Abstract
Campylobacter jejuni is well known for synthesizing ganglioside mimics within the glycan component of its lipooligosaccharide (LOS), which have been implicated in triggering Guillain-Barré syndrome. We now confirm that this pathogen is capable of synthesizing a much broader spectrum of host glycolipid/glycoprotein mimics within its LOS. P blood group and paragloboside (lacto-N-neotetraose) antigen mimicry is exhibited by RM1221, a strain isolated from a poultry source. RM1503, a gastroenteritis-associated strain, expresses lacto-N-biose and sialyl-Lewis c units, the latter known as the pancreatic tumor-associated antigen, DU-PAN-2 (or LSTa). C. jejuni GC149, a Guillain-Barré syndrome-associated strain, expresses an unusual sialic acid-containing hybrid oligosaccharide with similarity to both ganglio and Pk antigens and can, through phase variation of its LOS biosynthesis genes, display GT1a or GD3 ganglioside mimics. We show that the sialyltransferase CstII and the galactosyltransferase CgtD are involved in the synthesis of multiple mimic types, with LOS structural diversity achieved through evolving allelic substrate specificity.
Keywords: Carbohydrate Biosynthesis, Carbohydrate Chemistry, Carbohydrate Structure, Glycoconjugate, Glycolipids, Campylobacter, Ganglioside, Guillain-Barre, Lipooligosaccharide, Lipopolysaccharide
Introduction
Seminal investigations by Yuki et al. (1) and Aspinall et al. (2) established that certain strains of Campylobacter jejuni isolated from patients with Guillain-Barré syndrome (GBS)2 express lipooligosaccharide (LOS)-bound ganglioside mimics. A large body of work has subsequently lent support for the premise that anti-LOS antibodies elicited upon exposure to C. jejuni can in some instances cross-react with gangliosides, causing autoimmune disease (for recent reviews see Refs. 3, 4).
The LOS biosynthesis genes in C. jejuni are localized to a hypervariable locus. To date, there have been 19 classes identified through genotyping (5, 6). Although all strains carry several common essential genes that direct the synthesis of the inner core and lipid A, each locus class is defined by a unique genetic composition and arrangement. Several appear to have arisen through recombination events between strains. The LOS loci in C. jejuni, as a group, are of a mosaic nature with varying levels of similarity.
C. jejuni strains belonging to classes “A,” “B,” and “C,” which will be referred to collectively as the ABC group, have received considerable attention because they carry genes encoding sialic acid-processing enzymes and glycosyltransferases (GTases), which are necessary to direct the synthesis of ganglioside-like glycans (7). The arrangements of genes in these loci are presented in Fig. 1. Genetic profiling of isolates from a variety of sources indicates that a majority of strains carry ABC loci, but up to 30–40% fall outside this group and likely do not express ganglioside mimics (5, 6, 8). Representative LOS structures of these “non-ganglioside” strains remain largely uncharacterized. Some non-ABC loci carry sialic-acid processing genes and sialyltransferases, which indicates that LOS-sialylation is not confined to ganglioside-like structures (Fig. 1). In a majority of non-ABC strains, the functions of the GTases that catalyze outer core synthesis have not been elucidated, and it is difficult to make any structural predictions based on their LOS genes.
FIGURE 1.
Organization of C. jejuni LOS loci. To date, all strains known to express LOS-bound ganglioside mimics have been identified as belonging to the ABC group. They possess NeuAc-associated genes as well as cgtA and cgtB, which synthesize the Gal-β1,3-GalNAc-β1,4-Galβ ganglioside glycan backbone. Classes D and F, which are almost identical to each other, possess none of the genes involved in the synthesis of ganglioside mimics. M and R appear to be recombinant classes generated from A and D/F and carry NeuAc-associated genes. We have determined the LOS structures from strains belonging to classes F (RM1221), M (RM1503), and R (GC149). The functions of LOS genes, where known, are indicated (7, 15, 32–34). Some genes labeled “common” and “NeuAc-associated with” are also GTases.
In this study, we report three LOS structures from representative strains that have non-ABC loci. These newly determined structures considerably expand upon our existing understanding of LOS-associated glycan mimics exhibited by C. jejuni. We also show how GTases found in multiple loci types have evolving substrate preferences to adapt to novel substrate acceptors, which serve to multiply the types of glycan mimics displayed by this bacterium.
MATERIALS AND METHODS
C. jejuni Growth and LOS Isolation
C. jejuni strain RM1221 was isolated in the United States from a retail chicken and was typed as Penner serotype HS:53. C. jejuni RM1503 was isolated from a gastroenteritis patient in Canada, and it is the HS:43 Penner serotype reference strain. C. jejuni GC149 was isolated from a Japanese patient who had gastroenteritis and subsequently developed GBS as a post-infectious complication. C. jejuni GC149 was typed as Penner serotype HS:31. Plated C. jejuni cells were grown on Mueller-Hinton agar under microaerophilic conditions at 37 °C. Suspension growth in liquid culture (Mueller-Hinton medium) was used to generate large scale biomass, following which the cells were dried, and crude LOS was isolated by phenol/water extraction (9). Anhydrous hydrazine was used for O-deacylation of the LOS (10). Acid-hydrolyzed core oligosaccharide (COS) was obtained by incubating LOS in 1% acetic acid for 90 min at 100 °C and recovered in the aqueous phase following centrifugation (8000 × g, 20 min). O,N-deacyl-LOS was prepared using 4 m KOH based on the method described by Holst et al. (11). Purification of delipidated LOS forms for NMR and MS characterization was achieved by size-exclusion chromatography with a Bio-Gel P-2 column (1 cm × 1 m) eluted with H2O or using anion-exchange chromatography (HiTrap Q HP column) on an AKTA explorer system (Amersham Biosciences) with subsequent removal of contaminating NaCl with HiTrap desalting columns (Amersham Biosciences).
NMR Characterization of C. jejuni LOS and Glycosyl Derivatives
NMR spectra were obtained with COS and O,N-deacyl-LOS material dissolved in 100% D2O. To visualize sialylated species, spectra were obtained with O-deacyl-LOS, which was dispersed in perdeuterated SDS micelles (50 mm, containing 1 mm d4-EDTA). Data were acquired on Varian instruments operating at 500 and 600 MHz and processed using Bruker Topspin software. Standard 1H-1H homonuclear NOESY, COSY, TOCSY, and DQF-COSY correlation and 1H-13C heteronuclear HSQC, HMQC, and HMBC correlation spectra were used to obtain complete proton and carbon resonance assignments for the COS component of the LOS in each strain, to confirm the identity of constituent monosaccharide units, and to establish inter-residual linkage positions. These experiments are described in detail by Gilbert et al. (7). To facilitate resonance assignments of COS material, and to confirm specific GTase involvement in LOS biosynthesis, we chemoenzymatically synthesized glycoside derivatives resembling the LOS structures (see cloning and in vitro GTase activity protocols). These derivatives were completely assigned and characterized using homo- and heteronuclear spectra described above.
MS Analysis of C. jejuni LOS
Mass spectra were acquired with material isolated from colonies grown on plated media, as well as LOS purified from cells grown in suspension. Capillary electrophoresis-MS and multiple step (MSn) fragmentation experiments were performed using a CE system (Prince Technologies, The Netherlands) coupled to a 4000 QTRAP mass spectrometer (Applied Biosystems/MDS Sciex, Canada) as outlined previously (12). Intact LOS analysis was performed using electrophoresis-assisted open tubular LC-MS, and performed as described previously (13). Methylation analysis was carried out using the NaOH/DMSO/CH3I procedure as described by Ciucanu and Kerek (14). Methyl derivatives were characterized by gas-liquid chromatography MS using a Hewlett-Packard chromatograph equipped with a 30-m DB-17 capillary column (180 °C to 230 °C at 2.5 °C/min). MS was done in the electron impact mode and recorded on a Varian Saturn II mass spectrometer.
Cloning and Expression of cgtD and cstII
Construct CJL-99 expressing a fusion of the Escherichia coli maltose-binding protein (MalE, without the leader peptide) and CgtD from RM1221 has been described previously (15). GC149 cgtD (GenBankTM accession number AY962325) and RM1503 cstII (GenBankTM accession number EF140720) were amplified using Advantage 2 polymerase (Clontech). The following primers were used to amplify cgtD from GC149: CJ-722 (5′-CAAAGAAGGGCATATGAAGCAAGAAATAGTAG-3′, 33-mer, NdeI site underlined) and CJ-765 (5′-AAGGCTGTCGACTTAAATTAAATATTTTTCCAT-3′, 33-mer, SalI site underlined). The following primers were used to amplify cstII from RM1503: CJ-720 (5′-CAAGGAAATACATATGAAAAATAATATTATTATTGCTGG-3′, 39-mer, NdeI site underlined) and CJ-737 (5′-TAGCTTGTCGACAATATTATTTTTATAATATTTCTTGAG-′, 39 mer, SalI site underlined). These two primers amplified the region encoding amino acids 1–265 of cstII from C. jejuni RM1503. The PCR products were digested with NdeI and SalI and cloned in pCWori+(−lacZ) containing the sequence encoding MalE and the thrombin cleavage site, giving constructs CJL-167 (MalE-CgtD from GC149) and CST-119 (MalE-CstII from RM1503). E. coli AD202 strains containing either construct CJL-99, CJL-167, or CST-119 were grown in 2YT medium containing 0.2% dextrose and 150 μg/ml ampicillin. The cultures were incubated at 37 °C until A600 = 0.35, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and then incubated overnight at 25 °C. The cells were broken using an Avestin C5 Emulsiflex cell disruptor (Avestin, Ottawa, Canada). The MalE-Cst-II or MalE-CgtD fusions were purified by affinity chromatography on amylose resin following the manufacturer's instructions (New England Biolabs, Beverly, MA).
In Vitro GTase Reactions and Synthesis of LOS-like Glycosyl Derivatives
6-(5-Fluorescein-carboxamido)hexanoic acid succinimidyl ester (FCHASE)-labeled oligosaccharides were prepared as described previously (16). α-2,3- and α-2,8-sialyltransferase activities were assayed as described previously (17). The assay mixture for CstII activity included 0.5 mm acceptor, 1 mm CMP-NeuAc, 50 mm Hepes (pH 7.5), and 10 mm MgCl2. CgtD activity was assayed with 0.5 mm acceptor, 1 mm UDP-Gal, 50 mm Hepes (pH 7.5), and 10 mm MnCl2. Reactions were performed at 37 °C for 5 min, stopped with acetonitrile (25% final), and then analyzed by CE (16). One unit of activity is defined as the amount of enzyme producing 1 μmol of product/min. Quantitation of reactions was performed using the MDQ 32 Karat software (Beckman, CA).
RESULTS
The complete core oligosaccharides from C. jejuni strains RM1221, RM1503, and GC149 were characterized, and the structures are presented in Fig. 2. LOS material was drawn from plated cells for analytical characterization by MS, as well as suspension cultures in which preparative quantities were obtained using the phenol/water extraction procedure (9). The first three subsections under “Results” provide descriptions of structural data obtained for each strain obtained through a combination of MS and NMR techniques that are summarized in Figs. 3–6 and in supplemental Tables S1–S7. The final subsection under “Results” describes the characterization of CstII and CgtD, glycosyltransferases that are involved in the synthesis of multiple glycan mimic types.
FIGURE 2.
COS structures for C. jejuni strains RM1221, RM1503, and GC149. Colony-specific MS analysis of GC149 LOS indicates that this strain expresses GT1a and GD3 ganglioside mimics in addition to the novel glycan structure elucidated, and this is due to phase variable expression of cgtA (orf5, β1,4-GalNAc-transferase) and cgtD (orf16, α1,4-galactosyltransferase).
FIGURE 3.
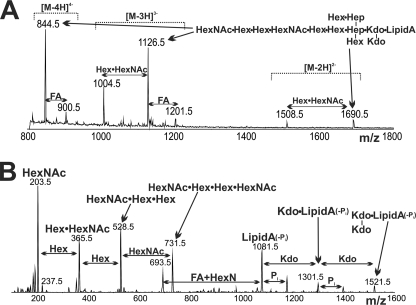
MS analysis of C. jejuni RM1221 LOS. A, negative ion mode scan of pure O-deacyl-LOS. We see glycoforms that weigh 366 Da (Hex·HexNAc) less than the predominant species, as well as molecules that carry one additional N-linked fatty acid (FA). B, positive mode MS/MS spectrum produced from the fragmentation of the major species that shows the presence of a HexNAc·Hex·Hex·HexNAc unit, and its related fragmentation ions.
FIGURE 4.
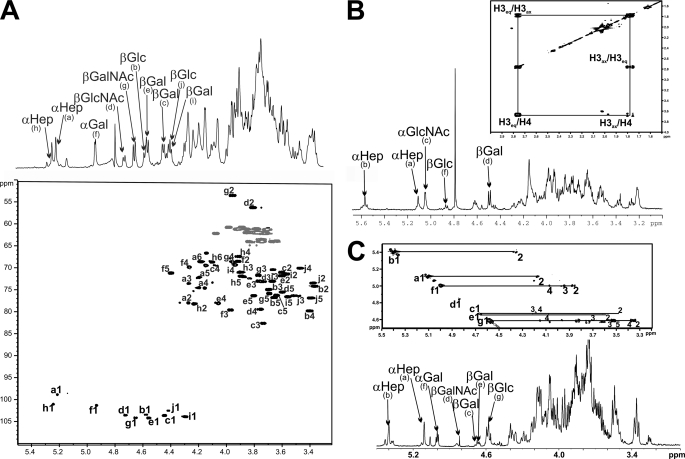
NMR spectra of C. jejuni LOS. All residues are labeled alpha-numerically based on the structures presented in supplemental Tables S2, S4, and S7. A, two-dimensional HSQC spectrum of RM1221 COS with all CH protons labeled, and a one-dimensional 1H spectrum overlay with anomeric protons indicated (CH2 protons are not labeled in the HSQC spectrum due to excessive overlap of signals.) B, one-dimensional 1H spectrum of RM1503 COS with anomeric protons labeled. Inset shows a two-dimensional COSY spectrum of O-deacyl-LOS with the H3ax/H3eq/H4 spin network of NeuAc that is α2,3-linked to the distal Gal residue. C, one-dimensional 1H spectrum of GC149 COS showing anomeric protons and a two-dimensional TOCSY overlay with correlations between anomeric and ring protons are indicated. Note that correlations with the anomeric resonance of residue d are partially suppressed due to its proximity to the HDO signal.
FIGURE 5.
MS analysis of C. jejuni RM1503 LOS. A, negative ion mode scan of intact LOS with the principal glycoform labeled. Additionally, there is evidence of variability in the presence of phosphoethanolamine and glycyl modifications, as well as ion pairing due to shortened acyl chains (by two –CH2– groups). B, positive mode MS/MS spectrum of acid-hydrolyzed LOS indicates that there is a Hex·HexNAc extension from the core, in addition to the presence of a phosphoethanolamine-modified Hep residue.
FIGURE 6.
MS analysis of LOS from the novel C. jejuni GC149 substrain. A, negative ion mode scan of intact LOS with the principal glycoform labeled. There is evidence of variability in the presence of phosphoethanolamine modifications, as well as ion pairing due to shortened acyl chains (by two –CH2– groups). B, positive ion mode scan of acid-hydrolyzed LOS show ions at m/z 1492.0 and 1783.2 representing ammonium adducts of nonsialylated and sialylated forms of the Hex-extended GM1-like structure, respectively. C, further evidence for the Hex-extended species is obtained from a positive mode MS/MS spectrum, which shows the presence of a fragment ion at m/z 690.2 corresponding to a Hex·Hex·HexNAc·Hex unit.
C. jejuni RM1221 LOS Displays Paragloboside and P Blood Group Antigen Mimics
MS analysis of de-O-acylated LOS from RM1221 in negative ion mode (Fig. 3A and supplemental Table S1) showed related doubly, triply, and quadruply charged ions at m/z 1690.5, 1126.5, and 844.5, respectively, which correspond to a species with a mass of ∼3382.3 Da. This corresponds to a de-O-acyl-lipid A component containing a standard GlcN3N-GlcN backbone (160.2 + 161.2 Da), three 3-hydroxymyristate chains (3 × 226.4 Da), and two phosphate groups (2 × 80.0 Da), summing to a mass of 1178.4 Da. The remaining mass of the COS is consistent with a glycoform containing six Hex (6 × 162.1 Da), two HexNAc (2 × 203.2 Da), two Hep (2 × 192.2), and two 3-deoxy-d-manno-2-octulosonic acid (2 × 220.2 Da) residues (total COS mass = 2221.9 Da). We also observed prominently related doubly and triply charged ions at m/z 1508.5 and 1004.5, respectively (Fig. 3A), from a molecule weighing 366 Da less than the predominant species, demonstrating that a subset of RM1221 glycoforms lack a terminal Hex·HexNAc extension. We performed MS/MS analysis of O-deacyl-LOS in positive ion mode because this yields higher quality fragmentation patterns than seen in negative ion mode. The MS/MS spectrum (Fig. 3B) showed ions at m/z 731.5, 528.5, 365.5, and 203.5, which is a pattern resulting from a HexNAc·Hex·Hex·HexNAc tetrasaccharide unit, which is fragmented to lose HexNAc initially, followed by the loss of 2 Hex units (Fig. 3B).
Because of the unusual composition and large size of the RM1221 COS, we performed carbohydrate linkage analysis, which serves as a helpful guide, in addition to NMR analysis for identifying the types and linkage position of constituent monosaccharides. GC-MS analysis of methylated residues suggest the presence of terminal Glc, terminal Gal, 4-linked Gal, 4-linked Glc, 3-linked Gal, 2-linked Hep, terminal GalNAc, 2,3,4-linked Hep, and 4-linked GlcNAc in the approximate ratios of 2:1:1:2:2:1:0.25:1:0.5 (data not shown).
NMR spectra of O,N-deacyl RM1221 LOS gave rise to three α-anomeric signals that were assigned to two αHep residues (a, 5.21 ppm and h, 5.24 ppm), which are found in all C. jejuni cores, as well as αGal (f, 4.93 ppm) (Fig. 4A and supplemental Table S2). Residues a and h were assigned as αHep units based on the strong intraresidual NOE from H1 to H2 and due to the lack of transfer of magnetization beyond H2 from H1 in 1H-1H TOCSY experiments (because of small J1,2 couplings) (Table S2). Residue f was assigned as αGal based on a strong H1-H2 NOE and observable JH,H correlations in TOCSY spectra between H1 and other ring protons up to H4, both characteristics of residues in the α-galactopyranose configuration (TOCSY transfer is not carried to H5 or H6 due to small J4.5 couplings). As shown in Fig. 4A, there are seven β-anomeric resonances from the remaining βGalNAc (g, 4.65 ppm), βGlcNAc (d, 4.72 ppm), βGlc (b, 4.57; j, 4.42 ppm) and βGal (c, 4.44; e, 4.56; i, 4.29 ppm) residues of the COS. Residues b and j were assigned as βGlc based on H1 to H3 and H1 to H5 NOEs, observable JH,H correlations in TOCSY spectra from H1 to H6, as well as an upfield chemical shift for their H2 atoms (3.36 and 3.38 ppm for residues b and j, respectively) (supplemental Table S2). Residue d also has intraresidual H1 to H3 and H1 to H5 NOEs and H1 to H6 JH,H correlations in TOCSY spectra; however, its C2 chemical shift is upfield (at 56.3 ppm, see supplemental Table S2 and Fig. 4A), consistent with N-acetylation and its assignment as βGlcNAc (18). Residues c, e, and i had H1 to H3 and H1 to H5 NOEs, as well as observable JH,H correlations from H1 to H4, consistent with assignment as βGal. Residue g had similar intraresidual NOE and TOCSY profiles to βGal residues, but its C2 chemical shift was found to be upfield (at 53.5 ppm) (supplemental Table S2), leading to its assignment as a βGalNAc unit.
The linkage between residues was established using 1H-13C HMBC spectra, which provide the necessary three-bond correlation between atoms across glycosidic bonds, and confirmed through the observation of inter-residual 1H-1H NOEs, which correlate neighboring residues via through-space connectivities, as well as carbon chemical shift comparisons for atoms participating in glycosidic linkages versus typical values for non-glycosidically linked atoms in analogous residues. For instance, we were able to determine that g (assigned as βGalNAc) was linked to f (assigned as αGal) at carbon 3 (see structure in supplemental Table S2). This was established based on the following three observations: 1) an HMBC cross-peak between the anomeric proton of residue g (at 4.65 ppm) and C3 of residue f (at 79.6 ppm); 2) an inter-residual NOE between the anomeric proton of residue g and H3 proton of residue f; and finally, 3) a shift downfield for C3 of residue f (79.6 ppm) (>8 ppm) in comparison with C3 of αGal in monosaccharide form (19). This strategy was uniformly utilized to establish the linkage position of the residues of the COS. For RM1221 COS, we observed the following three-bond connectivities between atoms across glycosidic bonds to confirm the linkage of assigned residues: H1 (g)–C3 (f); H1 (f)–C4 (e); H1 (e)–C4 (d); H1 (d)–C3 (c); H1 (c)–C4 (b); H1 (b)–C4 (a); H1 (h)–C3 (a); H1 (i)–C2 (h); and H1 (j)–C2 (a) (see supplemental Table S2).
In summary, the RM1221 COS structure was determined to have a Gal-β1,4-GlcNAc-β1,3-Gal-β1,4-Glcβ extension from heptose, which is identical to paragloboside, or the lacto-N-neotetraose (LNnT) antigen (Fig. 2). RM1221 LOS species found to have a mass of ∼3016 Da (Fig. 3A) express terminal LNnT. The major RM1221 species is further extended by GalNAc-β1,3-Galα linked to the terminal Gal of LNnT, to mimic the terminal tri- and disaccharide units of the P (globotetraose) and Pk (globotriose) blood group antigens, respectively. The Galα1,4 linkage to LNnT also mimics the P1 blood group antigen. The proposed structure is supported by binding studies with anti-P1/Pk and anti-LNnT antibodies, where specific colony isolates were shown to express one of these two antigens or both simultaneously (data not shown).
C. jejuni RM1503 LOS Displays a Type I Sialyl-Lewis c Mimic
Analysis of intact LOS from RM1503 by MS in the negative ion mode shows the presence of related triply and quadruply charged ions at m/z 1297.2 and 972.7, respectively, which correspond to a species with a mass of ∼3894.2 Da (Fig. 5A and supplemental Table S3). This corresponds to a lipid A component containing a standard GlcN3N-GlcN backbone (160.2 + 161.2 Da), four 3-hydroxymyristate chains (4 × 226.4 Da), two palmitate chains (2 × 238.4 Da), two phosphoethanolamines (2 × 123.05 Da), and two phosphate groups (2 × 80.0 Da), summing to a mass of ∼2127.7 Da. The composition of the remaining COS is consistent with a glycoform containing two Hex (2 × 162.1 Da), one HexNAc (203.2 Da), two Hep (2 × 192.2 Da), two 3-deoxy-d-manno-2-octulosonic acid (2 × 220.2 Da), one phosphoethanolamine (123.05 Da), and one NeuAc (291.3 Da) (total COS mass = 1784.5 Da). We also performed MS analysis of O-deacyl-LOS and found species with masses that fully support the proposed COS composition (supplemental Table S3). The MS spectra showed ion pairs with mass differences of 28 Da (2 methylene groups), due to variation in acyl chain length, as well as species weighing an additional ∼57 Da due to the presence of glycine (Fig. 5A), which we have previously confirmed is regularly added to the C. jejuni LOS core (13). MS/MS fragmentation of the major COS glycoform derived from acid-hydrolyzed LOS material, performed in positive mode, showed ions at m/z 1255.3, 1093.4, and 931.3, which corresponds to a Hex·HexNAc-extended COS that loses successive Hex residues (Fig. 5B). In addition, we observed ions at m/z 728.4, 536.3, and 316.1 due to further fragmentation of the glycan resulting in successive losses of HexNAc and Hep residues, leaving a phosphoethanolamine-substituted Hep species.
NMR spectra of RM1503 COS revealed the presence of three α-anomeric protons, two which were assigned αHep residues (a, 5.11 and b, 5.57 ppm) and a third from αGlcNAc (c, 5.05 ppm) (Fig. 4B and supplemental Table S4). Residues a and b were assigned as αHep based on NOEs between H1 and H2 and due to the lack of transfer of magnetization beyond H2 from H1 in TOCSY experiments (supplemental Table S4). The assignment of the αGlcNAc residue was based on an H1-H2 NOE, observable JH,H correlations in TOCSY spectra between H1 and ring protons through to H6, as well as an upfield chemical shift for its C2 atom (54.2 ppm) (18). Two β-anomeric protons seen in Fig. 4B were assigned to βGal (d, 4.49 ppm) and βGlc (f, 4.87 ppm) residues. Both d and f showed H1 to H3 and H1 to H5 NOEs. The assignment of f as βGlc was based on TOCSY connectivities from H1 through to H6, as well as an upfield chemical shift for its H2 resonance (3.37 ppm), whereas for residue d, only correlations between H1 and ring protons through to H4 were observed, leading to its assignment as βGal. Linkages between assigned residues were established using 1H-13C HMBC, and 1H-1H NOESY spectra, as well as carbon chemical shift comparisons, as outlined for the structure determination of RM1221 described earlier. We observed the following 3-bond HMBC connectivities across glycosidic bonds to confirm the linkage of assigned residues: H1 (d)–C3 (c); H1 (c)–C2 (b); H1 (f)–C4 (a); and H1 (b)–C2 (a). The spectra also showed phosphoethanolamine 1H signals at 4.14 and 3.30 ppm, and we note that the chemical shift for C6 of Hep (a) is shifted downfield (74.3 ppm) consistent with its link with phosphate and in agreement with previous studies that have shown 6-linked phosphoethanolamine in the analogous Hep residue of C. jejuni LOS cores (20, 21).
We also obtained spectra of sialylated species by dispersing of O-deacyl-LOS in perdeuterated SDS micelles (Fig. 4B, inset). As expected, the quality of spectra did not permit complete characterization of the oligosaccharide in this medium; however, we were able to assign resonances of the terminal NeuAc-α2,3-Galβ disaccharide (supplemental Table S4), with the C3 carbon chemical shift of the galactose residue clearly shifted downfield (at 76.4 ppm) as a result of its linkage with NeuAc. In accord with MS and enzymology data (see below), there was no evidence of disialyl units. In summary, the structure of RM1503 LOS was found to contain a lacto-N-biose (LNB) unit that is α-linked to the distal Hep of the core (Fig. 2), which upon sialylation corresponds to the sialyl-Lewis c antigen, also know as DU-PAN-2 (or LSTa).
C. jejuni GC149 Expresses a Ganglio/Pk Antigen Hybrid Structure
The characterization of the LOS from C. jejuni GC149, which was cultured from a GBS patient, presented an analytical challenge. When material from plated cells was analyzed by MS, we observed the presence of multiple glycoforms, and the variability could not be ascribed simply to standard substitutions or derivatization. We determined that the masses of some species were consistent with cores containing GD3 and GT1a mimics (refer to supplemental Table S5 for a complete summary of observed glycoforms). This was not surprising given that the “R” class LOS locus carried by GC149 is almost identical to that of class A strains OH4382 and OH4384, which express these ganglio structures. In addition, we observed a group of ions that were from sialylated species and that yielded MS/MS fragmentation patterns incompatible with that of a standard ganglio-oligosaccharides. Through a process of colony screening, we were able to isolate GC149 sub-strains that grew with homogeneous LOS structures, containing either the GD3 mimic, the GT1a mimic, or the novel species. GC149 has phase-variable homopolymeric G-tracts in two genes encoding outer core GTases as follows: cgtA (orf5), which encodes a β1,4-N-GalNAc-transferase that extends the proximal sialylated Gal; and cgtD (orf16), which encodes an α1,4-galactosyltransferase (Fig. 1, function described in detail below). We noticed that the GC149 sub-strain with a phased off cgtA (10-G tract) expressed a GD3 mimic, consistent with its inability to extend the outer core beyond the proximal Gal (Fig. 2). When cgtA is phased on (9-G tract) and cgtD (10-G tract) is phased off (i.e. cgtA+/cgtD−), a GT1a mimic is synthesized. When cgtA is phased on (9-G tract) and cgtD (9-G tract) is also phased on (i.e. cgtA+/cgtD+), the novel LOS species is expressed (Fig. 2). We subsequently characterized the novel LOS species from cgtA+/cgtD+ GC149.
MS analysis of intact LOS from this novel sub-strain indicates that it possesses a GM1-like structure that is extended by a single hexose. A negative ion mode scan of intact LOS (Fig. 6A and supplemental Table S6) showed related triply and quadruply charged ions at m/z 1364.3 and 1023.1, respectively, which correspond to a species with a mass of ∼4095.4 Da. This is consistent with a lipid A component containing a standard GlcN3N-GlcN backbone (160.2 + 161.2 Da), four 3-hydroxymyristate chains (4 × 226.4 Da), two palmitate chains (2 × 238.4 Da), two phosphoethanolamines (2 × 123.05 Da), and two phosphate groups (2 × 80.0 Da), summing to a mass of 2127.7 Da. The remaining mass of the COS is consistent with a glycoform containing four Hex (4 × 162.1 Da), one HexNAc (1 × 203.2 Da), two Hep (2 × 192.2), two 3-deoxy-d-manno-2-octulosonic acid (2 × 220.2 Da), and one NeuAc (291.3 Da) (total COS mass = 1985.7 Da). The extracted mass spectrum of acid-hydrolyzed COS in positive ion mode (Fig. 6B) gave rise to an ion at m/z 1492.0 which corresponds to an ammonium adduct of the proposed Hex-extended GM1-like COS. We also observe an ammonium adduct for a sialylated form of this molecule (m/z = 1783.2) that is detectable due to a small proportion of COS molecules that did not lose this acid-labile moiety during hydrolysis. MS/MS analysis of acid-hydrolyzed LOS in positive ion mode provides additional strong evidence for a Hex-extended GM1-like structure (Fig. 6C). We clearly see a Hex·Hex·HexNAc·Hex fragment ion (m/z = 690.2), that is liberated upon fragmentation of the glycan.
NMR spectra of acid-hydrolyzed cgtA+/cgtD+ GC149 LOS showed the presence of three α-anomeric protons, two which were assigned αHep residues (a, 5.11 and b, 5.40 ppm) and a third from αGal (f, 5.00 ppm) (Fig. 4C and supplemental Table S7). Residues a and b were assigned as αHep units based on intraresidual NOEs from H1 to H2 and due to the lack of transfer of magnetization beyond H2 from H1 in TOCSY spectra (supplemental Table S7). The assignment of f as αGal was based on an H1-H2 NOE and observable JH,H correlations in TOCSY spectra between H1 and ring protons through to H4. The four visible β-anomeric protons (Fig. 4C) were assigned to βGal (c, 4.68 ppm; e, 4.67 ppm), βGlc (g, 4.59 ppm, and βGalNAc (d, 4.84 ppm) residues. All β-anomers showed H1 to H3 and H1 to H5 NOEs. The assignment of g as βGlc was based on clear TOCSY connectivities from H1 through to H5, as well as an upfield chemical shift for its H2 resonance (3.36 ppm). Residues c and e had TOCSY correlations between H1 and ring protons through to H4 leading to assignment as βGal, whereas residue d was assigned as βGalNAc based on its upfield C2 chemical shift (51.7 ppm) and similar H1 to H4 TOCSY correlations (Fig. 4C). The connections between assigned residues were established primarily based on NOESY spectra, as well as HMBC connectivities and carbon chemical shift comparisons. We observed the following inter-residual NOE connectivities across glycosidic bonds to determine the linkage positions of the residues: H1 (f)–H4 (e); H1 (e)–H3 (d); H1 (d)–H4 (c); H1 (c)–H3 (b); H1 (b)–H3 (a); H1 (g)–H4 (a).
The COS structure of the novel C. jejuni GC149 sub-strain was elucidated to be an α1,4-Gal extended GM1a oligosaccharide, which in addition to its ganglioside similarity mimics the terminal disaccharide of the Pk antigen. To help verify the proposed structure, we synthesized preparative quantities of Gal-α1,4-GM1a-S-phenyl using recombinant CgtD from GC149, the enzyme that catalyzes the hexose extension of the GM1-oligosaccharide structure (see below), and we confirmed that the 1H and 13C chemical shifts for this derivative were similar to those of analogous residues in GC149 COS at the nonreducing end (supplemental Table S8).
Evolving Substrate Specificity of CgtD and CstII
CgtD is an α1,4-galactosyltransferase (15) that is involved in the synthesis of the core structures of both RM1221 and GC149 despite the fact that they are markedly different (i.e. P-antigen versus Gal-GM1a). To help evaluate their catalytic similarity, we cloned and functionally expressed the CgtD alleles from these two strains. CgtDGC149 was found to efficiently catalyze αGal transfer to Lac-, LacNAc-, and GM1a-FCHASE substrates, with a slight preference for GM1a, its in vivo substrate (Table 1). CgtDRM1221 transferred αGal to Lac and LacNAc, but there was a complete absence of GTase activity with GM1a. GC149 has a class R locus that was created through recombination of classes D and A (6). Early class R strains likely had an incompatible substrate/GTase pair (i.e. GM1a/CgtD) until accumulated mutations to CgtD produced an allele capable of catalyzing α-Gal transfer to the GM1a-like LOS substrate (note that CgtDGC149 and CgtDRM1221 share 67.3% protein sequence identity). Later generations could thereby synthesize a novel LOS structure and a new form of glycan mimic.
TABLE 1.
Substrate specificity of the galactosyltransferase CgtD from RM1221 and GC149, measured using FCHASE-reducing end glycan derivatives (at 0.5 mm) and determined by CE
CgtD from RM1221 promotes efficient α1,4-Gal transfer to LacNAc (and Lac), matching its LOS substrate, but it is not capable of promoting transfer to a terminal Galβ1,3-GalNAcβ acceptor (GM1a), the preferred substrate for CgtD in GC149.
| Acceptor (−FCHASE) | Specific activity |
||
|---|---|---|---|
| CgtD-RM1221 | CgtD-GC149 | ||
| units/mg | |||
| Lac- | Galβ-1,4-Glc- | 0.40 | 0.54 |
| LacNAc- | Galβ-1,4-GlcNAc- | 0.60 | 0.51 |
| GM1a- | Galβ-1,3-GalNAcβ-1,4-[NeuAcα-2,3-]-Galβ-1,4-Glc- | 0.00 | 0.75 |
A similar set of circumstances was likely at play with the sialyltransferase CstII present in RM1503. In strains with ABC loci, the function of this enzyme is well documented because of its role in the synthesis of ganglioside mimics (7). Mono- (α2,3-) and bifunctional (α2,3-/α2,8-) alleles that have been characterized so far have been shown to sialylate Gal-β1,4-Glcβ or Gal-β1,3-Hepα acceptors to generate GM3/GD3 mimics and Gal-β1,3-GalNAcβ to generate GM1/GD1a/GT1a mimics (7). RM1503 is a class “‘M” strain that synthesizes an LNB oligosaccharide acceptor. We assayed recombinant MalE-CstIIRM1503 and found that it had greater than 5-fold more activity with LNB than with any of the acceptors produced by class ABC strains (Table 2). Pairwise alignments showed that CstIIRM1503 shares 86–91% protein sequence identity with other CstII sequences available in GenBankTM (data not shown). Currently, it is not possible to predict which amino acid substitutions are responsible for its adaptation to an acceptor with a Lewis-type (I) linkage. CstIIRM1503 also has very low activity with 3′-sialyllactose (Table 2), which indicates that it is effectively a mono-functional version of CstII (i.e. without α2,8-sialyltransferase specificity).
TABLE 2.
Substrate specificity of the sialyltransferase CstII from RM1503 measured using FCHASE-reducing end glycan derivatives (at 0.5 mm) and determined using CE
CstII from RM1503 promotes LNB sialylation with greater than 5-fold more efficiency than terminal Gal-β1,3-GalNAcβ (GM1a) and Gal-β1,4-Glcβ (Lac) acceptors, which are natural substrates for the enzyme in strains expressing ganglioside mimics.
| Acceptor (−FCHASE) | Specific activity | |
|---|---|---|
| milliunits/mg | ||
| LNB- | Galβ-1,3-GlcNAc- | 111.9 |
| GM1a- | Galβ-1,3-GalNAcβ-1,4-[NeuAcα-2,3-]-Galβ-1,4-Glc- | 17.5 |
| Lac- | Galβ-1,4-Glc- | 16.8 |
| LacNAc | Galβ-1,4-GlcNAc- | 6.6 |
| 3′Sialyl-Lac- | NeuAcα-2,3-Galβ-1,4-Glc- | 0.1 |
DISCUSSION
C. jejuni strains within the ABC LOS loci group have the necessary genes to synthesize ganglioside mimics and can trigger the production of autoreactive antibodies leading to GBS. Genomic cataloguing of the LOS loci from a large collection of environmental and clinical isolates (both GBS- and non-GBS-associated) indicate a majority belong to the ABC group, suggestive of a selective advantage in displaying ganglioside-like outer cores (5, 6, 8, 22). Nevertheless, over 30% of strains carry non-ABC loci and likely do not display ganglioside mimics. The hypervariability within the LOS locus has led to innumerable genetic arrangements within this bacterium, and to date there are 16 known non-ABC classes (6). Some are ABC-related, carrying for instance genes that direct LOS sialylation, although others show no similarity and synthesize different cores (Fig. 1).
The LOS from C. jejuni RM1221, RM1503, and GC149 offer insight into the structural properties that characterize non-ganglioside strains. A recurring theme is host glycan mimicry. Although this is not a new observation among mucosal pathogens, the range of human mimicry displayed by C. jejuni appears unrivaled among bacterial pathogens. As summarized in Fig. 7, this organism has the capability of expressing the following: 1) the ganglio glycan series; 2) the lacto-N-neotetraose/paragloboside series; 3) P blood group antigens; and 4) lacto-N-biose and Lewis type (I) linkages. Undoubtedly there are other forms to be found.
FIGURE 7.
Glycan mimics expressed by C. jejuni in its LOS and the GTases that direct their synthesis. Ganglio series mimics have been found in a number of strains linked to GBS. Note that disialylation of the proximal Gal in the ganglio series has only been observed in absence of Galβ1,3-GalNAcβ1,4-elongation i.e. the NeuAc residue shown in gray has been observed only in the GD3 mimic. C. jejuni can also synthesize Lewis type-I (RM1503) mimics, paragloboside, and P blood group antigens (RM1221), as well as “hybrid” forms such as the structure exhibited by GC149 which has similarity to ganglio and Pk antigens. This strain can, through phase variation, also express complete GT1a or GD3 mimics.
It should be noted that an LOS class is not always associated with a specific Penner serotype, because Penner typing is based on capsule structures whose synthesis is dependent on a genome locus distinct from that governing LOS biosynthesis (23). Consequently, the structures reported in this work are not expected to be representative of the Penner types of the studied strains (HS:31 for GC149, HS:43 for RM1503, and HS:53 for RM1221). Parker et al. (6) have sequenced the LOS locus of another HS:31 strain (GB24) and two other HS:53 strains (RM1508 and RM3435), and their loci differed from those found in GC149 and RM1221, respectively.
Paragloboside (LNnT) and P-antigen mimicry has been observed previously in the LOS of Haemophilus and Neisseria spp., with some known examples of sialyl-LNnT as well (24–27). It is noteworthy that phase-variable expression of outer core GTases in RM1221 can result potentially in different proportions of cells expressing terminal P antigen (globoside), P1 antigen, or LNnT. We have been able to isolate colonies expressing these antigens with anti-P1/Pk and anti-LNnT monoclonal antibodies binding to colonies on nitrocellulose.3 Lewis blood group mimics are well documented components of the O-chain in Helicobacter pylori LPS (28, 29). Although type I-fucosylated Lewis units (i.e. Lewis a/b) are common human antigens, sialyl-Lewis c, which we observed in C. jejuni RM1503, has only been detected in gastrointestinal tumors and is referred to clinically as DU-PAN-2. This antigen may have a broader distribution in other organisms colonized by this bacterium.
The expression of surface glycan mimics by pathogenic bacteria is thought to be driven by the need to evade an immune response, such as complement fixing, as well as to act as ligands that hijack host-cell receptors and/or lectins during adhesion and colonization (30). It is interesting to note that although ganglioside-like structures have been well documented and studied in relation to their involvement in pathogenic auto-antibody generation, a defining role in gastrointestinal colonization and cell invasion has yet to be established in the many years since their discovery. This owes partly to limited LOS structural data among strains that are not representative of ABC loci types. These newly discovered LOS-associated glycan mimics should help delineate structure-dependent roles governing the pathogenesis of this bacterium.
There are several forms of genome-level modifications that give rise to phenotypic differences in LOS expression by C. jejuni (17). These include macroscopic gene rearrangements that produce different LOS classes, as well as polynucleotide tracts and point mutations that vary the expression of, or inactivate, LOS biosynthesis genes. Accumulated point mutations also give rise to allelic forms of GTases that have altered acceptor specificities and in some cases functionality. We have previously characterized allelic variants of CstII, CgtA, and CgtB (7, 17, 31), but within the context of ABC class strains. C. jejuni RM1221 and GC149 have very different LOS loci and synthesize different types of glycan mimics, both of which require the galactosyltransferase CgtD. The class R locus, carried by GC149, appears to have arisen from a fusion of A and D classes, the latter of which carries a CgtD allele that can transfer αGal to Lac and LacNAc (Table 1). Initially, class R strains would have been unable to express a CgtD variant that participated in COS synthesis, until its substrate specificity was broadened to include GM1a-like structures through mutations in later generations. A similar scenario could be envisioned for CstII in RM1503. Its class M locus is thought to have also arisen following fusion of classes A and D, although with a rearrangement quite different from that of class R. The strong preference of CstIIRM1503 for LNB-type substrates over ganglio-oligosaccharides synthesized by the ABC strains also appears to have been a substrate-driven phenomenon in descending generations from those that initially acquired this locus.
With respect to its LOS structure, GC149 can be envisioned as a transitional strain. It possesses an ABC-like locus and can synthesize GT1a and GD3 ganglioside mimics. The presence of CgtD and its ability to transfer to a ganglio-oligosaccharide substrate enables this strain to display the αGal-1,4-Galβ antigen, a terminal disaccharide that is frequently expressed by mucosal pathogens, presumably because of its abundance in human tissue. In GC149, cgtD and cgtA exhibit phase variable expression that enables the switching from one mimic type to another over a short generation time span.
CONCLUSIONS
We have provided new insight into the diversity of C. jejuni LOS structures that mimic mammalian cell-surface glycans. Ganglioside-like structures, which are expressed most frequently, have been implicated in the development of GBS. P blood group- and LNnT-related antigens appear in up to 10–15% of strains based on the abundance of class D and F loci, although sialylated non-ganglioside LOS expression appears limited to less than 5% of strains (5). Novel C. jejuni LOS structures associated with antigenic mimicry provide opportunities for further research relevant to colonization of different hosts, virulence, and food safety.
Acknowledgements
We thank Dr. Warren Wakarchuk for critical review of the manuscript and Tom Devecseri for help with the figures.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S8.
R. E. Mandrell, A. H. Bates, M. Gilbert, and C. T. Parker, unpublished data.
- GBS
- Guillain-Barré syndrome
- COS
- core oligosaccharide
- GTase
- glycosyltransferase
- LNB
- lacto-N-biose
- LNnT
- lacto-N-neotetraose
- LOS
- lipooligosaccharide
- FCHASE
- 6-(5-fluorescein-carboxamido)-hexanoic acid succinimidyl ester
- CE
- capillary electrophoresis.
REFERENCES
- 1. Yuki N., Taki T., Inagaki F., Kasama T., Takahashi M., Saito K., Handa S., Miyatake T. (1993) J. Exp. Med. 178, 1771–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aspinall G. O., McDonald A. G., Pang H., Kurjanczyk L. A., Penner J. L. (1994) Biochemistry 33, 241–249 [DOI] [PubMed] [Google Scholar]
- 3. Rinaldi S., Willison H. J. (2008) Curr. Opin. Neurol. 21, 540–546 [DOI] [PubMed] [Google Scholar]
- 4. Yuki N. (2010) Methods Mol. Biol. 600, 51–65 [DOI] [PubMed] [Google Scholar]
- 5. Parker C. T., Horn S. T., Gilbert M., Miller W. G., Woodward D. L., Mandrell R. E. (2005) J. Clin. Microbiol. 43, 2771–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parker C. T., Gilbert M., Yuki N., Endtz H. P., Mandrell R. E. (2008) J. Bacteriol. 190, 5681–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert M., Brisson J. R., Karwaski M. F., Michniewicz J., Cunningham A. M., Wu Y., Young N. M., Wakarchuk W. W. (2000) J. Biol. Chem. 275, 3896–3906 [DOI] [PubMed] [Google Scholar]
- 8. Godschalk P. C., Heikema A. P., Gilbert M., Komagamine T., Ang C. W., Glerum J., Brochu D., Li J., Yuki N., Jacobs B. C., van Belkum A., Endtz H. P. (2004) J. Clin. Invest. 114, 1659–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westphal O., Jann K. (1965) Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 10. Holst O., Broer W., Thomas-Oates J. E., Mamat U., Brade H. (1993) Eur. J. Biochem. 214, 703–710 [DOI] [PubMed] [Google Scholar]
- 11. Holst O., Thomas-Oates J. E., Brade H. (1994) Eur. J. Biochem. 222, 183–194 [DOI] [PubMed] [Google Scholar]
- 12. Li J., Richards J. C. (2007) Mass Spectrom. Rev. 26, 35–50 [DOI] [PubMed] [Google Scholar]
- 13. Dzieciatkowska M., Brochu D., van Belkum A., Heikema A. P., Yuki N., Houliston R. S., Richards J. C., Gilbert M., Li J. (2007) Biochemistry 46, 14704–14714 [DOI] [PubMed] [Google Scholar]
- 14. Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 15. Houliston R. S., Bernatchez S., Karwaski M. F., Mandrell R. E., Jarrell H. C., Wakarchuk W. W., Gilbert M. (2009) Glycobiology 19, 153–159 [DOI] [PubMed] [Google Scholar]
- 16. Wakarchuk W. W., Cunningham A. M. (2003) Methods Mol. Biol. 213, 263–274 [DOI] [PubMed] [Google Scholar]
- 17. Gilbert M., Karwaski M. F., Bernatchez S., Young N. M., Taboada E., Michniewicz J., Cunningham A. M., Wakarchuk W. W. (2002) J. Biol. Chem. 277, 327–337 [DOI] [PubMed] [Google Scholar]
- 18. Duus J., Gotfredsen C. H., Bock K. (2000) Chem. Rev. 100, 4589–4614 [DOI] [PubMed] [Google Scholar]
- 19. Hobley P., Howarth O., Ibbett R. N. (1996) Magn. Reson. Chem. 34, 755–760 [Google Scholar]
- 20. Aspinall G. O., McDonald A. G., Raju T. S., Pang H., Moran A. P., Penner J. L. (1993) Eur. J. Biochem. 213, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 21. Aspinall G. O., McDonald A. G., Raju T. S., Pang H., Kurjanczyk L. A., Penner J. L., Moran A. P. (1993) Eur. J. Biochem. 213, 1029–1037 [DOI] [PubMed] [Google Scholar]
- 22. Koga M., Gilbert M., Takahashi M., Li J., Koike S., Hirata K., Yuki N. (2006) J. Infect. Dis. 193, 547–555 [DOI] [PubMed] [Google Scholar]
- 23. Karlyshev A. V., Linton D., Gregson N. A., Lastovica A. J., Wren B. W. (2000) Mol. Microbiol. 35, 529–541 [DOI] [PubMed] [Google Scholar]
- 24. Jennings H. J., Johnson K. G., Kenne L. (1983) Carbohydr. Res. 121, 233–241 [DOI] [PubMed] [Google Scholar]
- 25. Mandrell R. E. (1992) Infect. Immun. 60, 3017–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mandrell R. E., Apicella M. A. (1993) Immunobiology 187, 382–402 [DOI] [PubMed] [Google Scholar]
- 27. Moran A. P., Prendergast M. M., Appelmelk B. J. (1996) FEMS Immunol. Med. Microbiol. 16, 105–115 [DOI] [PubMed] [Google Scholar]
- 28. Aspinall G. O., Monteiro M. A., Pang H., Walsh E. J., Moran A. P. (1996) Biochemistry 35, 2489–2497 [DOI] [PubMed] [Google Scholar]
- 29. Chan N. W., Stangier K., Sherburne R., Taylor D. E., Zhang Y., Dovichi N. J., Palcic M. M. (1995) Glycobiology 5, 683–688 [DOI] [PubMed] [Google Scholar]
- 30. Mandrell R. E., Apicella M. A., Lindstedt R., Leffler H. (1994) Methods Enzymol. 236, 231–254 [DOI] [PubMed] [Google Scholar]
- 31. Bernatchez S., Gilbert M., Blanchard M. C., Karwaski M. F., Li J., Defrees S., Wakarchuk W. W. (2007) Glycobiology 17, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 32. Gilbert M., Parker C. T., Moran A. P. (2008) in Campylobacter (Nachamkin I., Szymanski C. M., Blaser M. J. eds) 3rd Ed., pp. 483–504, American Society for Microbiology, Washington, D. C [Google Scholar]
- 33. Houliston R. S., Endtz H. P., Yuki N., Li J., Jarrell H. C., Koga M., van Belkum A., Karwaski M. F., Wakarchuk W. W., Gilbert M. (2006) J. Biol. Chem. 281, 11480–11486 [DOI] [PubMed] [Google Scholar]
- 34. Linton D., Karlyshev A. V., Hitchen P. G., Morris H. R., Dell A., Gregson N. A., Wren B. W. (2000) Mol. Microbiol. 35, 1120–1134 [DOI] [PubMed] [Google Scholar]