Abstract
The two major components of the Eubacteria Sec-dependent protein translocation system are the heterotrimeric channel-forming component SecYEG and its binding partner, the SecA ATPase nanomotor. Once bound to SecYEG, the preprotein substrate, and ATP, SecA undergoes ATP-hydrolytic cycles that drive the stepwise translocation of proteins. Although a previous site-directed in vivo photocross-linking study (Mori, H., and Ito, K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16159–16164) elucidated residues of SecY needed for interaction with SecA, no reciprocal study for SecA protein has been reported to date. In the present study we mapped residues of SecA that interact with SecY or SecG utilizing this approach. Our results show that distinct domains of SecA on two halves of the molecule interact with two corresponding SecY partners as well as with the central cytoplasmic domain of SecG. Our data support the in vivo relevance of the Thermotoga maritima SecA·SecYEG crystal structure that visualized SecYEG interaction for only one-half of SecA as well as previous studies indicating that SecA normally binds two molecules of SecYEG.
Keywords: ATPases, Membrane Biogenesis, Membrane Proteins, Membrane Trafficking, Molecular Motors, Protein Cross-linking, Protein Translocation, Protein-Protein Interactions, Photocross-linking
Introduction
Sec-mediated protein translocation takes place utilizing the evolutionarily conserved translocon, SecYEG, in prokaryotes and Sec61αβγ in eukaryotes (1). These channel-forming transmembrane complexes (2) cooperate with different cytosolic partners to provide distinct pathways for protein topogenesis. In bacteria, SecYEG typically interacts with the signal recognition particle-ribosome pathway for the co-translational integration of integral membrane proteins, whereas its interaction with the SecB and SecA pathway is generally reserved for the post-translational translocation of periplasmic and outer membrane proteins as well as for the biogenesis of integral membrane proteins containing sizable periplasmic domains that need to traverse the plasma membrane (3, 4).
SecA cooperates with the SecB chaperone to target preproteins to SecYEG. To carry out this function, SecA contains regions specific for binding signal peptides on preproteins, the SecB chaperone and SecYEG (for review, see Ref. 5). Once preproteins are bound to the SecA·SecYEG complex, they are translocated by ATP-driven conformational cycles of the SecA nanomotor that have been previously referred to as the SecA membrane insertion/de-insertion cycle based on evidence suggesting that portions of SecA may penetrate transmembrane regions of SecYEG (6–8) (for an alternate view, see Ref. 9). Although SecA and SecYE proteins are sufficient to catalyze protein translocation, the presence of SecG protein significantly stimulates both in vivo and in vitro protein translocation, particularly at low temperatures (10, 11). SecG has been shown to enhance the SecA membrane insertion/de-insertion cycle through its coupled topology inversion, although this latter proposal has been challenged by the normal translocation behavior of a topologically fixed form of SecG (12–16). Although significant progress has been made recently in defining those regions of SecA that are responsible for SecB and signal peptide recognition, elucidation of those regions of SecA that are responsible for SecYEG binding and activation has remained a more difficult task (17–20).
A variety of different approaches have been taken to elucidate regions of SecA and SecYEG that are important for complex formation as well as activation. SecY forms the bulk of the translocon channel complex, and it is composed of 10 transmembrane (TM1-TM10),2 six cytoplasmic (C1-C6), and five periplasmic domains (P1-P5) (2, 21). Although initial genetic studies revealed that C5 and C6 are important for the SecA-activation function of SecY, single amino acid substitutions within these regions or even a small deletion within C6 had no effect on SecA binding (6, 22, 23). Similar results were found for substitution mutations at the highly conserved Arg-357 residue within C5 as well as Glu-176 within TM4 (24). By contrast, alterations within TM10 or TM7 (Ile-408 to Asn or Ile-278 to Cys, respectively) that resulted in signal sequence suppressor phenotypes did give rise to an increased affinity of SecA for SecY (25, 26). However, given the location of these residues within the interior of SecY (2), it seems more plausible that this latter result is due to an altered SecY conformation that imparts an increased SecA affinity.
Peptide affinity blotting and peptide library-scanning approaches have also been utilized to map regions of SecYEG that specifically bind to SecA (24, 27–29). These studies indicated that the initial interaction of SecA with SecYEG occurs largely through cytoplasmically exposed regions, as five of six cytoplasmic domains of SecY (all except C3) as well as the sole cytoplasmic domain of SecE were implicated in SecA binding (29). Additional studies employing alanine mutagenesis showed that multiple substitutions within C1, C2, or C6 of SecY resulted in a decreased affinity for SecA. Furthermore, the identification of SecA-binding peptides within transmembrane and even periplasmic domains of SecY (TM3, P3, TM4, TM5, TM8, and TM9), SecE (TM2), or SecG (TM2) was interpreted as supporting previous studies that identified portions of SecA that insert into the membrane during its ATP-driven protein translocation cycle (7, 8, 30–33). However, given the artificial nature of this approach as well as major differences in the number of SecY-binding peptides identified by different laboratories (e.g. compare Refs. 24, 28, and 29), these results should be interpreted with caution until more physiological studies can be completed.
Given the limitations inherent in genetic or biochemical approaches that employ mutant systems or purified complexes, respectively, in vivo cross-linking approaches offer an attractive alternative by which to study native systems. Recently a site-directed in vivo photocross-linking approach was developed by Schultz and co-workers (for review, see Ref. 34) that allows for detailed protein-protein interaction maps to be constructed. For this purpose p-benzoylphenylalanine, a photo-reactive phenylalanine derivative (35), is incorporated at engineered amber codons within a plasmid-borne target gene utilizing the appropriate cloned Methanococcus jannaschii amber suppressor tRNA and tRNA synthetase genes. The benzophenone group of pBPA reacts with nearby C-H bonds upon excitation at 350–365 nm to produce covalent cross-links between interacting partner proteins. This approach has been utilized previously to map SecA-interacting regions within the cytoplasmic domains of SecY. Of 53 residues tested within SecY, SecA-specific photocross-links were detected within C2, C4, C5, and C6 (36).
In the present study we have continued to utilize this powerful in vivo approach to define SecA regions that interact with SecY or SecG proteins. Our results show that distinct domains on two halves of the SecA molecule interact with two corresponding SecY partners as well as with the central cytoplasmic domain of SecG protein that contains the TLF motif (37). Our data also support the in vivo relevance of the recently published Thermotoga maritima SecA·SecYEG crystal structure that visualized SecYEG interaction for only the carboxyl-terminal half of SecA (38).
EXPERIMENTAL PROCEDURES
Chemicals, Media, Strains, and Plasmids
pBPA1 was purchased from Bachem, whereas LB (Miller) broth and agar were obtained from EMD Chemicals and Difco, respectively. Most other chemicals were obtained from Sigma or a comparable supplier and were reagent quality or better. Plasmid mutations were made by the QuikChange (Stratagene) method utilizing oligonucleotide primers (Integrated DNA Technologies) designed on the Agilent website, and mutations were verified by DNA sequence analysis (University of Pennsylvania DNA Sequence Facility). Escherichia coli BLR(λDE3) [F−ompT hsdS (rB− mB−) gal dcm Δ(srl-recA)306::Tn10 (TetR)] was obtained from Stratagene. pSup-pBpARS-6TRN is a p15A chloramphenicol resistant plasmid encoding the Methanocaldococcus jannaschii amber suppressor tRNA/tRNA synthetase system that allows for the efficient incorporation of pBPA into amber codons and was obtained from Peter Schultz (Scripps) (39). Plasmids encoding an amino-terminal myc-tagged SecY or carboxyl-terminal myc-tagged SecG, pCDFT7secYmycEG or pCDFT7secEGmyc, respectively, are CloDF13 streptomycin-resistant plasmids that are compatible with both colE1- and p15A-derived plasmids, and they were constructed as follows. To PCR amplify the secYEG genes from pET610 (40) and attach an myc tag onto the 5′ end of secY, forward and reverse primers, 5′-GGGAATTCCATATGGAAGAACAGAAACTCATCTCCGAAGAGGACCTGGCTAAACAACCGGGATTAGATTTTCAAAGTGCCAAAG-3′ and 5′-CCGCTCGAGTTAGTTCGGGATATCGCTGGTC-3′, respectively, were used in a PCR reaction. The appropriate PCR fragment was gel-purified using the QIAquick (Qiagen) method, digested with NdeI and XhoI (New England Biolabs), and the secYmycEG DNA fragment was cloned into a pCDFDuet-1 vector (Novagen) that had been similarly digested and gel-purified. After ligation, transformation, and purification, the resulting plasmid, pCDFT7secYmycEG, was verified both by restriction enzyme mapping and DNA sequence analysis. pCDFT7secYEGmyc was constructed from pT7CDFT7secYmycEG by first deleting the secY myc tag and then by adding an myc tag onto the 3′ end of secG utilizing QuikChange and appropriate oligonucleotide primers. The secA and secG amber mutations were constructed in pT7secA-his (colE1, ampicillin resistant) (41) and pCDFT7secYEGmyc, respectively. In addition, initial studies to set up photocross-linking controls employed a secY amber mutation at codon 434, which was constructed in pET610. For photocross-linking studies with secA amber alleles, BLR(λDE3) derivatives containing three plasmids, pT7secA-his with appropriate amber alleles, pCDFT7secYmycEG, and pSup-pBpARS-6TRN, were constructed by transformation. For comparable studies with secG amber alleles, BLR(λDE3) derivatives containing three plasmids, pCDFT7secYEGmyc with appropriate amber alleles, pT7secA-his, and pSup-pBpARS-6TRN were constructed similarly.
In Vivo Photocross-linking
Strains were grown at 37 °C in 200 ml of LB supplemented with ampicillin (100 μg/ml) chloramphenicol (50 μg/ml), streptomycin (100 μg/ml), and pBPA (1 mm) to an A600 between 0.4 and 0.6, when isopropyl-β-d-thiogalactopyranoside was added to 1 mm, and growth was continued for an additional 2 h. The culture was chilled on ice, and all subsequent steps were done on ice or at 4 °C unless specified otherwise. Based on A600 readings, equivalent numbers of cells were sedimented at 8000 × g for 10 min at 4 °C, washed with ice-cold PBS (10 mm sodium phosphate, pH 7.2, 140 mm NaCl), and resuspended in 48 ml of PBS. 24 ml samples were transferred to 100 mm × 15-mm polystyrene Petri dishes and irradiated at 365 nm for 15 min on a bed of ice using a Rayonet 2000 UV cross-linker (Southern New England Ultraviolet Co.). Treated and control samples were sedimented at 13,000 × g for 10 min at 4 °C, washed with ice-cold 10 mm Tris-HCl, pH 8.1, and suspended in 6 ml of 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.25 mm PMSF, 1 mm dithiothreitol, 100 μg/ml RNase, 100 μg/ml DNase, 4 mg/ml lysozyme, and 10× protease inhibitor mixture (Sigma P2714). Cells were disrupted by one or two passages through the French press (Aminco) at 8000 lb/in2, and unbroken cells were removed by 2 repeated sedimentations at 13,000 × g for 20 min at 4 °C. Membranes were isolated by sedimentation at 240,000 × g for 50 min at 4 °C. The membrane pellet was resuspended in 6 ml of 0.2 m Na2CO3, pH 11.5, incubated on ice for 30 min with occasional mixing, and resedimented as above. The final carbonate-treated membrane pellet was solubilized in 0.2 ml of 5% SDS, 10 mm Tris-HCl, pH 8.0, 1 mm EDTA at 37 °C with occasional mixing for 2 h, when insoluble material was removed by sedimentation at 100,000 × g for 20 min at 4 °C. The concentration of total membrane protein was determined spectrophotometrically at A280, and all samples were adjusted to have an equivalent concentration of total membrane protein. An equal volume of sample buffer (2% SDS, 125 mm Tris-HCl, pH 6.8, 5% 2-mercaptoethanol, 15% glycerol, 0.005% bromphenol blue) was added to the final sample, and 100-μl aliquots were analyzed on 7.5% SDS-PAGE gels. To better separate high molecular weight complexes, 20-cm long SDS-PAGE gels were run at 50–60 V for 16 or 17 h, and the upper portion of the gel was collected for further analysis. Proteins were transferred to nitrocellulose membranes (VWR) by electroblotting (Bio-Rad Criterion blotter) at 100 V for 1 h, and membranes were blocked by overnight incubation in 20 mm Tris-HCl, pH 7.5, 140 mm NaCl supplemented with 10% nonfat dry milk. SecA·SecY cross-linked complexes were detected by Western blotting using SecA antisera (TANA Laboratories) or a rabbit polyclonal (Santa Cruz Biotechnology) or mouse monoclonal antibody (Genescript) against c-Myc as primary antibodies. Horseradish peroxidase-conjugated anti-rabbit (Molecular Probes) or anti-mouse (Genescript) secondary antibody was then employed, and immunocomplexes were visualized by enhanced chemiluminescence using SuperSignal West Pico (Pierce) and imaged using a Syngene Gelbox system. Initial optimizing of the photocross-linking reaction or screening of our secA amber collection was done using a smaller scale method. In this case 25-ml cultures were prepared, washed cells were resuspended in a 6 ml volume, and only 1-ml samples were subjected to photocross-linking for 3 min. After photocross-linking, cells were broken in a cup horn sonicator (Heat Systems Ultrasonics) at the highest setting using one or more bursts of 60 s each at 0 °C, and total membranes were isolated and analyzed similarly as described above.
Computer Analysis and Modeling
The LIGPLOT program that also contained the DIMPLOT program was obtained from Roman Laskowski (European Bioinformatics Group), and it was utilized to identify interacting residues of SecA and SecG proteins utilizing the T. maritima SecA·SecYEG structure (38). The program was set up in the directory path as an executable file, and a modified version of the PDB file (3DIN) containing only the SecA and SecG coordinates was made and saved in the same directory. The program was run in the command prompt as dimplot file.pdb B E (where the last two letters designate the chains in the PDB file of the two proteins). The output was given in a postscript format designating all interacting residues between the two proteins. To generate front-to-front and back-to-back models of SecA bound to a SecYEG dimer, a combination of PyMOL, DSviewerpro, and Coot were utilized. In both cases a second copy of SecYEG was manually docked onto the original SecA·SecYEG structure in either front-to-front or back-to-back orientations to form a mirror image with the corresponding SecYEG partner. In the latter case the third transmembrane helixes of the two SecE molecules were brought into proper proximity with one another (42).
RESULTS
SecA·SecY Interaction Studies
To define regions of SecA that interact with SecY, we utilized a site-directed in vivo photocross-linking approach whereby the photoreactive amino acid pBPA was incorporated into SecA at selected amber codons throughout the gene. To maximize our chances of locating regions of SecA·SecY interaction, we relied on our prior study where cysteine scanning and sulfhydryl accessibility approaches were combined to construct an in vivo membrane topology map of SecA (33). Because productive labeling in this latter case appears to have utilized the protein-conducting channel in order for the sulfhydryl-labeling reagent to gain access to SecA (i.e. the sulfhydryl reagent utilized is ordinarily membrane impermeable), we reasoned that many of these periplasm-accessible SecA residues that are within fluid contact of the channel should also be proximal to SecY. As such, they should serve as an ideal starting place for positioning amber codons within secA. Furthermore, because the 26 SecA cysteine residues that we have now converted to amber codons are reasonably dispersed throughout the secA gene, this approach also provides good coverage of the different SecA regions.
We constructed strains with three compatible plasmids for this study. First, we constructed secA amber alleles in pT7secA-his, where secA expression is regulated by the T7 promotor, and SecA protein is synthesized with a carboxyl-terminal hexahistidine tag that has been shown previously not to affect its function (see “Experimental Procedures”) (41). Second, we constructed a c-Myc-tagged version of SecY, pCDFT7secYmycEG, as, our anti-SecY peptide antibody was of low affinity and did not allow for good detection of the SecY cross-linked species, whereas our SecA antisera often visualized multiple SecA cross-linked species (data not shown). This latter result is not surprising given the size and complexity of SecA protein and its potential for interacting with multiple partners both of cytoplasmic as well as membrane origin. The combination of these two plasmids, pT7secA-his and pCDFT7secYmycEG, overproduced the SecA·SecYEG complex severalfold, and the overproduced complex has been shown to be functional by a number of criteria (see e.g. Ref. 43). Finally we utilized pSup-pBpARS-6TRN encoding the highly expressed M. jannaschii amber suppressor tRNA and tRNA synthetase genes that is needed for efficient incorporation of pBPA at amber codons (39).
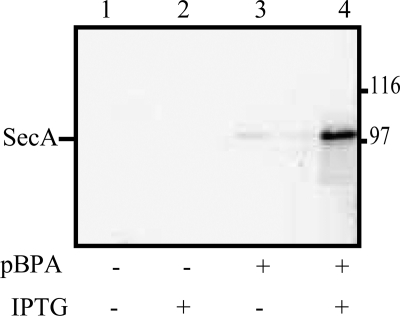
To check for specific and efficient incorporation of pBPA at amber codons, a representative strain carrying a secA amber mutation at codon 59, secA59(Am), was grown in the presence or absence of pBPA, and SecA protein production was monitored by Western blotting. No SecA protein was detected when cells were grown in the absence of pBPA, whereas full-length SecA was detected upon inclusion of pBPA and isopropyl-β-d-thiogalactopyranoside (IPTG) in the media (Fig. 1). This result indicates that pBPA incorporation into plasmid-derived, full-length SecA occurred with reasonable efficiency.
FIGURE 1.
Incorporation of pBPA into SecA protein. The secA59(Am) mutant was grown in the presence or absence of pBPA and/or isopropyl-β-d-thiogalactopyranoside (IPTG) as indicated. Cell cultures were grown and analyzed as described under “Experimental Procedures.” Western blotting with Ni+2-conjugated horseradish peroxidase is shown. The position of SecA protein is given.
We next turned to optimizing the photocross-linking step. For this purpose we utilized a Rayonet RPR-100 photoreactor, as this long wavelength UV source has a narrow spectrum at the appropriate wavelength and a consistent spectral intensity over a large surface area (44). This precaution minimizes the photo-damage of proteins that occurs at shorter wavelengths of UV light, and it promotes more consistent cross-linking efficiencies when processing multiple samples within the reaction chamber. We then utilized the secY434(Am) mutant as a photocross-linking calibration standard, as this position within SecY has been shown previously to promote efficient photocross-linking with SecA (36). The secY434(Am) mutant was subjected to a time course of UV irradiation and analyzed by Western blotting utilizing SecA antisera. Similar to the previous study, we detected a prominent UV-dependent band of an appropriate size for the SecA·SecY complex (supplemental Fig. S1). Because the intensity of this band was already maximal after 15 min of UV exposure, we utilized this dosage for the remainder of our study.
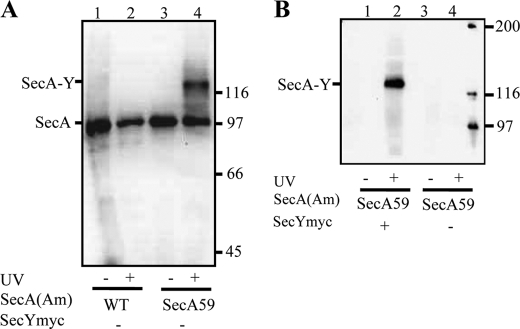
Reliable screening of our secA amber mutant collection by photocross-linking required us to not only use our three-plasmid expression system for specific detection of the relevant SecA·SecY cross-linked complex, but also we found it necessary to utilize subcellular fractionation of UV-irradiated cells to specifically enrich for integral membrane proteins. The addition of this step to our procedure enhanced its sensitivity for detection of less efficient cross-linking for certain of our mutants (shown below). Furthermore, the fractionation procedure eliminates cytoplasmic and peripheral membrane-associated SecA complexes, which could complicate the analysis. Thus, we were able to avoid resorting to immunoprecipitation or affinity purification procedures, as these latter methods can complicate comparisons due to a high degree of variability in sample recovery. Results from our optimized system are shown in Fig. 2 for the secA59(Am) mutant. A UV-dependent SecA59-SecY band was visualized with both SecA and c-Myc antibodies (Fig. 2, panel A, lanes 3 and 4, and panel B, lanes 1 and 2, respectively), and no such species was detected if the secA gene lacked an amber codon (Fig. 2A, lanes 1 and 2) or if the strain lacked the c-Myc-tagged copy of secY (Fig. 2B, lanes 3 and 4).
FIGURE 2.
Specificity of SecA·SecY photocross-linking. The indicated secA and secY plasmid-bearing strain was grown and subjected to photocross-linking (+) or not (−), and cells were analyzed as described in “Experimental Procedures.” Western blotting utilized either SecA antisera (A) or c-Myc antibody (B). The positions of SecA, the SecA·SecY cross-linked complex, and molecular weight markers are given. Only the upper portion of the 20-cm-long gel utilized for better separation of high molecular weight proteins was transferred by electroblotting.
Cross-linking results for our 26 secA amber mutants are summarized in Table 1, and the data are given in supplemental Fig. S2. 20 of 26 SecA residues tested were strongly or weakly positive for cross-linking to SecY protein, and these residues are located in 5 of 6 SecA domains. Two distinct electrophoretic mobilities of the SecA·SecY complexes were noted presumably based on the radius of gyration of the relevant cross-linked complex (supplemental Fig. S2D for examples of the faster and slower migrating SecA·SecY complexes). In addition, certain secA amber mutants (e.g. 530, 661, 833, and 858) generated two distinct SecA·SecY complexes, presumably because these residues could interact with more than one region of SecY during the translocation process. Mori and Ito (36) previously made a similar observation where they found that SecA cross-linked to residues within the C2 or C5 domains of SecY migrated more slowly than comparable SecA·SecY complexes cross-linked at residues within C4 or C6. Because their results were also obtained by in vitro photocross-linking with purified components, it is unlikely that such results are due to differential proteolysis.
TABLE 1.
In vivo photocross-linking of secA(Am) mutants to SecY
| E. coli secA(Am) residuea | T. maritima residueb | SecA domainc | X-linking resultd | Mobility of SecA·SecY complexe |
|---|---|---|---|---|
| 59 | 52 | NBD-I | + | F |
| 104 | 97 | − | NA | |
| 226 | 269 | PPXD | W | S |
| 233 | 276 | + | S | |
| 256 | 288 | W | S | |
| 265 | 297 | − | NA | |
| 300 | 325 | + | S | |
| 334 | 359 | − | NA | |
| 340 | 365 | + | F | |
| 344 | 369 | − | NA | |
| 350 | 375 | + | F | |
| 423 | 448 | NBD-II | W | F |
| 427 | 452 | + | F | |
| 447 | 472 | + | F | |
| 458 | 483 | + | F | |
| 518 | NA | W | F | |
| 530 | NA | + | F, S | |
| 597 | 596 | − | NA | |
| 600 | 599 | + | F | |
| 640 | 636 | HSD | + | S |
| 656 | 652 | + | F | |
| 661 | 657 | + | F, S | |
| 753 | 749 | HWD | − | NA |
| 833 | NA | CTD | + | F, S |
| 858 | NA | + | F, S | |
| 896 | NA | + | S |
a The indicated secA amber mutant was grown and subjected to photocross-linking analysis as described under “Experimental Procedures.”
b Indicates the homologous residue of E. coli SecA for T. maritima SecA.
c Indicates the SecA domain location of each Bpa-labeled SecA residue.
d + or − or W indicates that a positive or negative or weakly positive photocross-linkzng result was obtained for the indicated secA(Am) mutant based on the intensity of any SecA-SecY species observed. Representative data from a number of repetitions are shown in supplemental Fig. S2.
e F or S indicates faster or slower electrophoretic mobility of the indicated SecA·SecY complex; Rf values of 0.57 ± 0.03 and 0.36 ± 0.02, respectively, were obtained in this case relative to phosphorylase b, which was used as an internal standard. NA indicates “not applicable.”
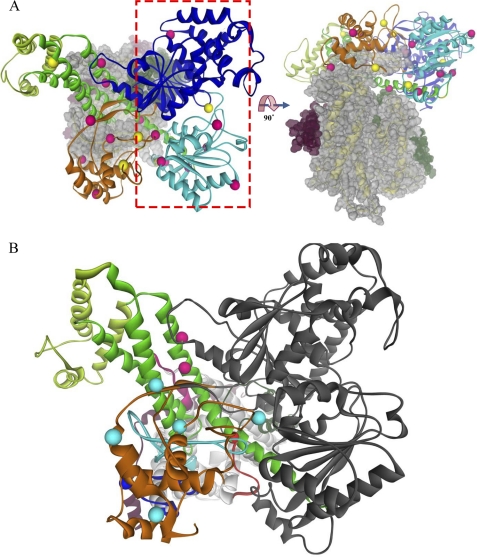
We mapped the 26 SecA residues examined in our study onto the recently published T. maritima SecA·SecYEG co-crystal structure (Fig. 3A) (38). In agreement with the earlier photocross-linking study of Mori and Ito (38), we found that many of the positive SecA residues within the PPXD and HSD domains were proximal to potential acceptors within SecY domains C2, C4, C5, and C6, particularly taking into account the distance between the Cα carbon and the cross-linkable carbonyl of pBPA of SecA (6.7 Å) along with potential side chain distances to the Cα carbon of the SecY acceptor residue (Fig. 3B). Combined, these distances could exceed 10 Å for longer amino acid side chains of SecY. Furthermore, we expected that certain SecA residues would not be proximal to potential SecY acceptor residues, as unlike protein crystallography, our approach is capable of capturing different dynamic SecA·SecYEG states throughout the protein translocation cycle.
FIGURE 3.
Location of SecY-reactive SecA residues on the T. maritima SecA·SecYEG crystal structure. SecA domains are depicted as colored ribbons as follows: dark blue, NBD-I; cyan, NBD-II; orange, PPXD; green, HSD; light green, HWD. SecYEG is depicted in space filling (A) or ribbons (B) as follows; SecY is in gray or yellow, SecE is in magenta, and SecG is in dark green. A, the SecA·SecYEG complex is viewed from the cytosol (left) or the side (right). SecA residues that were positive or negative for cross-linking to SecY are shown as pink or yellow balls, respectively. The red rectangle outlines the NBD-I-NBD-II half of SecA for which logical SecY acceptors are missing. B, shown is the proximity of SecY-reactive SecA residues within the PPXD-HSD-HWD half of SecA to SecY cytosolic domains. The cytosolic domains of SecY are colored as follows; C2 is in dark red, C4 is in cyan, C5 is in dark blue, and C6 is in pink. SecY-reactive SecA residues are colored according to their most proximal SecY cytosolic domain, which was determined utilizing PyMOL with the appropriate script.
We found that the positive and negative residues within the NBD-I and NBD-II domains of SecA generally showed no proximity to SecYEG (Fig. 3A, see the red rectangle or the side view), the exceptions being SecA residues 597 and 600 that were proximal to the SecY C2 domain. All other residues within this region of SecA were greater than 20 Å from potential SecY acceptor residues. This result is consistent with the observation that a second copy of SecYEG was lost from the T. maritima SecA·SecYEG complex during its purification before crystallization as well as previous cross-linking studies that found that the two halves of SecA (i.e. NBD-I-NBD-II and PPXD-HSD-HWD-CTL) each bind a distinct SecYEG protomer (36, 38, 45). Thus, taken as a whole, our data not only support the in vivo relevance of the T. maritima SecY-SecYEG crystal structure for the E. coli Sec system, but also they further extend such studies by defining the second SecY-interacting region of SecA that is missing from the T. maritima SecA·SecYEG structure.
To explicitly test the hypothesis that two SecYEG protomers interact with one molecule of SecA in vivo, we created the double mutant strain, secA59(Am)/896(Am), that contained amber mutations in the two halves of SecA (NBD-I and CTD, respectively), and we tested its ability to form a double cross-link between the two SecYEG protomers and SecA. Although the secA59(Am) and secA896(Am) single mutants each showed a single UV-dependent band with a mobility appropriate for a SecA1·SecY1 complex, the secA59(Am) 896(Am) double mutant showed two UV-dependent bands, with mobilities consistent with SecA1·SecY1 and SecA1·SecY2 complexes (Fig. 4A). Both bands were detected by probing with either c-Myc antibody or SecA antisera, confirming their identity as authentic SecA·SecY cross-linked species (data not shown). However, we cannot rule out the possibility that formation of the SecA1·SecY1 complex induces a conformational change that promotes cross-linking at one of these SecA residues to a third protein species, although we do not favor this more complicated hypothesis. This result represents the first in vivo support for a SecA1·SecY2 complex, which has been observed previously under in vitro conditions utilizing tandem-linked SecY molecules that have the potential to artificially induce this complex (45, 46).
FIGURE 4.
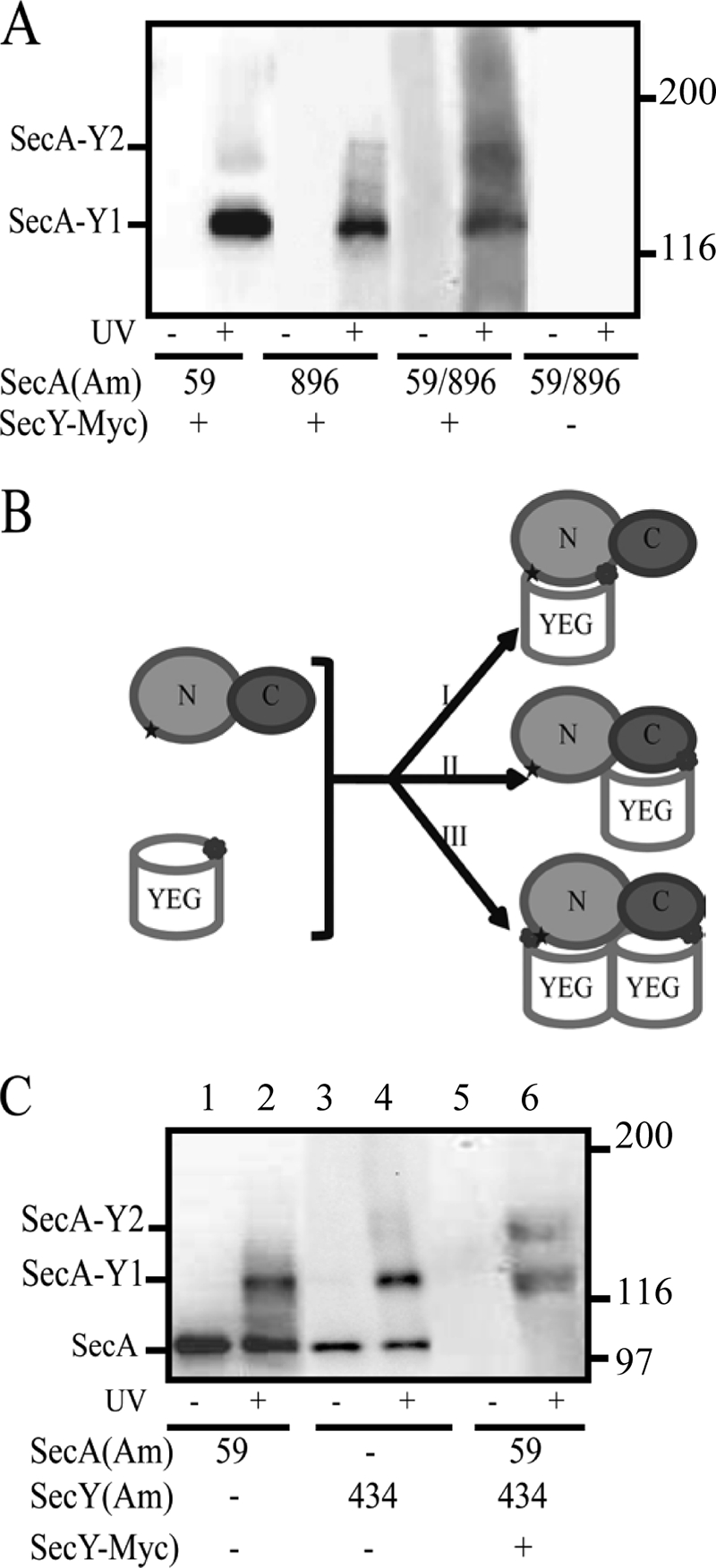
Photocross-linking analysis of secA or secA secY double amber mutant strains. A, the indicated single or double secA(Am) mutant strain was grown and subjected to photocross-linking (+) or not (−), and cells were analyzed by Western blotting utilizing c-Myc antibody as described under “Experimental Procedures.” SecA1-SecY1 and SecA1-SecY2 complexes are indicated. B, shown is a scheme depicting the generation of either (I and II) SecA1-SecY1 or (III) SecA1-SecY2 complexes for the secA59(Am) and secY434(Am) or secA59(Am) secY434(Am) double mutant strains, respectively. N and C designate the two halves of SecA protein comprised of NBD-I-NBD-II or PPXD-HSD-HWD-CTL, respectively, whereas YEG designates each SecYEG protomer. C, the indicated secA(Am) or secY(Am) single or double mutant strain was grown and subjected to photocross-linking (+) or not (−), and cells were analyzed by Western blotting utilizing either SecA antisera (lanes 1–4) or c-Myc antibody (lanes 5–6) as described under “Experimental Procedures.”
Based on the effect of sodium azide and secY mutations on SecA·SecY photocross-linking efficiency of wild type SecA or a SecA truncate, it was proposed that the SecY C4 and C5 domains are involved in more static binding of the amino-terminal two-thirds of SecA, whereas the SecY C6 domain is involved in a more dynamic interaction with the actively translocating carboxyl-terminal third of SecA (36). To test this idea, we constructed the double mutant strain, secA59(Am) secY434(Am), where SecA residue 59 should interact with the NBD-I-NBD-II-bound SecYEG copy, whereas a second copy of SecYEG with residue 434 within C6 should interact with the PPXD-HSD-HWD-CTL portion of SecA (Fig. 4B). Indeed, interaction of SecY residue 434 with the HSD domain of SecA is apparent in the T. maritima SecY-SecYEG structure (38). Although the secA59(Am) and secY434(Am) single mutants each showed a single UV-dependent band with a mobility appropriate for a SecA1·SecY1 complex, the secA59(Am) secY434(Am) double mutant showed two UV-dependent bands with mobilities consistent with SecA1·SecY1 and SecA1·SecY2 complexes (Fig. 4C). This result further supports the proposal that the two halves of SecA interact with distinct SecYEG partners.
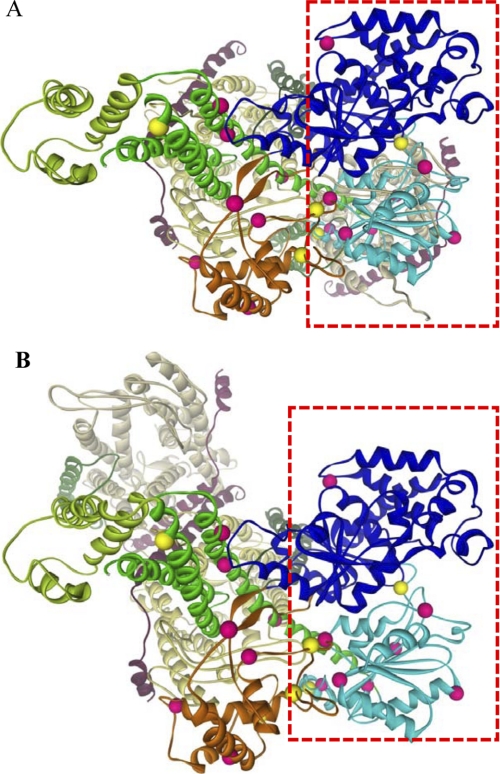
The orientation of the two SecYEG protomers within the dimer has remained a controversial matter with both front-to-front and back-to-back orientations proposed (for review, see Ref. 47). We mapped the 20 SecA residues that were positive for SecY photocross-linking on to front-to-front and back-to-back models of the SecA·SecYEG complex that were made in silico (Fig. 5). We found that our dataset was most consistent with the former model, as nearly all of the positive residues in both SecA halves were proximal to potential SecY acceptor residues in this case. The minority of SecA residues that were distal to potential SecY acceptor sites presumably reflect dynamic regions of SecA whose SecY interactions were not captured by this static front-to-front model. By contrast, potential SecY acceptor residues for the NBD-I-NBD-II half of SecA were not apparent in the back-to-back model. Thus, our data clearly favor the front-to-front SecYEG protomer orientation as a major SecA-bound species in vivo.
FIGURE 5.
Comparison of SecA·SecY photocross-linking results with front-to-front and back-to-back SecA·SecYEG models. Front-to-front (A) and back-to-back (B) models of SecA·SecYEG were generated from the T. maritima SecA·SecYEG crystal structure (38) as described under “Experimental Procedures.” SecA and SecYEG are depicted in colored ribbons as described in the Fig. 3 legend except that the two SecY molecules are shown in light yellow or light gray to better highlight the different orientations. SecA residues that were positive or negative for cross-linking to SecY are shown as pink or yellow balls, respectively. The red rectangle outlines the NBD-I-NBD-II half of SecA, whose SecY-reactive resides are better accommodated by the front-to-front orientation of the SecYEG homodimer.
SecA·SecG Interaction Studies
Although a number of genetic studies have strongly suggested a direct interaction between SecA and SecG proteins, biochemical proof of this proposal is generally lacking (13, 48–50). One group was able to cross-link these two proteins in vitro utilizing a relatively nonspecific hydrophobic cross-linking agent (14), but the relevance of this observation for the normal function of these proteins remains unclear. Given the availability of the T. maritima SecY-SecYEG crystal structure along with the success of our in vivo photocross-linking approach, we decided to also study SecA·SecG interaction.
We utilized the DIMPLOT program to identify potential SecA·SecG interacting residues on the T. maritima SecY-SecYEG structure (supplemental Fig. S3). This analysis allowed us to focus on a limited number of residues that were present at the SecA·SecG interface (Fig. 6A). All five SecG residues chosen are located in the central cytosolic loop of SecG protein according to previous topology studies (12), whereas the five SecA residues chosen are located within either the NBD-I or HSD domains.
FIGURE 6.
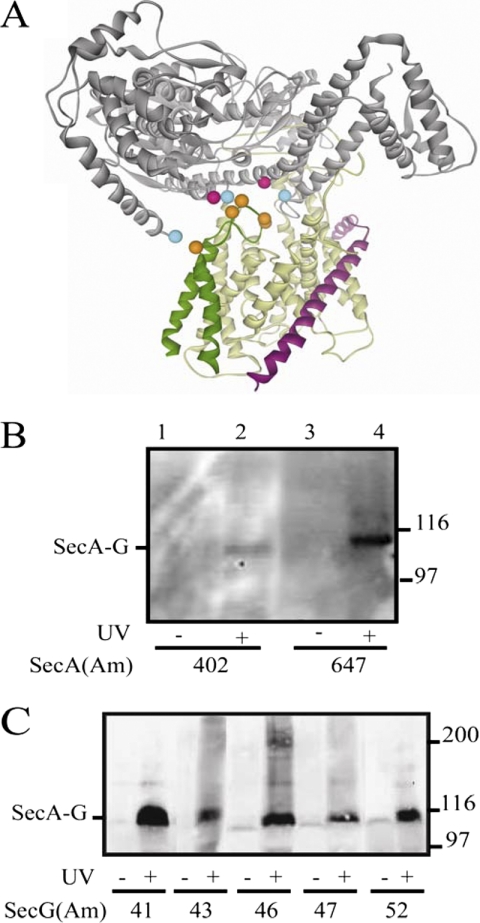
Photocross-linking analysis of secA and secG amber mutant strains. A, SecA is in gray, SecY is in yellow, SecE is in magenta, and SecG is in dark green. The location on the T. maritima SecA·SecYEG crystal structure (38) of residues tested for photocross-linking is depicted as colored balls; SecA residues 402 and 647 that gave positive results are colored in pink, SecA residues 2, 403, and 800 that gave negative results are colored in cyan, and SecG residues 41, 43, 46, 47, and 52 that all gave positive results are colored in orange. B and C, the indicated secA(Am) (B) or secG(Am) (C) mutant strain was grown and subjected to photocross-linking (+) or not (−), and cells were analyzed by Western blotting utilizing c-Myc antibody as described under “Experimental Procedures.” The positions of the SecA·SecG cross-linked complex and molecular weight markers are given.
To readily detect the SecA·SecG photocross-linked complex, we constructed plasmid pCDFT7secYEGmyc that produced SecG protein with a carboxyl-terminal c-Myc tag (see “Experimental Procedures”). The extreme carboxyl-terminal region of SecG has been previously shown to be nonessential for normal function (12). In vivo photocross-linking experiments were performed similarly to above using our three-plasmid system. We found that only two of the five SecA residues tested, 402 and 647, showed UV-dependent SecA·SecG complexes, although the former band was quite weak (Fig. 6B). SecA residues 2, 403, and 800 gave negative results. By contrast, all five SecG residues tested were positive, although different cross-linking efficiencies were also observed (Fig. 6C). Furthermore all SecA·SecG complexes detected were also positive when immunoblotted with SecA antisera (data not shown). As further controls, we noted that no SecA·SecG complex was observed in the secG52(Am) mutant lacking a c-Myc tag or in the secG84(Am) mutant (data not shown). SecG Trp-84 is located within its carboxyl-terminal periplasmic domain, and therefore, it would be expected to be inaccessible to SecA (12). These results provide the first demonstration of SecA·SecG interaction in vivo, and similar to our SecA·SecY study above, they are generally supportive of the T. maritima SecA·SecYEG structure as a good working model for E. coli SecA·SecG interaction despite the distant phylogenetic separation of these two protein complexes.
DISCUSSION
Given limitations inherent in genetic and biochemical approaches to studying SecA·SecYEG interaction, in the present study we have utilized an alternative methodology to address this problem, namely site-directed in vivo photocross-linking. We initially embarked on this study in the absence of a high resolution structure of the SecA·SecYEG complex that was published during the course of our work (38). Accordingly, we relied on our prior SecA membrane topology map to locate SecA residues that should be proximal to the SecYEG channel complex (33). We initially focused on the SecA·SecY interaction, as SecY constitutes the bulk of the protein-conducting channel and contains a number of cytosolic domains that have been suggested to bind SecA (2, 21). Indeed, we found that 20 of 26 SecA residues tested could be photocross-linked to SecY (Table 1). The observation that a minority of SecA residues tested were negative in our assay is not surprising as the geometry of the benzophenone group of pBPA relative to appropriate acceptor groups on neighboring SecY residues may be inappropriate for photocross-linking to occur in such cases, or alternatively, it may do so at an efficiency below our detection limit. By contrast, the sulfhydryl-labeling reagent that we utilized previously in our SecA membrane topology study, 3-(N-maleimido-propinyl)biocytin, is not subject to these limitations as it is freely diffusible and reacts with a higher yield.
We note the excellent agreement between on data (Table 1) and that of Cooper et al. (51), who utilized site-directed spin labeling of SecA to locate sites of SecYEG interaction based on the constraints in the mobility of the nitroxide probe upon SecA·SecYEG complex formation. SecYEG-interacting SecA residues identified in their study include 52, 220/221, 339/344, 350, 600/601/604/605/608, 641, and 661, which are identical or proximal to SecY-reactive SecA residues identified by us, namely 59, 226, 340, 350, 600, 640, and 661, respectively. In addition, we note that SecA residue 645, which was also identified as SecYEG-constrained, is proximal to our SecG-reactive SecA residue 647.
When our photocross-linking data for the PPXD-HSD-HWD half of SecA was mapped onto the T. maritima SecA·SecYEG structure we found that many of the positive SecA residues within this region were proximal to potential acceptors within SecY domains C2, C4, C5, and C6, particularly when the lengths of the potential reactive SecA and SecY amino acid side chains was taken into account (Fig. 3). The SecA residues that were found to be more distal to SecYEG, such as 226, 233, and 256, within PPXD, presumably represent more dynamic regions of SecA that undergo substantial movement during the various steps of the protein translocation cycle. In that regard a recent T. maritima SecA x-ray structure appeared in which the PPXD domain rotated away from the DEAD motor to open the presumed preprotein binding clamp of SecA for substrate loading (52), although this structure appears to represent an earlier translocation state than the T. maritima SecA·SecYEG structure that we utilized to model our data. Additional x-ray structures will be needed to more clearly resolve the more dynamic regions of SecA and SecYEG. That there is a good overall agreement between these two very different approaches for such phylogenetically distant SecA·SecYEG complexes validates the T. maritima SecA·SecYEG structure as a good working model for E. coli SecA·SecYEG interaction. Furthermore, it suggests that this particular state of the protein complex is either relatively abundant in vivo or that much of this portion of the SecA·SecY interface remains relatively static during the protein translocation cycle.
Our data indicates that the NBD-I-NBD-II half of SecA also interacts with SecYEG given a similar density of SecY-reactive amino acid residues within this region of the protein (Fig. 3). This result is consistent with the proposed loss of one of the two SecYEG protomers from the T. maritima SecA·SecYEG complex during its purification along with earlier reports which indicated that the two halves of SecA bind distinct copies of SecYEG (36, 38, 45). Given the limitations inherent in these earlier studies (e.g. the use of a tandem SecY molecule that might artificially generate the requisite SecA1:SecYEG2 complex), we decided to directly test this proposal through our double cross-linking experiments (Fig. 4). Indeed, based on appropriate gel mobility shifts, we were able to capture SecA·SecYEG complexes consistent with a 1:2 stoichiometry, thus supporting the presence of such complexes in vivo. Given the limitations inherent in our in vivo approach, however, we are currently unable to define the proportion of SecA·SecYEG complexes in vivo that have one or two SecYEG protomers or to follow any dynamic sequence of events in relation to the protein translocation cycle. Based on earlier in vivo photocross-linking results utilizing the SecA ATPase inhibitor sodium azide, it has been suggested that the C4 and C5 domains of SecY are involved in a more static, receptor-like binding of the NBD-I-NBD-II half of SecA, whereas the SecY C6 domain is involved in a more dynamic interaction with the PPXD-HSD-HWD half of SecA (36). This latter interaction appears to constitute, at least in part, the actively translocating portion of the SecA·SecYEG complex based on the double capture of a translocating preprotein to the two-helix finger HSD region of SecA as well as the channel region of SecYEG (53).
The orientation of the two SecYEG protomers within the homodimer has remained a controversial matter (for review, see Ref. 47). Although a cryoelectron microscopic study of SecYEG bound to a translating ribosome favored a front-to-front orientation for the two SecYEG protomers, a back-to-back orientation has been suggested based on structure-fitting in two-dimensional crystals as well as the ability of neighboring SecE molecules to be cross-linked to one another via helix 3, which would only approach one another in the back-to-back orientation (42, 54, 55). When the 21 SecY-reactive SecA residues were mapped onto front-to-front and back-to-back models of the SecA·SecYEG complex that we made in silico, we found that our dataset was most consistent with the former orientation (Fig. 5). Because our methodology presumably captures the most abundant SecA·SecYEG complexes within the cell independent of their translocation status, these results should be interpreted with caution. Clearly additional experiments, particularly those that capture SecA·SecYEG complexes with translocation intermediates, will be needed to clarify this matter further.
Given the importance of SecG in enhancing SecA function as well as the availability of the T. maritima SecA·SecYEG x-ray structure, we also decided to investigate SecA·SecG interaction utilizing a more structurally directed approach (Fig. 6). The five SecA-reactive SecG residues identified in our study reside in a relatively small, contiguous region of the cytosolic loop of SecG that has been shown to be critical for its function, particularly the TLF motif, located at residues 41–43 (37). Our results suggest that at least one important function of this region of SecG is to stabilize the binding of particular regions of SecA. It is tempting to speculate that this initial interaction may help to destabilize regions of SecA that allow for its membrane insertion, a step that SecG is known to facilitate (12, 13). In that regard the two SecG-reactive SecA residues, 402 and 647, are adjacent to Glu-400 and Arg-642, respectively. These latter two residues form an ionic pair that regulates coupling between SecA ATPase and protein translocation cycles (56). Association of SecG with this region of SecA may help to destabilize this ionic pair, thereby releasing an inhibitory interaction between the SecA DEAD motor and its proposed two-helix finger ratchet within the HSD domain of SecA (38, 57). This proposal would be consistent with the previously observed SecG-dependent stimulation of the SecA ATPase cycle and its coupling with the SecA membrane insertion and de-insertion cycle (7, 13). In that regard we have recently noticed that the secG null mutant shows a defect in the formation of a normal level of membrane-integrated SecA protein,3 consistent with this line of thinking.
In conclusion, our site-directed in vivo photocross-linking studies allowed us to construct maps of the SecA·SecY and SecA·SecG interfaces within living cells. Our data support the in vivo relevance in the E. coli system of the T. maritima SecA·SecYEG crystal structure that captured the interaction of the more distal half of SecA with one SecYEG protomer. They also reveal a series of contacts between the more proximal half of SecA containing the DEAD motor with a second copy of SecYEG. That SecA possesses such an extensive surface for binding two SecYEG partners should allow for exquisite control of the various steps needed to promote protein translocation, even for proteins with complex topologies that need SecA to cooperate with the signal recognition particle-ribosome pathway. Given the power of the current site-directed photocross-linking approach, additional studies can now be undertaken to address questions relating SecA·SecYEG structure with its mechanism of action.
Acknowledgments
We thank Aaron Olsen and Jessica Chukwu for construction of secA(Am) and secG(Am) mutations, respectively, as well as for technical assistance in the photocross-linking analysis. We also thank Swastik De and Richard Olson for help with the Pymol analysis and Sarah Auclair, Lorry Grady, Manju Hingorani, and Ishita Mukerji for intellectual input and encouragement during our study.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
S. Das and D. B. Oliver, unpublished observation.
- TM1-TM10
- first through tenth transmembrane domain of SecY
- pBPA
- p-benzoyl-phenylalanine
- C1-C6
- first through sixth cytoplasmic domains of SecY
- CTL
- carboxyl-terminal linker domain of SecA
- HSD
- helical scaffold domain of SecA
- HWD
- helical wing domain of SecA
- NBD-I
- nucleotide binding domain I of SecA
- NBD-II
- nucleotide binding domain II of SecA
- P1-P5
- first through fifth periplasmic domain of SecY
- PPXD
- preprotein cross-linking domain of SecA.
REFERENCES
- 1. Osborne A. R., Rapoport T. A., van den Berg B. (2005) Annu. Rev. Cell Dev. Biol. 21, 529–550 [DOI] [PubMed] [Google Scholar]
- 2. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 3. Mori H., Ito K. (2001) Trends Microbiol. 9, 494–500 [DOI] [PubMed] [Google Scholar]
- 4. Veenendaal A. K., van der Does C., Driessen A. J. (2004) Biochim. Biophys. Acta 1694, 81–95 [DOI] [PubMed] [Google Scholar]
- 5. Papanikou E., Karamanou S., Economou A. (2007) Nat. Rev. Microbiol. 5, 839–851 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto G., Yoshihisa T., Ito K. (1997) EMBO J. 16, 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Economou A., Wickner W. (1994) Cell 78, 835–843 [DOI] [PubMed] [Google Scholar]
- 8. Eichler J., Wickner W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5574–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Does C., Manting E. H., Kaufmann A., Lutz M., Driessen A. J. M. (1998) Biochemistry 37, 201–210 [DOI] [PubMed] [Google Scholar]
- 10. Nishiyama K., Hanada M., Tokuda H. (1994) EMBO J. 13, 3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanada M., Nishiyama K. I., Mizushima S., Tokuda H. (1994) J. Biol. Chem. 269, 23625–23631 [PubMed] [Google Scholar]
- 12. Nishiyama K., Suzuki T., Tokuda H. (1996) Cell 85, 71–81 [DOI] [PubMed] [Google Scholar]
- 13. Matsumoto G., Mori H., Ito K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagamori S., Nishiyama K., Tokuda H. (2002) J. Biochem. 132, 629–634 [DOI] [PubMed] [Google Scholar]
- 15. Sugai R., Takemae K., Tokuda H., Nishiyama K. (2007) J. Biol. Chem. 282, 29540–29548 [DOI] [PubMed] [Google Scholar]
- 16. van der Sluis E. O., van der Vries E., Berrelkamp G., Nouwen N., Driessen A. J. (2006) J. Bacteriol. 188, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel C. N., Smith V. F., Randall L. L. (2006) Protein Sci. 15, 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Breukink E., Nouwen N., van Raalte A., Mizushima S., Tommassen J., de Kruijff B. (1995) J. Biol. Chem. 270, 7902–7907 [DOI] [PubMed] [Google Scholar]
- 19. Gelis I., Bonvin A. M., Keramisanou D., Koukaki M., Gouridis G., Karamanou S., Economou A., Kalodimos C. G. (2007) Cell 131, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Auclair S. M., Moses J. P., Musial-Siwek M., Kendall D. A., Oliver D. B., Mukerji I. (2010) Biochemistry 49, 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akiyama Y., Ito K. (1987) EMBO J. 6, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taura T., Yoshihisa T., Ito K. (1997) Biochimie 79, 517–521 [DOI] [PubMed] [Google Scholar]
- 23. Chiba K., Mori H., Ito K. (2002) J. Bacteriol. 184, 2243–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van der Sluis E. O., Nouwen N., Koch J., de Keyzer J., van der Does C., Tampé R., Driessen A. J. (2006) J. Mol. Biol. 361, 839–849 [DOI] [PubMed] [Google Scholar]
- 25. van der Wolk J. P., Fekkes P., Boorsma A., Huie J. L., Silhavy T. J., Driessen A. J. (1998) EMBO J. 17, 3631–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manting E. H., Kaufmann A., van der Does C., Driessen A. J. (1999) J. Biol. Chem. 274, 23868–23874 [DOI] [PubMed] [Google Scholar]
- 27. Snyders S., Ramamurthy V., Oliver D. (1997) J. Biol. Chem. 272, 11302–11306 [DOI] [PubMed] [Google Scholar]
- 28. Robson A., Booth A. E., Gold V. A., Clarke A. R., Collinson I. (2007) J. Mol. Biol. 374, 965–976 [DOI] [PubMed] [Google Scholar]
- 29. Karamanou S., Bariami V., Papanikou E., Kalodimos C. G., Economou A. (2008) Mol. Microbiol. 70, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eichler J., Wickner W. (1998) J. Bacteriol. 180, 5776–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim Y. J., Rajapandi T., Oliver D. (1994) Cell 78, 845–853 [DOI] [PubMed] [Google Scholar]
- 32. van der Does C., den Blaauwen T., de Wit J. G., Manting E. H., Groot N. A., Fekkes P., Driessen A. J. (1996) Mol. Microbiol. 22, 619–629 [DOI] [PubMed] [Google Scholar]
- 33. Jilaveanu L. B., Oliver D. B. (2007) J. Biol. Chem. 282, 4661–4668 [DOI] [PubMed] [Google Scholar]
- 34. Wang L., Xie J., Schultz P. G. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 225–249 [DOI] [PubMed] [Google Scholar]
- 35. Kauer J. C., Erickson-Viitanen S., Wolfe H. R., Jr., DeGrado W. F. (1986) J. Biol. Chem. 261, 10695–10700 [PubMed] [Google Scholar]
- 36. Mori H., Ito K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bost S., Belin D. (1995) EMBO J. 14, 4412–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zimmer J., Nam Y., Rapoport T. A. (2008) Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryu Y., Schultz P. G. (2006) Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 40. van der Does C., de Keyzer J., van der Laan M., Driessen A. J. (2003) Methods Enzymol. 372, 86–98 [DOI] [PubMed] [Google Scholar]
- 41. Jilaveanu L. B., Zito C. R., Oliver D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7511–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaufmann A., Manting E. H., Veenendaal A. K., Driessen A. J., van der Does C. (1999) Biochemistry 38, 9115–9125 [DOI] [PubMed] [Google Scholar]
- 43. Douville K., Price A., Eichler J., Economou A., Wickner W. (1995) J. Biol. Chem. 270, 20106–20111 [DOI] [PubMed] [Google Scholar]
- 44. Farrell I. S., Toroney R., Hazen J. L., Mehl R. A., Chin J. W. (2005) Nature Methods 2, 377–384 [DOI] [PubMed] [Google Scholar]
- 45. Osborne A. R., Rapoport T. A. (2007) Cell 129, 97–110 [DOI] [PubMed] [Google Scholar]
- 46. Duong F. (2003) EMBO J. 22, 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rusch S. L., Kendall D. A. (2007) Biochim. Biophys. Acta 1768, 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramamurthy V., Dapíc V., Oliver D. (1998) J. Bacteriol. 180, 6419–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suzuki H., Nishiyama K., Tokuda H. (1998) Mol. Microbiol. 29, 331–341 [DOI] [PubMed] [Google Scholar]
- 50. Mori H., Sugiyama H., Yamanaka M., Sato K., Tagaya M., Mizushima S. (1998) J. Biochem. 124, 122–129 [DOI] [PubMed] [Google Scholar]
- 51. Cooper D. B., Smith V. F., Crane J. M., Roth H. C., Lilly A. A., Randall L. L. (2008) J. Mol. Biol. 382, 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zimmer J., Rapoport T. A. (2009) J. Mol. Biol. 394, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erlandson K. J., Miller S. B., Nam Y., Osborne A. R., Zimmer J., Rapoport T. A. (2008) Nature 455, 984–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. (2005) Nature 438, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Breyton C., Haase W., Rapoport T. A., Kühlbrandt W., Collinson I. (2002) Nature 418, 662–665 [DOI] [PubMed] [Google Scholar]
- 56. Mori H., Ito K. (2006) J. Biol. Chem. 281, 36249–36256 [DOI] [PubMed] [Google Scholar]
- 57. Vrontou E., Karamanou S., Baud C., Sianidis G., Economou A. (2004) J. Biol. Chem. 279, 22490–22497 [DOI] [PubMed] [Google Scholar]