the full-length protease. The molecule is rotated of 90° along the horizontal axis compared with A, and the color code is the same as in A. C, bound calcium ion drawn as a sphere and hepta-coordination indicated by dotted lines. Initial Fo − Fc electron density map contoured at 1.0 σ is displayed in blue.
Active Site Cleft
The active site cleft is shaped by three insertion loops, the 30-loop (138–144), the 60-loop (166–172), and the 140-loop (267–273), as illustrated in Fig. 2B. These loops were superimposed on those of digestive or coagulation/complement SPs (like trypsin, chymotrypsin, thrombin, or MASP1) and of Hd-PPAF-I.
The conformation of the 140-loop of Grass is such that it blocks access to the cleft, thus acting as a latch to prevent any accidental/nonspecific activity of the zymogen form. This function was described previously for Hd-PPAF-I (19). The 140-loop, which is known as the autolysis loop, was shown to undergo conformational changes upon activation (32). Thus, it may be possible that the 140-loops of both Grass and Hd-PPAF-I will adopt another conformation in the active proteases. The 30-loop of Grass is similar in size and conformation to that of Hd-PPAF-I. It is 5 residues longer than that of trypsin and 3 residues longer than that of MASP1. The 60-loop of Grass is 4 residues shorter than that of Hd-PPAF-I and is smaller than that of thrombin or MASP1 (6 and 14 residues, respectively). In general, the restricted specificity of proteases (for example in the coagulation) results from a deep and narrow active site cleft. Indeed, several studies highlighted the role of the 60-loop in regulating the specificity of the proteases by shielding the active-site pocket (33–35).
The sequence of the 30- and 60-loop of Grass was also compared with that of clip-SPs with a reported function (supplemental Fig. S2). No striking resemblance could be detected between the sequences of Grass and of the other clip-SPs. The 30-loops are rather homogeneous in size, and that of Grass is not different from the others. In contrast, the 60-loop of Grass is shorter than that of SPEs, Dm-Easter, or Ms-PAPs. Actually, the 60-loop of Grass resembles that of that of Hd-PPAF-III the most in size.
Activation Site Highly Resistant to Nonspecific Proteolysis
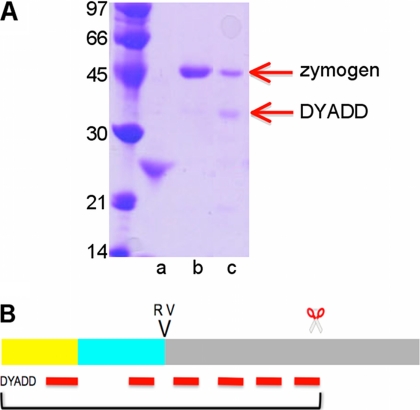
In Fig. 3A, SDS-PAGE analysis shows that Grass zymogen migrates at a molecular mass of 45 kDa. The endogenous activator of Grass in Drosophila is unknown; however, its cleavage site Arg118↓Val119 (P1–P′1 of the activation site) indicates that a trypsin-like activity is required. Hence, Grass zymogen was submitted to proteolysis by bovine and porcine pancreatic trypsin. This resulted in a decreased intensity of the band at 45 kDa and the appearance of a novel band at 30 kDa. Edman degradation showed that the sequence of the 30 kDa band matches the N terminus of Grass zymogen (DYAD), indicating that the hydrolysis did not occur at the activation site but within the catalytic domain. This was confirmed by peptide mass fingerprint analysis of this band. Indeed, the measured masses correspond to peptides that match the sequence between Asp27 and Lys251 of Grass (Fig. 3B). More drastic conditions resulted in a complete digestion of Grass zymogen.
FIGURE 3.
Limited proteolysis of Grass. A, digestion of Grass by trypsin, analyzed on a SDS-PAGE: Grass zymogen (lane b) and Grass digested by trypsin (lane c), and trypsin (lane a). A red arrow indicates the band that was sequenced, and the result of the N-terminal sequencing is indicated. B, schematic representation of Grass zymogen with the hydrolysis sites. Above the rectangles, two letters indicate the P1–P′1 activation site, and scissors indicate the site cleaved by trypsin. The color code is that of Fig. 2 (clip in yellow, linker in cyan, and catalytic domain in gray). Red lines stand for the fragments identified by peptide mass fingerprint.
A specific cleavage site for enterokinase was introduced in Grass by replacing the sequence FLSQR118 by DDDDK. Surprisingly, enterokinase did not cleave this mutant. Interestingly, a similar situation was already described for a Hd-PPAF-I mutant (19). The authors proposed that the depth of the active site cleft of enterokinase prevents its access to the activation site of Hd-PPAF-I.
Calcium Binding Loop (70-Loop)
An extra density was visible in the electron density map. By homology with Hd-PPAF-I (19), it was attributed to a calcium ion. It is hepta-coordinated with a pseudo-octahedral geometry involving the carboxylates of Glu179 (one oxygen) and Asp187 (two oxygens), the carbonyl oxygens of Thr184 and Arg181, and two molecules of water (Fig. 2C). These residues constitute the calcium binding loop (70-loop). The coordination is strictly identical to that found in the SP domain of Hd-PPAF-I (19).
75-Loop Prevents Access to the Activation Site
The 75-loop of Grass (188–197), which is a protuberance extending from the calcium binding loop (70-loop), folds into a hairpin. It is stabilized by a disulfide bridge between Cys188 and Cys197, two additional cysteines, compared with trypsin-like SPs (Fig. 2C). The 75-loop was described previously in Hd-PPAF-I and proposed by Piao et al. to restrict the access of the activation site (19). The high resistance of Grass and its mutant to activation is consistent with this hypothesis. Approximately half of the Drosophila clip-SPs display a 75-loop, which may vary in length and sequence. Grass contains a sequence of four positively charged residues, RKKK, that is highly conserved in the 12 species of Drosophila (RK(K/R/E/T)K). This positively charged patch may constitute a binding interaction module with a negatively charged ligand.
The superposition of Grass and Hd-PPAF-I (supplemental Fig. S3A) highlights slightly divergent conformations of the activation loops and the 75-loops, as depicted in supplemental Fig. S4. The activation loops do not superimpose from residues 114–123 of Grass and 104–114 of Hd-PPAF-I. In Hd-PPAF-I, the two loops stand very close to each other (4.9 Å between Cα of Ile110 and of Gly186) and the side chain (atom Nζ) of Lys109 (K109↓I110LNG cleavage site) is engaged in a hydrogen bond with the Val188 main chain carbonyl. Thus, the Lys109 side chain is not accessible to any activating protease. In Grass, the activation loop and the 75-loop are more distant (15 Å between Cα of Arg118 and of Gln190) due to the presence of a symmetry-related molecule. The spatial arrangement of the two loops in Grass may reflect the loop opening motion necessary for the activation process.
Clip Domain of Grass
The clip domain of Grass (residues 27–90) adopts an α/β mixed fold consisting of two helices, α1 (residues 49–61) and α2 (67–76), and an antiparallel distorted β-sheet made of four strands, β1 (29–34), β2 (37–43), β2B (78–81), and β3 (84–89) (Fig. 4A). The numbering of the secondary structure elements (α1, α2, β1, β2, and β3) is that used for the published structures of the two Ms-PAP2 clip domains (36). Grass contains an additional strand (β2B), which interacts with the β3 strand. The two helices are antiparallel and are almost perpendicular to the β-sheet. Three disulfide bridges (Cys32–Cys88, Cys42–Cys78, and Cys48–Cys89) stabilize the β-sheet, Cys48 being the only cysteine that is not located on a β-strand. The structure of the clip of Grass was compared with those of Ms-PAP2 (PDB codes 2IKD and 2IKE), which were determined by NMR (Fig. 4B). Because of a flexible linker between the two domains, they were considered as two separate entities for the resolution of the structures. The overall fold of the three clip domains is similar, notably with the presence of two antiparallel helices. Superimposition of Grass and Ms-PAP2 clips show that 26 and 30 residues among 56 and 54 have topologically equivalent position (with distance between the superimposed residues Cα atoms less than 1.5 Å), giving a structural similarity of 46 and 55% for clip1 and clip2, respectively. The major difference occurs in the region between residues 78 and 86. In Grass, this region is structured into a small sheet (β2B–β3) whereas in Ms-PAP2, the corresponding regions (54–62 and 113–120) form nonstructured loops that fold back toward the α2 helices. The conformation observed in Grass is likely due to the presence of the catalytic domain that contacts the tip of the loop (residues 81–84).
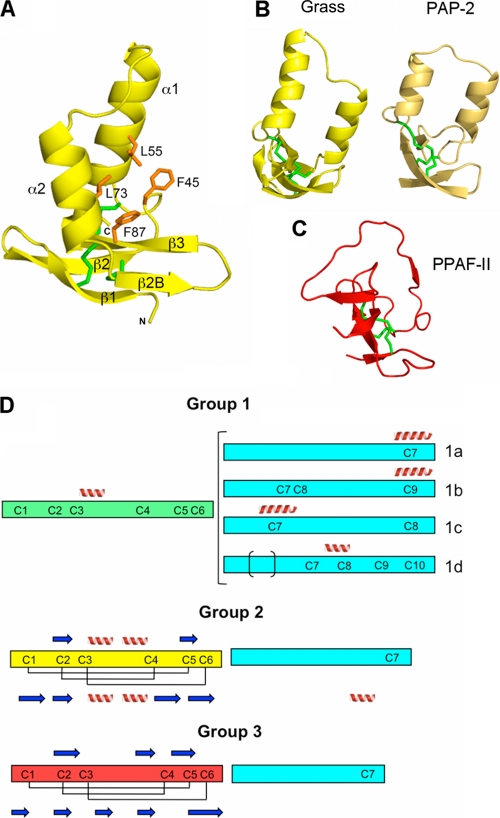
FIGURE 4.
Clip domain of Grass and the classification of the clips into three groups. A, structure of the clip domain in ribbon representation. The orientation is slightly different from in Fig. 2A, for more clarity on the β-strands. The hydrophobic residues are represented as orange sticks. The cysteines are colored in green, and the α-helices and the β-strands are annotated. B, overall structure of clip domains of Grass (in yellow) and Ms-PAP2 (in light brown). A rotation of approximately 90° was applied along the vertical axis, compared with A. C, overall structure of PPAF-II clip domain of group 3, in the same orientation as in B. D, schematic organization of the three groups of clip domains. The clip domain and the linker sequences are represented as rectangles of different colors: cyan for the linkers, green for clip domain of group 1, yellow for clip domain of group 2, and red for group 3. α-Helices and β-strands predicted by the Jpred program are represented as spirals and arrows above the rectangle, and the experimentally determined disulfide bridges and secondary structures are represented below the rectangle.
Despite their different folds, we also compared the clips of Grass and of Hd-PPAF-II (18), the latter being composed of an irregular β-sheet and devoid of α-helix. The three disulfide bridges do superimpose, as do, partially, the β-strands. The main divergence occurs in the region between Cys3 and Cys4, which is a long loop in Hd-PPAF-II (Fig. 4C).
A central hydrophobic cavity was described for Hd-PPAF-II and Ms-PAP2 clips. It was proposed to be involved in the binding of proPO. This cavity is constituted of residues Tyr72, Val78, and Val111 of Hd-PPAFII and Leu26, Ala32, Val63 and Ile86, Leu92, and Val121 for clips 1 and 2 of Ms-PAP2, respectively. In Grass, a similar hydrophobic cavity, composed of Phe45, Leu55, Leu73, and Phe87 (Fig. 4A), could constitute a binding site for a yet undefined protein.
Organization of Domains in Clip-SPs
The clip domain of Grass is located opposite the activation loop and contacts the C-terminal α-helix (residues 366–374) of the SP domain through residues Tyr28, Ser47, and Asn82. The linker of Grass superimposes well over that of Hd-PPAF-I (residues 98–113 for Grass and 88–103 for Hd-PPAF-I), which clearly indicates that the clip of Hd-PPAF-I is likely to be located in a similar position as in Grass (supplemental Fig. S3A). Strikingly, despite a completely different organization between Grass and PPAF-II domains (supplemental Fig. S3B), PPAF-II has an N-terminal extension (residues 22–35) that forms an α-helix (residues 22–35) located at the same position as the α-helix (residues 98–106) of the linker of Grass. In contrast to what occurs in Grass, the clip domain of Hd-PPAF-II (residues 57–114) is tethered to the SP domain with a large interface. Moreover a paired β-strand between residues 94–96 (named β2-1) of the clip and the activation loop (150–151) totally prevents any hydrolysis of the 150–151 amide bond.
Structure-based Classification of Clip-SPs
Jiang and Kanost (1) have proposed a classification of the clip-SPs into two classes according to the length of the sequence separating Cys3 and Cys4 in the clip domain. The proteases of the first group display 15–17 residues between the two cysteines whereas those of the second group have 22–24 residues. This method failed to place Grass in any of the two groups as it has an unusually long sequence (29 residues) between Cys3 and Cys4 (16). Because Grass was the focus of our study, we undertook a new classification, taking into account the sequence and the structural data of the clip domains but also of the SP domains. Clip-SPs sequences were retrieved from the studies on the insects H. diomphalia, T. molitor, M. sexta, B. mori, and D. melanogaster. For Drosophila, annotated clip-SPs of unknown function were also considered.
A sequence alignment of the catalytic domains of clip-SPs of known function was made with ClustalW program. They were partitioned into two groups (supplemental Fig. S2). The major divergence between the two groups appears to be in the 75-loop, and we propose therefore to use it as a marker for the classification. This 75-loop is easily detectable because it is delineated by two supernumerary cysteines. It should be noted that the 75-loop is always associated with the calcium binding loop (70-loop), for which two residues (Glu179 and Asp187) are strictly conserved within the group.
Independently, an analysis of known structures of clip domains combined with secondary structure prediction using the Jpred program (37) was also performed. This enabled us to segregate the clip domains into three groups (Fig. 4D). Members of group 1 are predicted to have one helix between Cys3 and Cys4 and one helix centered on a cysteine of the linker. A detailed analysis of group 1 reveals four subgroups depending on the number of cysteines in the linker (1a–1d). The first subgroup (1a) displays the classical clip-SP cysteine organization consisting of Clip (6 Cys) + linker (1 Cys) + SP (7 Cys). The three other subgroups (1b, 1c, and 1d) display additional cysteines within their linker. Group 2 contains clip domains for which two helices are predicted between Cys3 and Cys4. For this group, the prediction is confirmed by the three experimentally determined structures, those of Grass and of Ms-PAP2 (36). The clip domains that are devoid of any helical secondary structure form group 3. From our analysis, this group contains only inactive clip-SPHs. Again, the crystal structure of Hd-PPAF-II clip consolidates the prediction made using Jpred program. Interestingly, a correlation can be established between the classifications of the catalytic domains and of the clip domains. Indeed, clip-SPs bearing a 75-loop always have a clip domain of group 2.
DISCUSSION
During the last decade, an increasing number of studies have investigated the proteolytic cascades involved in the immune responses of invertebrates. On one hand, large insects like T. molitor, B. mori, or M. sexta have shown that the proteolytic cascades are composed of two clip-SPs, one penultimate and one terminal. Some pathways include a third apical protease, which is not a clip-SP but a modular SP (Fig. 1). On the other hand, genetic studies of the Drosophila immune response have highlighted the role of several SPs in the activation of the Toll receptor. Buchon et al. (9) proposed a more complex model of a proteolytic cascade with at least four members: ModSP, an unknown SP, Grass, and SPE. A fourth SP is necessary for this model because ModSP could not cleave Grass, in vitro. This result is in accordance with the probable specificity pocket of ModSP. This pocket is predicted to be constituted of Leu557, Ala593, and Thr604 (12) and therefore, is unlikely to accommodate basic Arg118 constituting the activation site of Grass.
In the present study, we have determined the crystal structure of the full-length clip-SP Grass of D. melanogaster. The catalytic domain of Grass resembles that of chymotrypsin-like serine proteases with distinctive features that include a deep active site cleft and an activation site located on a protruding loop whose access is prevented by an additional loop (75-loop). The 75-loop is itself delineated by a disulfide bridge specific to the family.
We have also established a classification of the clip-SPs based on the 75-loop of the SP domain and on the conformation of the clip domain. This classification into two groups coincides with their position within the cascade. Indeed, the clip-SPs that are devoid of 75-loop and that have a clip of group 1 (Fig. 4D) are in penultimate position (like Dm-Snake, Ms-HP6, or Tm-SAE). The clip-SPs that have a 75-loop and a clip of group 2 (Fig. 4D) are terminal clip-SPs like Dm-Easter, Dm-SPE, Ms-HP8, or Tm-SPE. According to this classification, Grass should be a terminal protease. This assumption contradicts the current model for Drosophila immune response and gives rise to some questions.
The first question refers to the substrate of Grass. To our knowledge, terminal clip-SPs cleave three kinds of substrates: Spaetzle, proPO, and SPHs. A careful examination of the loops responsible of the substrate specificity reveals that the 60-loop of Grass stands apart from the clip-SPs of known function (supplemental Fig. S2) due to its small size. Interestingly, the 30- and 60-loops of Grass are similar in size to that of Hd-PPAF-III, the protease that cleaves Hd-PPAF-II, an inactive clip-SPH. This may give an indication of a similar substrate. Another question is the role of Grass in the activation of Dm-SPE. Several lines of evidence suggest that some proteolytic cascades are not strictly sequential and may be more complicated. Wang and Jiang (38) have shown that a minute amount of Ms-PAP1, a terminal proPO-activating protease, somehow leads to the activation of Ms-HP6 (a penultimate clip-SP), Ms-HP8, Ms-PAP1, and at least one clip-SPH in the presence of unknown plasma factors, that would be the substrates of Ms-PAP1 (Fig. 5, upper left). According to the authors, this indicates the existence of a positive feedback mechanism into the proPO activation system. The complexity of the proteolytic cascades is also illustrated with the model of H. diomphalia, where proPO, after cleavage by Hd-PPAF-I, requires the functional clip-SPH Hd-PPAF-II to become active. The functional form of Hd-PPAF-II is obtained by cleavage by Hd-PPAF-III, another terminal clip-SP (39) (Fig. 5, lower left).
FIGURE 5.
Schematic organization of the proteolytic cascades involving clip proteases. Upper left, proPO activation in M. sexta (16). Lower left, proPO activation in H. diomphalia (39). Right, model for Toll activation in Drosophila. The penultimate and terminal clip-SPs are colored according to their belonging to the groups defined in Fig. 4D. The clips are colored in green for group 1 or in yellow for group 2. The linker is in cyan. The catalytic domains are colored in purple or in gray, depending on the absence or the presence of the 75-loop. The letter in a white circle on the catalytic domain indicates the substrate specificity: T indicates trypsin-like; T with a line through it indicates different specificity. The letter in the linker stands for the P1 residue of the activation site. The N-terminal domains of ModSP are represented as a chain of beads. The letter on the top (F) is the P1 residue of the activation site. Blue arrows indicate an experimentally described direct interaction. A dashed black arrow indicates a hypothetical positive feedback mechanism (16). A dashed red line indicates a link demonstrated by epistasic studies (in D. melanogaster).
To reconcile a terminal position for Grass with genetic data (7, 9), we propose a novel model for Toll pathway activation, where the function of Grass, downstream of ModSP (Fig. 5, right) would be to cleave a regulatory plasma protein such as SPHs spheroid or sphinx (8). These inactive proteases may be cofactors necessary for the activation of SPE. This could explain the apparent position of Grass, upstream of SPE, as described by epistasic studies. Our analysis suggests a high level of complexity in the regulatory networks that control the activation of Drosophila innate immunity. Further studies demonstrating the existence of one or several intermediate clip-SPs acting upstream of Grass and of SPE will be necessary to refine our model.
Acknowledgments
We thank the European Synchrotron Radiation Facility at Grenoble and in particular the beamline ID14 staff for their assistance. We are grateful to the proteomic platform of the INRA in Tours (France) and in particular to Valerie Labas for peptide mass fingerprint analysis. We thank Annemarie C. Lellouch for careful reading of the revised manuscript and improvement of the English.
This work was supported by an Action Thématique et Incitative sur Programme of the Centre National de la Recherche Scientifique 2005–2009 (to A. R.) and by a grant from the Agence Nationale de la Recherche (to A. R. and J.-M. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table 1.
The atomic coordinates and structure factors (code 2XXL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SP
- serine protease
- Bm
- Bombyx mori
- Dm
- Drosophila melanogaster
- GD
- gastrulation-defective
- GNBP
- Gram negative-binding protein
- Hd
- Holotrichia diomphalia
- Ms
- Manduca sexta
- PGRP
- peptidoglycan recognition protein
- PAP
- prophenoloxidase-activating protein
- PDB
- Protein Data Bank
- PPAF
- prophenoloxidase-activating factor
- proPO
- prophenoloxidase
- PRR
- pattern recognition receptor
- SPE
- Spaetzle-processing enzyme
- SPH
- serine protease homolog
- Tm
- Tenebrio molitor.
REFERENCES
- 1. Jiang H., Kanost M. R. (2000) Insect Biochem. Mol. Biol. 30, 95–105 [DOI] [PubMed] [Google Scholar]
- 2. Krem M. M., Di Cera E. (2002) Trends Biochem. Sci. 27, 67–74 [DOI] [PubMed] [Google Scholar]
- 3. LeMosy E. K., Hong C. C., Hashimoto C. (1999) Trends Cell Biol. 9, 102–107 [DOI] [PubMed] [Google Scholar]
- 4. Han J. H., Lee S. H., Tan Y. Q., LeMosy E. K., Hashimoto C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9093–9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dissing M., Giordano H., DeLotto R. (2001) EMBO J. 20, 2387–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 7. El Chamy L., Leclerc V., Caldelari I., Reichhart J. M. (2008) Nat. Immunol. 9, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kambris Z., Brun S., Jang I. H., Nam H. J., Romeo Y., Takahashi K., Lee W. J., Ueda R., Lemaitre B. (2006) Curr. Biol. 16, 808–813 [DOI] [PubMed] [Google Scholar]
- 9. Buchon N., Poidevin M., Kwon H. M., Guillou A., Sottas V., Lee B. L., Lemaitre B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ligoxygakis P., Pelte N., Hoffmann J. A., Reichhart J. M. (2002) Science 297, 114–116 [DOI] [PubMed] [Google Scholar]
- 11. Gottar M., Gobert V., Matskevich A. A., Reichhart J. M., Wang C., Butt T. M., Belvin M., Hoffmann J. A., Ferrandon D. (2006) Cell 127, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim C. H., Kim S. J., Kan H., Kwon H. M., Roh K. B., Jiang R., Yang Y., Park J. W., Lee H. H., Ha N. C., Kang H. J., Nonaka M., Söderhäll K., Lee B. L. (2008) J. Biol. Chem. 283, 7599–7607 [DOI] [PubMed] [Google Scholar]
- 13. Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., Park B. J., Zhang R., Zhang J., Söderhäll K., Ha N. C., Lee B. L. (2008) J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 14. Gorman M. J., Wang Y., Jiang H., Kanost M. R. (2007) J. Biol. Chem. 282, 11742–11749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y., Jiang H. (2007) Insect Biochem. Mol. Biol. 37, 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. An C., Ishibashi J., Ragan E. J., Jiang H., Kanost M. R. (2009) J. Biol. Chem. 284, 19716–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katsumi Y., Kihara H., Ochiai M., Ashida M. (1995) Eur. J. Biochem. 228, 870–877 [DOI] [PubMed] [Google Scholar]
- 18. Piao S., Song Y. L., Kim J. H., Park S. Y., Park J. W., Lee B. L., Oh B. H., Ha N. C. (2005) EMBO J. 24, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piao S., Kim S., Kim J. H., Park J. W., Lee B. L., Ha N. C. (2007) J. Biol. Chem. 282, 10783–10791 [DOI] [PubMed] [Google Scholar]
- 20. Leone P., Bischoff V., Kellenberger C., Hetru C., Royet J., Roussel A. (2008) Mol. Immunol. 45, 2521–2530 [DOI] [PubMed] [Google Scholar]
- 21. Mishima Y., Quintin J., Aimanianda V., Kellenberger C., Coste F., Clavaud C., Hetru C., Hoffmann J. A., Latgé J. P., Ferrandon D., Roussel A. (2009) J. Biol. Chem. 284, 28687–28697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 23. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 24. Navaza J. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Leone P., Roussel A., Kellenberger C. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 26. Roussel A., Cambillau C. (1991) in Silicon Graphics Geometry Partners Directory (Graphic S. ed) p. 86, Silicon Graphics Mountain View, CA [Google Scholar]
- 27. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Bricogne G. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 37–60 [DOI] [PubMed] [Google Scholar]
- 30. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bode W., Schwager P., Huber R. (1978) J. Mol. Biol. 118, 99–112 [DOI] [PubMed] [Google Scholar]
- 32. Pasternak A., White A., Jeffery C. J., Medina N., Cahoon M., Ringe D., Hedstrom L. (2001) Protein Sci. 10, 1331–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobó J., Harmat V., Beinrohr L., Sebestyén E., Závodszky P., Gál P. (2009) J. Immunol. 183, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 34. Li W., Johnson D. J., Esmon C. T., Huntington J. A. (2004) Nat. Struct. Mol. Biol. 11, 857–862 [DOI] [PubMed] [Google Scholar]
- 35. Rezaie A. R., Yang L. (2005) Thromb. Haemost. 93, 1047–1054 [DOI] [PubMed] [Google Scholar]
- 36. Huang R., Lu Z., Dai H., Velde D. V., Prakash O., Jiang H. (2007) Biochemistry 46, 11431–11439 [DOI] [PubMed] [Google Scholar]
- 37. Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y., Jiang H. (2008) Insect Biochem. Mol. Biol. 38, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim M. S., Baek M. J., Lee M. H., Park J. W., Lee S. Y., Soderhall K., Lee B. L. (2002) J. Biol. Chem. 277, 39999–40004 [DOI] [PubMed] [Google Scholar]