Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium causing chronic infections in cystic fibrosis patients. Such infections are associated with an active type VI secretion system (T6SS), which consists of about 15 conserved components, including the AAA+ ATPase, ClpV. The T6SS secretes two categories of proteins, VgrG and Hcp. Hcp is structurally similar to a phage tail tube component, whereas VgrG proteins show similarity to the puncturing device at the tip of the phage tube. In P. aeruginosa, three T6SSs are known. The expression of H1-T6SS genes is controlled by the RetS sensor. Here, 10 vgrG genes were identified in the PAO1 genome, among which three are co-regulated with H1-T6SS, namely vgrG1a/b/c. Whereas VgrG1a and VgrG1c were secreted in a ClpV1-dependent manner, secretion of VgrG1b was ClpV1-independent. We show that VgrG1a and VgrG1c form multimers, which confirmed the VgrG model predicting trimers similar to the tail spike. We demonstrate that Hcp1 secretion requires either VgrG1a or VgrG1c, which may act independently to puncture the bacterial envelope and give Hcp1 access to the surface. VgrG1b is not required for Hcp1 secretion. Thus, VgrG1b does not require H1-T6SS for secretion nor does H1-T6SS require VgrG1b for its function. Finally, we show that VgrG proteins are required for secretion of a genuine H1-T6SS substrate, Tse3. Our results demonstrate that VgrG proteins are not only secreted components but are essential for secretion of other T6SS substrates. Overall, we emphasize variability in behavior of three P. aeruginosa VgrGs, suggesting that, although very similar, distinct VgrGs achieve specific functions.
Keywords: Bacteria, Gene Knockout, Membrane Trafficking, Protein Export, Protein Secretion, Bacteriophage, Hcp, Pseudomonas aeruginosa, Type VI Secretion System, VgrG
Introduction
Protein secretion systems in bacterial pathogens are virulence weapons allowing bacteria to successfully infect the host (1, 2). Secreted proteins are essential for nutrients or iron acquisition, to evade the immune system, to subvert host cell signaling, and to replicate within macrophages: in other words to survive and exploit the hostile environment of the host.
Secretion in Gram-negative bacteria has been catalogued in a series of systems, called type I to type V (3, 4). Each type shows a difference in the components involved and in the molecular mechanisms. Type I, type II, and type V usually deliver proteins onto the bacterial cell surface or into the surrounding environment. In some cases, such as the type II-dependent exotoxin A from P. aeruginosa (5), the protein has the capacity to subsequently self-translocate into eukaryotic cells. The type III secretion system (T3SS) of many bacterial pathogens is responsible for the delivery of effectors into eukaryotic cells. This yields a spectacular host cell response, resulting, for example, in complete reorganization of the cytoskeleton, thus promoting or avoiding bacterial uptake (6–8). Another system with similar properties found in many pathogens devoid of T3SS is the type IV secretion system (T4SS) (9). T3SS and T4SS deliver effectors that share similarities with eukaryotic proteins (such as Ser/Thr kinases) and mimic activities of endogenous cellular proteins. These secretion systems provide the pathogen with a unique functional interface with eukaryotic cells to subvert the host response (10).
A few years ago, a novel secretion system (type VI or T6SS) was identified in Vibrio cholerae (11) and in Pseudomonas aeruginosa (12). It was revealed that about 15 genes encoding this system are present in the genome of hundreds of bacteria (13) and that their function is extremely relevant for bacterial pathogenesis and host cell survival. In many cases, these genes are induced in vivo.
P. aeruginosa is the third most commonly isolated nosocomial pathogen, and is lethal for chronically infected cystic fibrosis patients. The genome of this bacterium contains at least three gene clusters encoding T6SS components (12, 14), which are now named H1-T6SS to H3-T6SS (15). Importantly, it was reported that mutants affected in T6SS genes are attenuated in a rat lung model of chronic infection (16). Since then, studies have shown that proteins secreted by the T6SS essentially belong to two families, namely Hcp and VgrG. Hcp is a small protein, which forms hexamers. It was proposed that the function of Hcp is to form nanotubes on the bacterial surface (17) and that these tubes may allow transport of other T6SS-dependent effector proteins. VgrGs contain two domains, which are related to proteins constituting the bacteriophage tail (namely gp5 and gp27) (18). It has been proposed that the VgrG proteins form trimeric complexes that could be used as puncturing devices to perforate membranes and allow the passage of proteins or macromolecular complexes. In some cases, VgrGs have a C-terminal extension. Bioinformatic analysis showed that some C-terminal domains display homologies with adhesins, protease or actin cross-linking protein (11). Few data are available about the role/function of VgrG proteins, and the only detailed reports concern VgrG1 from V. cholerae (19) and Aeromonas hydrophila (20). In V. cholerae, VgrG1 has a C-terminal actin cross-linking domain and is translocated into host cells, in particular upon internalization of the bacteria by macrophages. The success of this process is important for survival within macrophages and thus for successful infection. In V. cholerae, two other VgrG proteins, VgrG2 and VgrG3, have been identified. VgrG2 does not carry a C-terminal extension, whereas VgrG3 contains a peptidoglycan-binding domain.
Little has been experimentally demonstrated about the function of these VgrG proteins. We showed that in the PAO1 P. aeruginosa genome, 10 distinct vgrG-like genes could be identified. It is unclear why so many vgrG genes are found in the genome of one single bacterium. In this study, we characterized three P. aeruginosa VgrGs, namely VgrG1a (PA0091), VgrG1b (PA0095), and VgrG1c (PA2685). The corresponding vgrG genes are co-regulated with the H1-T6SS cluster, whose expression is controlled by RetS (12). Here, we clearly establish that all three VgrGs are secreted. However, we show that they have different behavior in terms of secretion and multimerization and suggest that they may fulfill specific functions within the T6SS process.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa mutants were constructed as described previously (21). Briefly, 500-bp DNA fragments contiguous to the 5′ (up) and 3′ (down) ends of the target gene were PCR-amplified by using PAK chromosomal DNA template and two pairs of oligonucleotides (Up5′/Up3′ and Down5′/Down3′) (Table 2). The fused up/down fragment was obtained by overlapping PCR, cloned into pCR2.1 (Invitrogen), sequenced (GATC, Konstanz, Germany), and subcloned into pKNG101 suicide vector (22). We constructed the plasmids pHL9, pHL6, pHL13, pHM60, pHL15, and pKNG-ΔclpV3, respectively, allowing the deletion of vgrG1a/PA0091, vgrG1b/PA0095, vgrG1c/PA2685, clpV1/PA0090, hcp1/PA0085, and a partial deletion of clpV3/PA2371 (region corresponding to the Asn218–Ala632 domain of ClpV3), respectively. The plasmids were maintained in the Escherichia coli CC118λpir strain and conjugated in P. aeruginosa strains. The structure of the mutants in which the double recombination events occurred, resulting in the non-polar deletion of the gene of interest, were verified by PCR with external primers. We also engineered strains with plasmid insertion interrupting the clpV2 or clpV3 gene. A pair of primers (either 5′-ATCCTGGCCCTGCTACGC-3′ and 5′- CGTCGATGAACAGGATGGTC-3′ or 5′-TCATTACGTAGAGATCGAGCACCT and 5′-CTGCGCCGGCGGCAGAACGCCGACGCC-3′) was used to PCR amplify a 0.5-kb internal fragment of clpV2 and a 0.56-kb internal fragment of clpV3, respectively. The respective fragments were cloned into pCR2.1, and recombinant plasmids were verified by sequencing. The resulting plasmids, pCR2.1clpV2::bla and pCR2.1clpV3::bla, were introduced into P. aeruginosa PAKΔretS by electroporation, and carbenicillin-resistant clones were isolated. The position of the insertion, which resulted either in the disruption of clpV2 at codon 55 or of clpV3 at codon 218, was verified by PCR. We also engineered a triple mutant, resulting from deletion in clpV1 and clpV3 genes, combined with an insertion in the clpV2 gene (Table 1).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source/Reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| Bl21 DE3 | E. coli B dcm ompT hsdS(rB−mB−) gal λDE3 | Laboratory collection |
| TG1 | F′: lacIq Tn10 mcrA Δ(mrr-hsdRMS-mcrBC) supEΔ(lac-proAB) thi hsdRΔ5 (F′: traΔ36proA+B+ZΔM15) | Laboratory collection |
| CC118(λpir) | Host strain for pKNG101 replication; Δ(ara-leu) araD ΔlacX74 galE galK-phoA20 thi-1 rpsE rpoB argE (Am) recA1 Rfr(λpir) | Laboratory collection |
| P. aeruginosa | ||
| PAK | Wild-type prototroph | Laboratory collection |
| PAKΔretS | retS deletion mutant | Ref. 27 |
| PAKΔretSΔvgrG1a | vgrG1a deletion mutant | This work |
| PAKΔretSΔvgrG1b | vgrG1b deletion mutant | This work |
| PAKΔretSΔvgrG1c | vgrG1c deletion mutant | This work |
| PAKΔretSΔvgrG1aΔvgrG1b | vgrG1a/vgrG1b double mutant | This work |
| PAKΔretSΔvgrG1aΔvgrG1c | vgrG1a/vgrG1c double mutant | This work |
| PAKΔretSΔvgrG1bΔvgrG1c | vgrG1b/vgrG1c double mutant | This work |
| PAKΔretSΔvgrG1aΔvgrG1bΔvgrG1c | vgrG1a/vgrG1b/vgrG1c triple mutant | This work |
| PAKΔretSΔclpV1 | clpV1 deletion mutant | This work |
| PAKΔretSΔhcp1 | hcp1 deletion mutant | This work |
| PAKΔretS clpV2::bla | clpV2 insertion mutant, ApR | This work |
| PAKΔretSclpV3::bla | clpV3 insertion mutant, ApR | This work |
| PAKΔretSΔclpV1 clpV2::bla ΔclpV3 | clpV1/clpV2/clpV3 triple mutant, ApR | This work |
| Plasmids | ||
| pRK2013 | Tra+Mob+, KmR | Laboratory collection |
| pETDEST-42 | Gateway expression vector tac promoter, ApR | Invitrogen |
| pCR2.1 | TA cloning vector, ApR, KmR | Invitrogen |
| pHM10 | pETDEST-42 containing vgrG1a-V5His6 | This work |
| pHM11 | pETDEST-42 containing vgrG1b-V5His6 | This work |
| pCR2.1clpV2::bla | pCR2.1-containing 50–550 nucleotides of clpV2 | This work |
| pCR2.1clpV3::bla | pCR2.1-containing 589–1151 nucleotides of clpV3 | This work |
| pKNG101 | Suicide vector, sacB, StrR | This work |
| pHM60 | pKNG101-ΔclpV1 mutator | This work |
| pHL9 | pKNG101-ΔvgrG1a mutator | This work |
| pHL6 | pKNG101-ΔvgrG1b mutator | This work |
| pHL13 | pKNG101-ΔvgrG1c mutator | This work |
| pHL15 | pKNG101-Δhcp1 mutator | This work |
| pKNG-ΔclpV3 | pKNG101-ΔclpV3 mutator | This work |
| pMMB67HE | Broad-host-range vector, IncQ ptac lacZα ApR | Laboratory collection |
| pHM14 | pMMB67HE containing vgrG1b-V5His6 | This work |
| pBBR1MCS-4 | Broad host range, ApR | Laboratory collection |
| pBBRladS | pBBR1MCS-4 containing ladS | Ref. 23 |
a ApR, ampicillin-resistant; StrR, streptomycin-resistant; KmR, kanamycin-resistant.
TABLE 2.
Oligonucleotides used in this study
| Deleted gene/oligonucleotide | Oligonucleotide sequencea |
|---|---|
| clpV1 | |
| Up5′ | GGAAGCTTTACTACTACACCGGCCAC |
| Up3′ | CGCGGATCCATACTCACTCATGTTCCT |
| Down5′ | CGCGGATCCTTCGCCGAGGCCGAGTGA |
| Down3′ | AAAGAATTCCCGGCTGATGAAGCTGAA |
| clpV2 | |
| 5′ | ATCCTGGCCCTGCTACGC |
| 3′ | CGTCGATGAACAGGATGGTC |
| clpV3 | |
| Up5′ | TCATTACGTAGAGATCGAGCACCT |
| Up3′ | CTGCGCCGGCGGCAGAACGCCGACGCC |
| Down5′ | CGGCAGAACGCCGACGCCCTCTACGGC |
| Down3′ | CTTGTAGCGTTCCCGCAGTTTTTC |
| vgrG1A | |
| Up5′ | AACACCCTGATCCTGCTCAC |
| Up3′ | TCAGCCCTTCGCTTGCATCAATCC |
| Down5′ | ATGCAAGCGAAGGGCTGAGGCGG |
| Down3′ | ACCCAAGGTCCTCAACTCCT |
| vgrG1B | |
| Up5′ | AATACCTGGCGCAGCATC |
| Up3′ | TCAGTTCTGAAGTGCCATGAAATCAT |
| Down5′ | ATGGCACTTCAGAACTGAAGCGGC |
| Down3′ | GTCGAGCCCCTGGTTGTAG |
| vgrG1C | |
| Up5′ | GAACGGCTTCGGCAGGTAGTC |
| Up3′ | GATATCGACATTCAGTGCCGTCGCGAAAGG |
| Down5′ | CCTTTCGCGACGGCACTGAATGTCGATATC |
| Down3′ | AGCCCTCCTGCGTCTGCAGTT |
| hcp1 | |
| Up5′ | TCGTGCACAAGAAGAACACC |
| Up3′ | TCAGGCCTGAACAGCCATCTTTCC |
| Down5′ | ATGGCTGTTCAGGCCTGATGAGC |
| Down3′ | AGTCGATGGCGGTGGAGTAG |
a Oligonucleotides are presented in the orientation 5′–3′.
The vgrG1b gene, encoding a V5/His-tagged version of VgrG1b (see below), was recloned from pHM11 into the broad host range plasmid pMMB67HE, yielding pHM14. This plasmid as well as the previously described pBBRladS encoding the ladS gene (23) were conjugated by triparental mating using pRK2013 into appropriate P. aeruginosa strains.
Production of Antibodies Directed against VgrG1a and VgrG1b
Briefly, vgrG1a and vgrG1b genes from the P. aeruginosa gene collection (24) were transferred by LR recombination (Invitrogen) from the pDONR shuttle vector to the pETDEST-42 expression vector. The plasmids pHM10 and pHM11, allowing production of VgrG1a-V5His6 and VgrG1b-V5His6, respectively, were transformed into E. coli BL21 (DE3). The resulting strains were cultured in LB medium at 37 °C under agitation in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 6 h. Inclusion bodies containing the recombinant proteins were isolated from bacterial pellets by sonication on ice in lysis buffer (10 mm Tris-HCl, pH 8, 1 mm PMSF). Inclusion bodies were collected by centrifugation at 3000 × g for 10 min, washed with lysis buffer twice, and dissolved in 8 m urea, pH 8, for 1 h at room temperature. Samples were spun for 1 h at 70,000 × g at 4 °C, and supernatants were applied to Ni2+-NTA columns for purification by imidazole gradients. Eluted proteins were dialyzed against 10 mm Tris, pH 8, and injected into rabbits for production of antibodies (Eurogentec).
Hcp1 and Tse3 antibodies were raised in rabbits by injecting purified peptides (Eurogentec). The peptides used were VKGESKDKTHAEEID and GPVKYGWNIRQNVQA for Hcp1 or AYAGNPRNWPRDNVRA and EQRDQTRDEDAKDYQ for Tse3.
Preparation of Supernatant from P. aeruginosa Culture
P. aeruginosa strains were grown overnight in TSB and subcultured to an A600 of 0.1 until early stationary phase at 37 °C under agitation. Cells were separated from culture supernatants by centrifugation at 4000 × g at 4 °C. Cells were resuspended in 1× Laemmli buffer (25). Proteins from the culture supernatant were precipitated with 10% TCA. Protein pellets were washed in 90% acetone, air-dried, and resuspended in 1× Laemmli buffer for analysis by SDS-PAGE. Samples were boiled at 95 °C for 10 min before SDS-PAGE unless indicated otherwise.
SDS-PAGE and Immunoblotting
For SDS-PAGE analysis, cell extracts or 10× concentrated culture supernatants were loaded onto polyacrylamide gels containing SDS followed by transfer to nitrocellulose membrane at 0.03 Amp/cm2. Following transfer, membranes were blocked overnight in blocking buffer (5% milk powder, 0.1% Tween 20 in Tris-buffered saline, pH 8.0). Polyclonal antibodies against VgrG1a, VgrG1b, or Hcp1 were used at a dilution of 1:1000, or anti-Tse3 antibodies were used at 1:250. Monoclonal anti-V5 antibody (Invitrogen) was used at a dilution of 1:5000. Monoclonal antibodies against the β subunit of RNA polymerase (Neoclone) and β-lactamase (QED Bioscience) were used at a dilution of 1:1000. Secondary antibodies conjugated to horseradish peroxidase were used at a dilution of 1:5000. Western blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and the LAS3000 Fuji Imager. Intensity of the VgrG1a band observed in Western blots was quantified using the freeware program ImageJ (available on the National Institutes of Health Web site).
Bioinformatic Analyses
P. aeruginosa protein sequences were retrieved from the Pseudomonas Genome Database. VgrG proteins encoded on the genome of P. aeruginosa were identified through an NCBI BLAST search using the sequence of VgrG1a. The phylogenetic tree of P. aeruginosa VgrG proteins was created using the freeware program Phylogeny (available on the World Wide Web). Secondary structure prediction was carried out using the PSIPRED Protein Structure Prediction Server.
The amino acid sequence of VgrG1a was used to perform a structural-based homology prediction using Phyre (available on the World Wide Web) (26). Two known crystal structures were found to present structural homology to VgrG1a. These structures correspond to Protein Data Bank accession codes 2P5Z and 1K28. The precision of the structural prediction is estimated by Phyre to be 100%. This estimated precision takes into account several factors, such as conservation of amino acid sequence, physicochemical properties of aligned residues, and conservation of secondary structural elements.
RESULTS
Characterization of the VgrG1a Protein
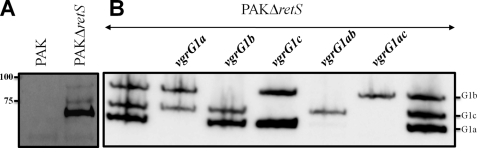
In order to study VgrG secretion in P. aeruginosa, we developed antibodies directed against VgrG1a. VgrG1a is coded by the PA0091 gene on the P. aeruginosa genome and was previously named VgrG1 (15). This gene is located within the H1-T6SS cluster (previously HSI-I). H1-T6SS genes have been shown to be up-regulated in the absence of the sensor kinase RetS (27–29). Here we demonstrate that overproduction of LadS (23), a sensor antagonistic to RetS, induces expression of H1-T6SS genes and results in an active machinery as shown by Hcp1 secretion (supplemental Fig. S1). However, further experiments were carried out in a retS mutant background (12, 27). Analysis of whole cell extracts of PAK and PAKΔretS by immunoblotting using antibodies directed against VgrG1a revealed three bands that were specifically detected in PAKΔretS when compared with PAK (Fig. 1A). These bands correspond to a molecular mass of 93, 82, and 72 kDa. In order to analyze whether all of the bands were VgrG1a-related, we engineered a deletion mutant of the vgrG1a gene in the PAKΔretS background (Table 1). Immunoblot analysis of cell extracts of PAKΔretSΔvgrG1a demonstrated that only the lowest of the three bands (72 kDa) was absent and hence corresponded to VgrG1a (Fig. 1B). This suggested that the two other bands are proteins recognized by VgrG1a antibodies.
FIGURE 1.
Identification of VgrG1a, VgrG1b, and VgrG1c proteins. A, whole cell extracts of PAK and the isogenic mutant PAKΔretS were analyzed by immunoblotting. VgrG proteins were detected using antibodies directed against VgrG1a. Molecular mass standards (kDa) are indicated on the left. B, same as in A, but extracts from different vgrG mutants (indicated above) have been analyzed. All strains are PAKΔretS derivatives. The positions of VgrG1a (G1a), VgrG1b (G1b), and VgrG1c (G1c) are indicated on the right.
Characterization of Additional VgrG1 Proteins
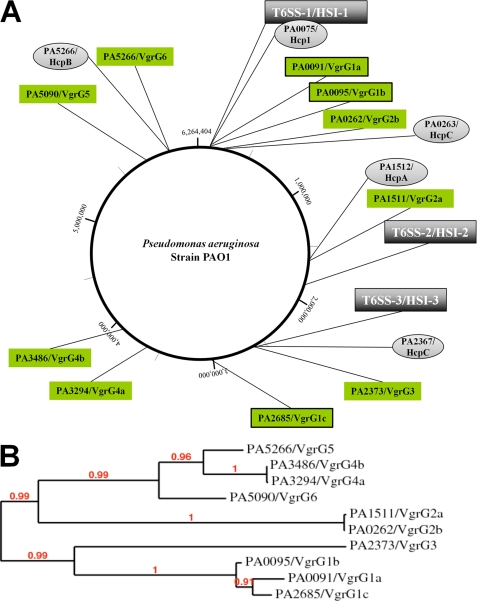
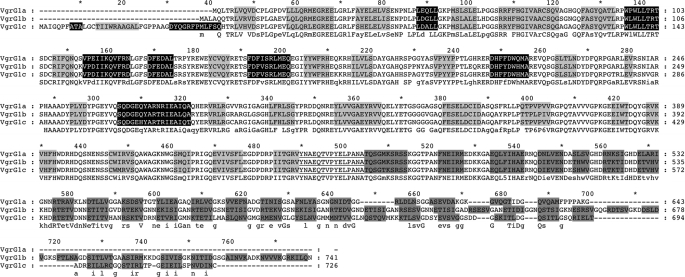
Putative protein candidates for cross-reactivity are other VgrG proteins with high similarity at the amino acid sequence level. We used the deduced amino acid sequence of the vgrG1a gene to perform a BLAST search against the genome sequence of PAO1, identifying 10 genes, scattered all over the chromosome, encoding putative VgrG proteins (Fig. 2A). The PA0095 gene is located within the H1-T6SS cluster and was shown to be up-regulated in a PAKΔretS genetic background (27). We called it PA0095 vgrG1b because it is located in the vicinity of the vgrG1a gene (PA0091). Furthermore, we used the amino acid sequence of all P. aeruginosa VgrGs to establish a phylogenetic tree (Fig. 2B) (30). Interestingly, VgrG1a and VgrG1b fell within the same branch of the tree, indicating that they are closely related (71% similarity, 64% identity). Furthermore, PA2685 is also found within the same branch and was thus called VgrG1c, although it was previously named VgrG4 (15) (73% similarity, 66% identity with VgrG1a) (Fig. 2B). The sequence alignment presented in Fig. 3 clearly shows the similarity between VgrG1a, VgrG1b, and VgrG1c. In addition, secondary structure prediction showed the succession of β-strands within the C-terminal region, which is characteristic of the gp5 structure of the T4 bacteriophage (18) (Fig. 3). It should be noted that, despite the various lengths of the C termini, these consist only of β-strand repeats and are thus exclusively gp5-like. VgrG1a, VgrG1b, and VgrG1c contain 20, 33, and 26 β-strands, respectively. A model of the predicted three-dimensional structure of VgrG1a is shown in supplemental Fig. S2. In order to assess whether the two additional bands detected by VgrG1a antibodies (Fig. 1B) correspond to VgrG1b and VgrG1c, vgrG1b and vgrG1c deletion mutants were constructed in PAKΔretS. Furthermore, multiple vgrG deletions within a single strain were also obtained (see “Experimental Procedures” and Table 1). Remarkably, we observed that the 82 kDa band corresponds to VgrG1c because it is absent in vgrG1c and vgrG1ac mutants, and the 93 kDa-band corresponds to VgrG1b because it is absent in vgrG1b and vgrG1ab mutants (Fig. 1B).
FIGURE 2.
P. aeruginosa VgrG proteins. A, the chromosome design was generated using Geneious version 5.1 (available on the World Wide Web). Distribution of the vgrG and hcp genes on the PAO1 genome is shown. Relative positions of vgrG genes (named G1a to G6) are indicated with green flags, and hcp genes are shown with gray flags. Flags representing genes of interest in the study have been boxed (vgrG1a, vgrG1b, and vgrG1c). In all cases, the corresponding PA number is indicated (39). The positions of the type VI secretion (T6SS) gene clusters are flagged in dark gray. Three T6SS clusters are identified, H1-T6SS, H2-T6SS, and H3-T6SS (15). B, phylogenetic analyses of P. aeruginosa (PAO1) VgrG proteins using Phylogeny.fr. Graphical representation of the inferred tree is shown. VgrG1a, VgrG1b, and VgrG1c are clustered at the bottom of the tree.
FIGURE 3.
Sequence alignment of VgrG1a, VgrG1b, and VgrG1c. Residues predicted to fold into α-helices and β-strands are shaded in black and gray, respectively. Two levels of gray are used; light and dark gray indicate β-strands predicted to have structural homology to the N-terminal domain of C3393 (gp27-like domain of VgrG in E. coli) and the C-terminal domain of gp5, respectively (see supplemental Fig. S2 for details). The shown secondary structure prediction corresponds to the consensus between psipred, jnet, and sspro programs. The sequence consensus is shown below aligned sequences, where letters in uppercase and lowercase correspond to residue conservation in all three or in two of three sequences, respectively. The underlined residues correspond to the predicted coil region where the gp5 domain starts.
ClpV1-dependent Secretion of VgrGs
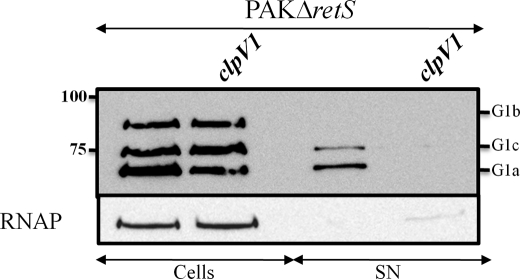
We analyzed whether the VgrG1a/b/c proteins were secreted into the extracellular medium. VgrG1a and VgrG1c were detected in significant amounts in the culture supernatant, although quite a large amount accumulated within the cells (Fig. 4). On the contrary, VgrG1b was not detectable in culture supernatants using VgrG1a antibodies. However, when antibodies directed against VgrG1b were used, secretion of VgrG1b was readily observed (Fig. 5, lane 1). In all cases, secretion was not the result of cell lysis because the cytoplasmic protein RNA polymerase (RNAP)4 was detected in the cell fraction but not in culture supernatants (Figs. 4 and 5).
FIGURE 4.
ClpV1-dependent secretion of VgrG proteins. Whole cell extracts (Cells) and proteins from culture supernatants (SN) of PAKΔretS and the clpV1 mutant derivative (indicated above each lane) were analyzed by immunoblotting. Cytosolic RNAP was detected using an antibody directed against its β subunit. VgrG proteins were detected using antibodies directed against VgrG1a. All strains are PAKΔretS derivatives. The positions of VgrG1a (G1a), VgrG1b (G1b), and VgrG1c (G1c) are indicated on the right. Molecular mass standards (kDa) are indicated on the left.
FIGURE 5.
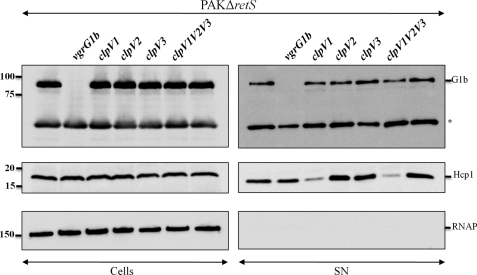
T6SS-independent secretion of VgrG1b. Whole cell extracts (Cells) and proteins from culture supernatants (SN) of PAKΔretS, vgrG1b, and clpV mutant derivatives (clpV1, clpV2, clpV3, or triple clpV1V2V3; indicated above each lane) were analyzed by immunoblotting. VgrG1b proteins were detected using antibodies directed against VgrG1b. Hcp1 was detected with anti-Hcp1 antibodies. Cytoplasmic RNAP was detected using an antibody directed against its β subunit. The positions of VgrG1b (G1b), Hcp1, and RNAP are indicated on the right. The band marked with an asterisk represents an uncharacterized protein recognized by antibodies directed against VgrG1b. Molecular mass standards (kDa) are indicated on the left.
We then tested whether the secretion of the VgrG proteins was H1-T6SS-dependent. A deletion mutant in clpV1, a gene encoding a key component of the H1-T6SS, was constructed (Table 1). In the clpV1 mutant, secretion of both VgrG1a and VgrG1c was not detectable (Fig. 4), which confirmed previous data obtained by Hood et al. (15). Interestingly, the level of VgrG1a in the cells of a clpV1 mutant seemed reduced (Fig. 4). A reduction of 54 ± 14% in the intensity of the VgrG1a band (as determined with ImageJ in 10 independent experiments) in PAKΔretSΔclpV1 mutant as compared with PAKΔretS confirmed this initial observation. VgrG1c or VgrG1b levels remained unaffected in the PAKΔretSΔclpV1 mutant (Figs. 4 and 5). This could suggest that VgrG1a is unstable in the absence of ClpV1. Alternatively, the clpV1 mutation may have a polar effect on the expression of vgrG1a. Surprisingly, using VgrG1b-specific antibodies, we observed that VgrG1b secretion was ClpV1-independent and thus H1-T6SS-independent (Fig. 5). We engineered mutants in clpV genes found in the other two T6SS clusters (clpV2 and clpV3) and observed that VgrG1b secretion was still effective in these mutants (Fig. 5). Furthermore, using a strain in which mutations in clpV1, clpV2, and clpV3 were combined, rendering all three P. aeruginosa T6SS inactive, VgrG1b secretion was still not affected (Fig. 5). Thus, VgrG1b secretion is independent of the three known T6SS clusters.
Relationship between VgrG1b and H1-T6SS
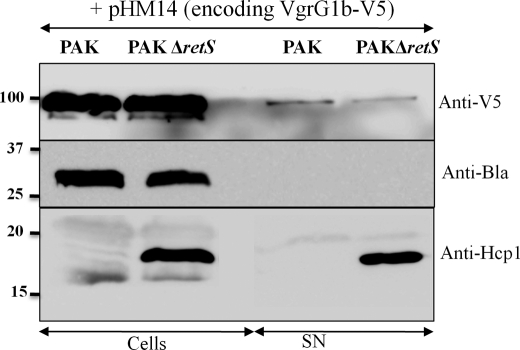
Under “ClpV1-dependent Secretion of VgrGs,” we showed that VgrG1b was recovered in the supernatant of a retS mutant and that its secretion was unaffected by deletion of clpV1, clpV2, or clpV3 (Fig. 5), suggesting that VgrG1b was secreted independently of any P. aeruginosa T6SS. To support the hypothesis of T6SS-independent VgrG1b secretion, secretion of VgrG1b was analyzed in PAK, a strain in which the H1- to H3-T6SS genes are not induced. The recombinant plasmid pHM14, encoding V5/His-tagged VgrG1b, was introduced in both PAK and the isogenic retS mutant. Recombinant VgrG1b was detectable in both cells and culture supernatant using anti-V5 antibody in both PAK and PAKΔretS (Fig. 6). However, it is interesting to note that no difference was observed between PAK (no active T6SSs) and the retS mutant (active H1-T6SS), suggesting that the weak secretion of VgrG1b is T6SS-independent. Monitoring the localization of periplasmic β-lactamase, encoded by the pMMB67HE plasmid, showed that there is no leakage of this enzyme into the supernatant (Fig. 6). Thus, detectable VgrG1b secretion in the culture supernatant was not a result of cell lysis or periplasmic leakage but due to a specific secretion process.
FIGURE 6.
Overproduction and secretion of VgrG1b in PAK. Secretion of VgrG1b was monitored in PAK and PAK ΔretS after production of V5/His-tagged VgrG1b from the plasmid pHM14. Cell fraction (Cells) and supernatant fraction (SN) were analyzed by immunoblotting using anti-V5 (V5) (top), anti-β-lactamase (Bla) (middle), and anti-Hcp1 antibodies (Hcp1) (bottom). Molecular mass standards (kDa) are indicated on the left.
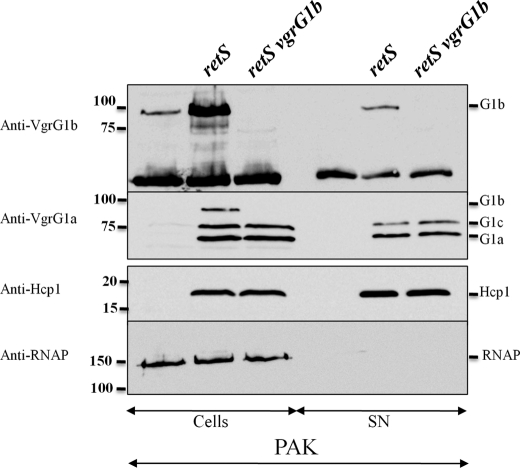
As for Hcp1 and VgrG1a/c, VgrG1b production was clearly induced in a retS mutant compared with the PAK strain (Fig. 7, top). To analyze if VgrG1b was required for a functional H1-T6SS, secretion of Hcp1 and VgrG1a/c was analyzed in a vgrG1b mutant (PAKΔretSΔvgrG1b). We confirmed that P. aeruginosa Hcp1 is secreted into the culture supernatant of PAKΔretS (Fig. 7, lane 2) and that it is VgrG1b-independent because it was also observed in the culture supernatant of the PAKΔretSΔvgrG1b mutant (Fig. 7, lane 3). Furthermore, neither VgrG1a nor VgrG1c secretion was affected by deletion of vgrG1b (Fig. 7, second panel). Overall, these results indicate that although vgrG1b is co-expressed with H1-T6SS genes, VgrG1b neither impacts secretion of components of the H1-T6SS nor requires H1-T6SS components for its own secretion.
FIGURE 7.
Deletion of vgrG1b does not affect Hcp1 and VgrG1a/c secretion. The H1-T6SS is not expressed in PAK but is induced in PAKΔretS. Cell and supernatant fractions of PAK, PAKΔretS (retS), and PAKΔretSΔvgrG1b (retS vgrG1b) were analyzed by immunoblotting using antibodies directed against VgrG1b (top panel), VgrG1a (second panel), Hcp1 (third panel), and the β subunit of RNAP (bottom panel). Molecular mass standards (kDa) are indicated on the left, and protein bands are identified on the right.
Secretion Dependence between Hcp1 and VgrG1a-c
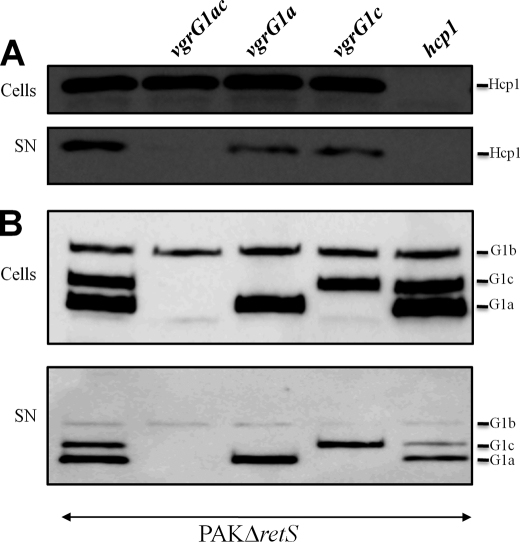
We further studied whether P. aeruginosa Hcp1 secretion was dependent on VgrG1a or VgrG1c. We observed that Hcp1 was still secreted by a single vgrG1a or vgrG1c mutant, but the secretion efficiency was reduced (Fig. 8A). However, when the vgrG1a mutation was combined with the vgrG1c mutation, Hcp1 secretion was abolished (Fig. 8A). In conclusion, VgrG1a and VgrG1c are essential for efficient Hcp1 secretion but can function independently. Indeed, we observed that one VgrG compensates for the absence of the other. This observation differs from the situation observed in V. cholerae (31). Notably, Hcp secretion is totally abolished in a V. cholerae vgrG2 mutant, whereas residual secretion is observed in a vgrG1 mutant. Interestingly, VgrG3 appears not to be required for V. cholerae Hcp secretion.
FIGURE 8.
VgrG-dependent secretion of Hcp1 and VgrGs. Whole cell extracts (Cells) and proteins from culture supernatants (SN) of PAKΔretS and PAKΔretS mutant derivatives vgrG1a, vgrG1c, double vgrG1a/vgrG1c (vgrG1ac), and hcp1 (indicated above each lane) were analyzed by immunoblotting. A, Hcp1 protein was detected using antibodies directed against Hcp1. B, VgrG proteins were detected using antibodies directed against VgrG1a. The positions of VgrG1a (G1a), VgrG1b (G1b), VgrG1c (G1c), and Hcp1 are indicated on the right.
Furthermore, we demonstrated that secretion of VgrG1a is not dependent on VgrG1c and vice versa (Fig. 8B). This is similar to V. cholerae, in which VgrG1 secretion is not dependent on VgrG2 and vice versa (31). Finally, VgrG1a and VgrG1c secretion is reduced but not abolished in an hcp1 mutant (Fig. 8B).
VgrGs Form Multimeric Complexes
VgrG proteins share similarity with the gp5 and gp27 bacteriophage components, which form a stable complex constituting the bacteriophage tail spike (31). We used the sequence of the VgrG1a protein from P. aeruginosa PAO1 to perform a structure-based homology prediction using several bioinformatics tools and databases (26, 32). The results clearly indicated that VgrG1a (643 amino acids) is composed of distinct domains and that a trimer is the biologically relevant oligomer (supplemental Fig. S2). The predicted VgrG1a structure is very similar to the T4 phage (gp5-gp27)3 structure reported previously (Protein Data Bank entry 1K28).
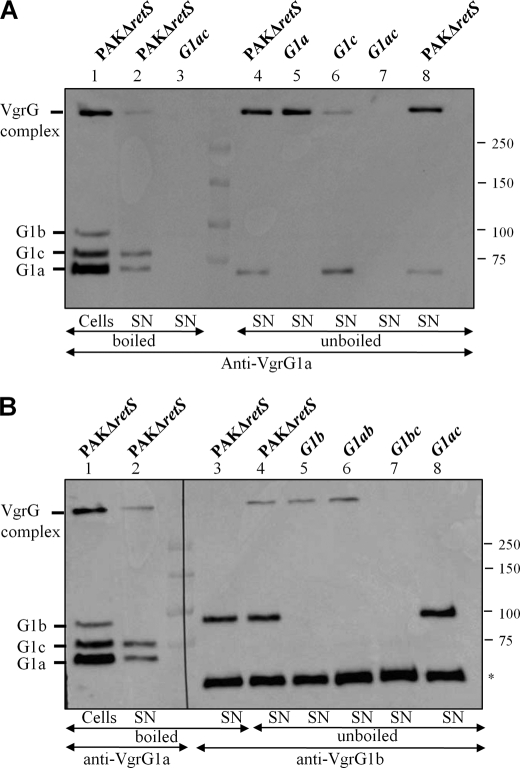
To analyze the relevance of this model, we investigated whether VgrG1a could form multimers in vivo. Supernatants from P. aeruginosa cultures were collected, and the protein content was analyzed by immunoblotting using anti-VgrG1a (Fig. 9A). A band with a high molecular weight, suggesting multimer formation, was detected as well as monomeric forms of VgrG1a and VgrG1c (Fig. 9A, lane 2). The complex seems to contain VgrG1a and/or VgrG1c because it was not detectable in a double vgrG1a/vgrG1c mutant (Fig. 9A, lane 3). This is in agreement with the previous observation by Hood et al. (15) showing that VgrG1c overproduction resulted in the occurrence of a large number of high molecular weight bands in the supernatant. Furthermore, when the samples were not boiled to reduce disruption of protein complexes, the amount of VgrG complex detected was significantly increased (Fig. 9A, lanes 4 and 8). The VgrG proteins are characterized by a C terminus containing octapeptide Val-Xaa-Gly-Xaa-Xaa-Xaa-Xaa-Xaa repeats similar to the C-terminal domain of the gp5 protein. It was previously reported that this gp5 domain forms a trimer with a β-helix structure (see supplemental Fig. S2) that is very stable and resistant to denaturation by SDS (33, 34). This is in agreement with the observed VgrG complex in our SDS-containing acrylamide gels (Fig. 9). In order to further characterize the complex, we performed similar experiments using isogenic vgrG1a and vgrG1c mutants. The complex was still detectable in the vgrG1a mutant supernatant, although the monomeric form of VgrG1a was no longer detectable (Fig. 9A, lane 5). In the vgrG1c mutant supernatant, the complex was also observed but in reduced quantities compared with the supernatant of the retS mutant (Fig. 9A, lane 6). The complex was no longer detectable (Fig. 9A, lane 7) in a double vgrG1a/vgrG1c mutant, suggesting that the complex contains both VgrG1a and VgrG1c. However, it is not possible to conclude whether VgrG1a and VgrG1c could form heteromultimeric complexes.
FIGURE 9.
Multimeric complexes formed by VgrG proteins. Cell extracts (Cells) and proteins from culture supernatants (SN) of PAKΔretS and PAKΔretS mutant derivatives vgrG1a (G1a), vgrG1b (G1b), vgrG1c (G1c), double vgrG1a/vgrG1b (G1ab), double vgrG1a/vgrG1c (G1ac), and double vgrG1b/vgrG1c (G1bc) (indicated above each lane where appropriate) were analyzed by immunoblotting. Samples were not boiled prior to analysis by SDS-PAGE and immunoblotting (unboiled) unless otherwise indicated (boiled). A, VgrG proteins were detected using antibodies directed against VgrG1a. B, VgrG proteins were detected using antibodies directed against VgrG1a or VgrG1b, as indicated. The positions of VgrG1a (G1a), VgrG1b (G1b), VgrG1c (G1c), and the VgrG complex are indicated on the left. The band marked with an asterisk is an uncharacterized protein recognized by antibodies directed against VgrG1b. Molecular mass standards (kDa) are indicated on the right.
Using VgrG1b antibodies, a complex of similar molecular weight was detected in the supernatant of the PAKΔretS strain (Fig. 9B), most efficiently in the unboiled sample versus the boiled sample (Fig. 9B, lane 4 versus lane 3). We observed that this complex was still detectable in a vgrG1b mutant (Fig. 9B, lane 5), suggesting that the antibodies could recognize either VgrG1a or VgrG1c although the monomeric form of these VgrGs remained undetectable. The complex fully disappeared in a double mutant vgrG1b/vgrG1c (Fig. 9B, lane 7) but was still present in a double mutant vgrG1a/vgrG1b (Fig. 9B, lane 6), which suggests that it essentially consists of VgrG1c. Finally, no complexes were detected in a double vgrG1a/vgrG1c mutant, which suggests that VgrG1b does not form complexes or that these complexes are very unstable (Fig. 9B, lane 8). It should be noted that because the VgrG1b antiserum detects the VgrG1c complex but not its monomeric form, it may recognize conformational epitopes.
VgrG-dependent Secretion of Tse3
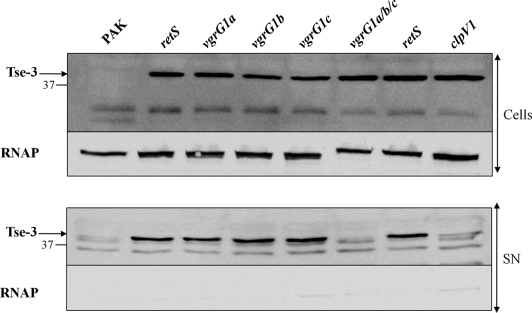
It was recently shown that the H1-T6SS from P. aeruginosa is involved in the secretion of three novel substrates, non-VgrG effectors, named Tse1 to -3 (15). One of these, Tse2, was described as a toxin component arresting growth of prokaryotic and eukaryotic cells. It was shown that secretion of VSV-G-tagged Tse proteins is ClpV1-dependent. Here, we generated polyclonal antibodies against Tse3 and assessed whether secretion of chromosomally encoded Tse3 was dependent on VgrG1a, VgrG1b, or VgrG1c. Our data confirmed that Tse3 is clearly detected in the retS background (increased expression of H1-T6SS genes) but not in the parental PAK strain (Fig. 10). Furthermore, as we previously showed for Hcp1, none of the single mutations in vgrG1a, vgrG1b, or vgrG1c abolished Tse3 secretion (Fig. 10). However, when all three vgrG genes were deleted (vgrG1a/b/c), Tse3 was no longer detected in the supernatant, whereas it was still found in the cellular fraction. This demonstrated that Tse3 secretion is VgrG-dependent, but there are no clear signs of specificity for one or another VgrG.
FIGURE 10.
VgrG-dependent secretion of Tse3. Cell extracts (Cells) and proteins from culture supernatants (SN) of PAK, PAKΔretS (retS), and PAKΔretS mutant derivatives vgrG1A, vgrG1B, vgrG1C, and triple vgrG1a/vgrG1b/vgrG1c (vgrG1a/b/c) (indicated above each lane) were separated by SDS-PAGE and analyzed by immunoblotting. Tse3 protein was detected using specific polyclonal antibodies directed against Tse3 peptides. RNAP was detected using an antibody directed against its β subunit. The positions of Tse3, RNAP, and molecular mass standards (kDa) are indicated on the right.
DISCUSSION
The discovery of the T6SS shed new light on the strategy used by bacterial pathogens to successfully infect their host (11, 14). The T3SS and T4SS have been described as injecting bacterial effectors into host cells and consequently manipulating the host response for their own benefit (9). Because the T6SS was initially discovered as being specifically induced in vivo and because T6SS appeared to be activated upon host cell contact (11, 20), it was proposed that it could fulfill a similar function and that putative effector proteins should be associated with the T6SS.
Very little knowledge is available about the characterization of these effectors and about proteins secreted in a T6SS-dependent manner. It was determined that Hcp is a bacteriophage gp19 homologue and could form a tube as part of the T6SS machinery (18, 35). This tube could span the bacterial cell envelope and become cell surface-exposed, which would explain why Hcp is recovered from bacterial culture supernatants. VgrG proteins were also described as bacteriophage protein homologues. VgrGs are composite proteins, all containing a domain similar to gp5 and a domain similar to gp27. The association of gp5 and gp27 in the bacteriophage infection process results in the formation of a puncturing device, which sits on the tip of the gp19 tube and perforates the membrane, thus allowing phage DNA injection into the bacterial cell. Based on this model, VgrG proteins may sit at the tip of the Hcp tube and could be released into the extracellular medium. Only few proteins have been described to be secreted in a T6SS-dependent manner, such as RbsB from Rhizobium leguminosarum (36) or IglI from Francisella tularensis (37). It was shown recently that three proteins, Tse1 to -3, are substrates for the P. aeruginosa H1-T6SS (15). Among them, Tse2 is a toxin that is targeted to prokaryotic cells. Alternatively, effector domains have been identified at the C terminus of VgrG proteins, which are referred to as evolved VgrGs. The C-terminal domain may be an adhesin or an enzyme for example. Only two of these evolved VgrGs have been experimentally studied, VgrG1 from V. cholerae (19, 31) and from A. hydrophila (20). In both cases, it was shown that the activity associated with the VgrG C-terminal domain, ACD, or VIP-2, respectively, was involved in remodeling the actin cytoskeleton of the host cell.
Genome mining in P. aeruginosa identified three T6SS clusters (H1- to H3-T6SS). Furthermore, we showed here that 10 vgrG genes could be identified on the PAO1 chromosome. Previous work using mass spectrometry analysis showed that at least two P. aeruginosa VgrGs, VgrG1 (PA0091) and VgrG4 (PA2685), were not recovered in the supernatant of an H1-T6SS mutant (clpV1 mutant) and therefore could be substrate for the H1-T6SS machinery (15). The authors confirmed this observation by overproducing VSV-tagged versions of the VgrG proteins and by following secretion using standard Western blot analysis. We focused our work on VgrG1, which we renamed here VgrG1a, and generated antibodies directed against this protein to follow its fate when expressed from the chromosomal gene. This protein is a standard VgrG protein without any C-terminal extension. Here, we confirmed that VgrG1a is efficiently secreted, and we demonstrated that at least two other VgrGs are also secreted, VgrG1b and VgrG1c. It is important to recall that VgrG1a is only detected in the retS mutant in which the vgrG1a/PA0091 gene is up-regulated (30-fold) (27). The transcriptomic analysis revealed that vgrG1b/PA0095 (3.7-fold) and vgrG1c/PA2685 (4.3-fold) are also up-regulated in a retS mutant, which explains why they are detected in the same conditions. In contrast to vgrG1b, which is located within the vicinity of vgrG1a and the H1-T6SS gene cluster, vgrG1c is located at a different locus and not clustered with any T6SS genes (Fig. 2A). Not only is the production of these three VgrG proteins coordinated, but they are closely related when compared with the seven other VgrG proteins (Fig. 2B). VgrG1c/PA2685 corresponds to the previously described VgrG4 (15), but we propose to rename the VgrG proteins according to their proximity in the phylogenetic tree (Fig. 2B). We also observed that neither VgrG1b nor VgrG1c contain a C-terminal extension (Fig. 3). In summary, we showed that three P. aeruginosa VgrGs are secreted, but none of them could be considered as an effector protein.
Next we analyzed the T6SS-dependent secretion of these VgrG proteins. Although we confirmed that VgrG1a and VgrG1c secretion is drastically reduced in a clpV1 mutant, VgrG1b secretion seems to remain unaffected. Furthermore, VgrG1b is still secreted in strains containing a mutation in any of the three T6SS clusters, suggesting that VgrG1b secretion can occur in a T6SS-independent manner. The reason for such behavior is unclear, but T6SS-independent secretion of VgrG proteins has also been reported in F. tularensis (37).
It has been proposed that VgrG proteins form trimeric complexes similar to the (gp27)3(gp5)3 tail spike structure of the T4 phage. We confirmed here using a three-dimensional structure model that this applies to VgrG1a (supplemental Fig. S2). When we analyzed the multimeric state of the VgrG proteins isolated from the supernatant of P. aeruginosa cultures, we observed that both VgrG1a and VgrG1c are capable of assembling into a multimeric form. This complex disappears only when both vgrG1a and vgrG1c genes are deleted. However, it is noticeable that when analyzing samples from a vgrG1a mutant, only the multimeric form of VgrG1c is detected. In contrast, analyzing samples from the vgrG1c mutant indicates that whereas VgrG1a complex is detectable, a large amount of monomeric VgrG1a is also detectable, suggesting that the complex is less stable. The presence of VgrG1b complex could not be shown using anti-VgrG1b antibodies, which might suggest that, if formed, the VgrG1b complex is very unstable.
Although three P. aeruginosa VgrGs appeared co-regulated and highly similar at the amino acid sequence level, none of them showed similar behavior in terms of secretion or multimerization. This discrepancy might suggest that each of the three VgrGs fulfills an independent function or allows a significant level of specificity. For example, if we consider VgrGs as a puncturing device, non-evolved VgrGs might serve the secretion of yet unknown proteins with which they may interact specifically. For example, we hypothesized that VgrG1a is involved in the secretion of one of the Tse proteins, whereas VgrG1c may be required for the secretion of a different Tse. In this way, each complex, VgrG1a or VgrG1c, together with their cognate exoproteins, might sit independently or alternatively on the tip of the Hcp1 tube. As for the evolved VgrG, it will have a dual function, which is on the one hand to perforate the membrane and on the other hand to carry an effector or secreted protein. If this is true, we might expect that accessibility of Hcp1 to the external milieu would depend either on VgrG1a or VgrG1c. This is exactly what we observed because Hcp1 secretion was reduced in a vgrG1a or a vgrG1c mutant but was abolished in a double vgrG1a/vgrG1c mutant.
In conclusion, we point out the remarkable feature that although release in the extracellular medium might be a general feature of VgrG proteins, there is a lot more subtlety in the function of these components. The large number of vgrG genes in P. aeruginosa is probably not the result of a duplication but rather a gain in specificity for the system, which might modulate its secretion profile according to the situation encountered within the host. Future work will aim at better understanding the exact role played by the non-evolved VgrGs, which we suspect is not exclusively to provide a channel through the cell envelope but also to specifically interact with yet unknown secreted proteins, such as the recently characterized Tse1 to -3 proteins (15). Our data regarding Tse secretion showed that although Tse3 secretion is VgrG-dependent, all three VgrGs need to be absent to prevent Tse3 release, thus showing no sign of specificity in this case. We still need to develop a refined quantitative assay to rule out any VgrG specificity, and we will further address VgrG-dependent secretion of Tse1 and Tse2. For the time being, two categories of VgrGs can be described. The evolved VgrGs form one block, including the puncturing device and the secreted domain covalently bound at the C terminus. This will conceptually resemble an autotransporter system (38). The standard VgrGs, such as VgrG1a, VgrG1b, and VgrG1c, described in this study, contain the sole puncturing device but could possibly recognize a secreted substrate. If this turns out to be the case, it will conceptually resemble a two-partner system.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- RNAP
- RNA polymerase.
REFERENCES
- 1. Cossart P., Sansonetti P. J. (2004) Science 304, 242–248 [DOI] [PubMed] [Google Scholar]
- 2. Merrell D. S., Falkow S. (2004) Nature 430, 250–256 [DOI] [PubMed] [Google Scholar]
- 3. Durand E., Verger D., Rêgo A. T., Chandran V., Meng G., Fronzes R., Waksman G. (2009) Infect. Disord. Drug Targets 9, 518–547 [DOI] [PubMed] [Google Scholar]
- 4. Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. (2010) Int. J. Med. Microbiol. 300, 534–543 [DOI] [PubMed] [Google Scholar]
- 5. Voulhoux R., Taupiac M. P., Czjzek M., Beaumelle B., Filloux A. (2000) J. Bacteriol. 182, 4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parsot C. (2009) Curr. Opin. Microbiol. 12, 110–116 [DOI] [PubMed] [Google Scholar]
- 7. McGhie E. J., Brawn L. C., Hume P. J., Humphreys D., Koronakis V. (2009) Curr. Opin. Microbiol. 12, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hauser A. R. (2009) Nat. Rev. Microbiol. 7, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Llosa M., Roy C., Dehio C. (2009) Mol. Microbiol. 73, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galán J. E. (2009) Cell Host Microbe 5, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 14. Filloux A., Hachani A., Bleves S. (2008) Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 15. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Potvin E., Lehoux D. E., Kukavica-Ibrulj I., Richard K. L., Sanschagrin F., Lau G. W., Levesque R. C. (2003) Environ. Microbiol. 5, 1294–1308 [DOI] [PubMed] [Google Scholar]
- 17. Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Cell Host Microbe 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010) J. Bacteriol. 192, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vasseur P., Vallet-Gely I., Soscia C., Genin S., Filloux A. (2005) Microbiology 151, 985–997 [DOI] [PubMed] [Google Scholar]
- 22. Kaniga K., Delor I., Cornelis G. R. (1991) Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 23. Ventre I., Goodman A. L., Vallet-Gely I., Vasseur P., Soscia C., Molin S., Bleves S., Lazdunski A., Lory S., Filloux A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Labaer J., Qiu Q., Anumanthan A., Mar W., Zuo D., Murthy T. V., Taycher H., Halleck A., Hainsworth E., Lory S., Brizuela L. (2004) Genome Res. 14, 2190–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. (2004) Dev. Cell 7, 745–754 [DOI] [PubMed] [Google Scholar]
- 28. Goodman A. L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. (2009) Genes Dev. 23, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vincent F., Round A., Reynaud A., Bordi C., Filloux A., Bourne Y. (2010) Environ. Microbiol. 12, 1775–1786 [DOI] [PubMed] [Google Scholar]
- 30. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- 33. Arisaka F., Kanamaru S., Leiman P., Rossmann M. G. (2003) Int. J. Biochem. Cell Biol. 35, 16–21 [DOI] [PubMed] [Google Scholar]
- 34. Mesianzhinov V. V., Leiman P. G., Kostyuchenko V. A., Miroshnikov K. A., Sykilinda N. N., Shneider M. M. (2004) Biochemistry 69, 1190–1202 [DOI] [PubMed] [Google Scholar]
- 35. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bladergroen M. R., Badelt K., Spaink H. P. (2003) Mol. Plant Microbe Interact. 16, 53–64 [DOI] [PubMed] [Google Scholar]
- 37. Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J., Celli J., Arulanandam B. P., Klose K. E. (2009) Mol. Microbiol. 74, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winsor G. L., Van Rossum T., Lo R., Khaira B., Whiteside M. D., Hancock R. E., Brinkman F. S. (2009) Nucleic Acids Res. 37, D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]