Abstract
Phospholipase C (PLC) enzymes are an important family of regulatory proteins involved in numerous cellular functions, primarily through hydrolysis of the polar head group from inositol-containing membrane phospholipids. U73122 (1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione), one of only a few small molecules reported to inhibit the activity of these enzymes, has been broadly applied as a pharmacological tool to implicate PLCs in diverse experimental phenotypes. The purpose of this study was to develop a better understanding of molecular interactions between U73122 and PLCs. Hence, the effects of U73122 on human PLCβ3 (hPLCβ3) were evaluated in a cell-free micellar system. Surprisingly, U73122 increased the activity of hPLCβ3 in a concentration- and time-dependent manner; up to an 8-fold increase in enzyme activity was observed with an EC50 = 13.6 ± 5 μm. Activation of hPLCβ3 by U73122 required covalent modification of cysteines as evidenced by the observation that enzyme activation was attenuated by thiol-containing nucleophiles, l-cysteine and glutathione. Mass spectrometric analysis confirmed covalent reaction with U73122 at eight cysteines, although maximum activation was achieved without complete alkylation; the modified residues were identified by LC/MS/MS peptide sequencing. Interestingly, U73122 (10 μm) also activated hPLCγ1 (>10-fold) and hPLCβ2 (∼2-fold); PLCδ1 was neither activated nor inhibited. Therefore, in contrast to its reported inhibitory potential, U73122 failed to inhibit several purified PLCs. Most of these PLCs were directly activated by U73122, and a simple mechanism for the activation is proposed. These results strongly suggest a need to re-evaluate the use of U73122 as a general inhibitor of PLC isozymes.
Keywords: Enzyme Inhibitors, Phosphatidylinositol, Phospholipase C, Phospholipid Metabolism, Protein Drug Interactions, PLC Activation, Phospholipase C Activation, U73122, Lipase Activation, Sulfhydryl
Introduction
Phospholipase C (PLC)3 enzymes compose a family of proteins involved in the cellular turnover of inositol-containing phospholipids. These enzymes cleave the polar headgroup from membrane lipids such as phosphatidylinositol 4,5-bisphosphate (PIP2) to generate inositol 1,4,5-triphosphate and diacylglycerol, intracellular second messengers that mobilize intracellular calcium and activate protein kinase C enzymes, respectively. To date, 13 human PLC isozymes have been identified comprising six distinct and differentially regulated families (β1–4, γ1–2, δ1,3–4, ϵ, η1–2, and ζ) (1–3). They vary in molecular size from the ∼70-kDa PLCζ to the much larger ∼250-kDa PLCϵ, but they share a common core domain structure including the EF domain, the C2 domain, and the highly conserved catalytic core made up of two regions, commonly referred to as X and Y domains (4, 5). Given the highly conserved active site residues within all PLCs, it is not surprising that they all act predominantly on just two substrates, PIP and PIP2, with some catalytic activity toward PI (6). Therefore, PLC isozymes likely achieve selectivity in their physiological functions through either isozyme-specific receptor activation or through unique downstream signaling mechanisms. PLC-mediated signaling has been implicated in a number of critical cellular functions such as motility (7), migration (8), growth and differentiation (9, 10), as well as in the assembly and regulation of cell-cell junctions (11–13).
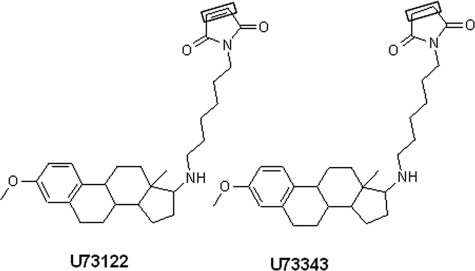
U73122 (1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione) (Fig. 1) is an aminosteroid first reported as an inhibitor of PLC-dependent processes in 1989 (14). Many studies have since reported that several isozymes of PLC are inhibited by U73122, but not by U73343, a close structural analog of U73122 containing N-alkylsuccinimide moiety in place of N-alkylmaleimide (15–19). These reports have established U73122 as the prototypical inhibitor of PLC enzymes. We have previously substantiated these reports and observed that in both Madin-Darby canine kidney and Caco-2 cells U73122, but not U73343, inhibits ATP-stimulated PLCβ activity in a concentration-dependent manner (19). However, several recent reports suggest that U73122 may not be selective for PLCs, as effects on numerous cellular proteins have been reported, including telomerase (20), 5-lipoxygenase (21), histamine H1 receptor (22), calcium channels (23), potassium channels (24), sarco/endoplasmic reticulum Ca2+-ATPase (25), phospholipase D (26), and PI-dependent as well as PI-independent exocytic processes (27), implying complex effects of this compound in whole cell systems.
FIGURE 1.
Structures of U73122 and U73343. The box highlights the only structural difference between the two compounds (i.e. maleimide in U73122 versus succinimide in U73343).
Consistent with the studies identifying alternative cellular targets for U73122, recent publications have questioned the use of this compound as a truly selective modulator of PLCs, implicating the electrophilic maleimide moiety for the lack of selectivity (Fig. 1) (28, 29). Maleimides are inherently reactive and readily interact with cellular thiols and amines; therefore, covalent modification of nucleophiles on other proteins provides a likely mechanism for the observed off-target effects. However, the fact remains that there are numerous reports of U73122 as an inhibitor of PLCs (14–17), and U73122 is widely used as a probe molecule to implicate the involvement of PLCs in signaling pathways and phenotypic cellular response (30–35). It is disconcerting that little work has been done to understand the interaction of this molecule with PLCs at the molecular level, considering that modulation of cellular functions by U73122 is often considered unequivocal evidence for involvement of PLCs in those cellular functions. In this study, interaction between U73122 and specific PLC isozymes was evaluated in a simple, cell-free, mixed micellar system. Surprisingly, these studies revealed that U73122 activates, not inhibits, the activity of several PLC isozymes, uncovering a novel and potentially general activation mechanism for lipases such as PLCs.
EXPERIMENTAL PROCEDURES
Materials
Dodecyl maltoside (DDM) was purchased from Fluka. [3H]PIP2 ([2-3H]myoinositol) (20 Ci/mmol) was purchased from American Radiolabeled Chemicals. PIP2 was purchased from Avanti Polar Lipids. U73122, U73343, N-ethylmaleimide, fatty acid-free bovine serum albumin (BSA), and all other reagents were purchased from Sigma unless otherwise indicated. Purified hPLCβ3 was generously provided by the laboratory of Dr. T. K. Harden (Department of Pharmacology, School of Medicine, University of North Carolina, Chapel Hill). hPLCβ2, hPLCγ1, and hPLCδ1 isozymes have been previously characterized (36, 37). Trypsin, thermolysin, and endoproteinase GluC for digestion of hPLCβ3 were from Promega, Fluka, and Roche Applied Science, respectively.
Mixed Micellar PLC Assay
The activity of hPLCs in cell-free systems was evaluated by an adaptation of previously published methods (38, 39). Briefly, for DDM mixed micellar assay, 3H-labeled (∼30,000 dpm) and unlabeled PIP2 (50 μm) were reconstituted in a 1 mm DDM solution (final assay concentration 0.5 mm) and mixed with assay buffer containing 40 mm HEPES (pH 7.4), 480 mm KCl, 40 mm NaCl, 8 mm EGTA, 4 mm MgCl2, and 7.6 mm CaCl2 in a final volume of 100 μl. Compounds at desired concentrations, and purified hPLCβ3 in 1% fatty acid-free BSA and 10 mm HEPES (pH 7.0), were subsequently added to the mixture. To initiate assays, samples were moved to a 37 °C water bath and incubated for 2–10 min such that less than 15% of total substrate was hydrolyzed. At designated times, reactions were stopped by addition of 750 μl of CHCl3/MeOH/HCl (40:80:1), followed by the addition of 100 μl of water, 250 μl of CHCl3, and 250 μl of 0.1 m HCl. Samples were vortexed and centrifuged at 3000 rpm for 10 min at 4 °C. The amount of [3H]inositol phosphates formed was measured by liquid scintillation counting of the upper phase (500 μl) in a Packard Tri Carb 4000 Series spectrophotometer.
For experiments in a cholate-mixed micellar assay, purified PLC isozymes were incubated with 10 μm U73122 in the presence of 3 μm cholate for 10 min at 32 °C. Subsequently, this mixture was added to [3H]PIP2 (∼30,000 dpm) and unlabeled PIP2 (50 μm) that were dried and resuspended in 0.5% cholate for a final volume of 60 μl. After incubation at 32 °C at time intervals between 0 and 10 min, reactions were stopped by the addition of 200 μl of 10% (v/v) trichloroacetic acid and 100 μl of 10 mg/ml BSA to precipitate uncleaved lipids and protein. After centrifugation of the reaction mixture, soluble [3H]inositol phosphates in the supernatant were quantified using liquid scintillation counting.
Time-dependent Activation of Human PLCβ3
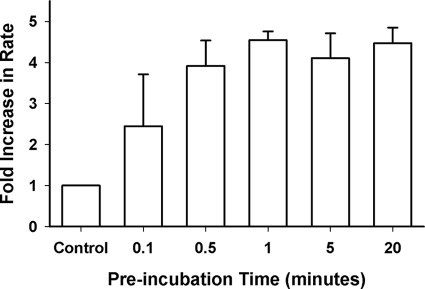
To further investigate the hypothesis that activation of hPLCβ3 by U73122 involves covalent alkylation of the enzyme, the effect of preincubation time on the observed activation was evaluated. hPLCβ3 was preincubated with U73122 in the absence of substrate and detergent. Excess U73122 was then removed with the addition of 1 mm glutathione, and the substrate, resuspended in detergent, was added to initiate the assay. Separate studies established that addition of 1 mm glutathione to 40 μm U73122 results in the immediate disappearance of U73122 as measured via LC/MS. It was further confirmed that the preformed U73122-glutathione conjugate had no direct effect on the activity of hPLCβ3.
To assess if the enzyme activation by U73122 is reversible or irreversible, hPLCβ3 was diluted in 1% fatty acid-free BSA and 10 mm HEPES, and mixed with equal volumes of assay buffer. U73122 or DMSO was added to the mixture, and samples were transferred to Microcon centrifugal filter devices (Ultracel YM-30 membrane, 30,000 NMWL, Millipore) and centrifuged at 4 °C, 14,000 rpm for 24 min. Retained concentrate was washed with 450 μl of cold HEPES/assay buffer mix without fatty acid-free BSA. Samples were centrifuged again, and the resuspended sample was used in mixed micellar PLC assays. Control measurements were performed in the absence of U73122.
Data Analysis
Data are expressed as the mean ± S.D. from three measurements unless indicated otherwise. Where indicated, statistical significance was assessed using a two-sample Student's t tests. Samples were assumed to have an unequal variance; significant differences were assigned at p < 0.05.
EC50 PLCβ3 Estimation
To accurately quantify the potency of U73122 for activating hPLCβ3 in DDM-mixed micelles, concentration-effect profiles were generated for each experiment (n = 4). The relationship between fold increase in rate for hPLCβ3 activity and U73122 concentration was fit to a standard Hill equation by nonlinear least squares regression (WinNonlin, version 4.1) to recover estimates for EC50(PLCβ3). Emax and E0 were fixed based on raw data within each experiment; an estimate was also generated for the sigmoidicity factor (γ).
Mass Spectrometry
Mass spectrometry of intact hPLCβ3 was performed using reversed-phase chromatography (40) coupled to a 6210 LC-MSD-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA). Ionization was achieved using electrospray ionization with a spray voltage of −4.0 kV with heated nitrogen (350 °C) serving as both a nebulizing (25 p.s.i.) and drying (12 liters/min) gas. The fragmentor was set at 280 V, and the instrument was set to detect ions from mass-to-charge 500 to mass-to-charge 2500. The mass-to-charge data were transformed to the mass domain using BioConfirm software from Agilent. For peptide sequencing, solutions of unmodified and modified hPLCβ3 were reduced using DTT, alkylated with iodoacetamide, and incubated in an ammonium bicarbonate buffer with trypsin, thermolysin, or endoproteinase GluC (enzyme:substrate = 1:20) overnight at 37 °C. The tryptic peptides were subsequently analyzed by nano-LC/MS/MS on a Dionex UltiMate nano-LC (Sunnyvale, CA) coupled to an ABI-Sciex (Toronto, Ontario, Canada) Q-Star Pulsar i mass spectrometer using both a trapping cartridge column (0.3 × 5 mm) and a PepMap (0.075 × 150 mm, 3 μm particle size) C18 column (Dionex). Mobile phase A contained 2% acetonitrile, and mobile phase B contained 95% acetonitrile, both with 0.2% formic acid. Following a 6-min load of sample onto the trapping cartridge at a flow rate of 25 μl/min with a mobile phase A and B ratio of 95:5, flow was then switched to back flush off the trapping cartridge and onto the nano-LC column at 0.2 μl/min. The percentage of mobile phase B was changed over 1 h from 5 to 40% with a linear gradient to adequately separate peptides, followed by a second linear gradient from 40 to 99% over the next 7 min. The results were processed using MASCOT (Matrix Sciences, London, UK) protein data base with the mass of U73122 addition created as a variable modification.
Model Generation to Elucidate the Activation Mechanism
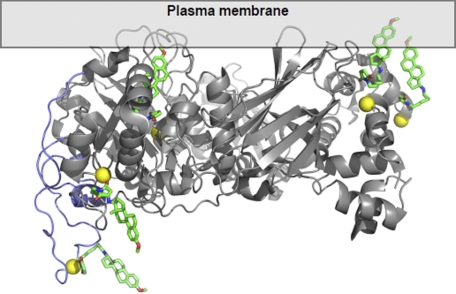
A set of coordinates for hPLCβ2 (Protein Data Bank code 2ZKM) was used to generate a model for illustrating the proposed mechanism of activation by U73122. A model X/Y linker was inserted into the 2ZKM structure to reflect the linker region that is present in hPLCβ3 but not in hPLCβ2. hPLCβ2 was used in lieu of hPLCβ3, because a crystal structure of hPLCβ3 was not available. Only five of the eight possible alkylation sites are shown, because other sites reside within the C-terminal domain that was not included in the protein used to obtain this crystal structure. The model is presented as Fig. 10.
FIGURE 10.
Illustration of the proposed mechanism for activation of PLC by U73122. The highly lipophilic U73122 (green, red, and blue) binds covalently to PLC (gray and purple ribbon structure) via cysteine residues (yellow) increasing the affinity of the protein for cell membranes. One or more U73122 molecules serve as lipid anchors for the modified protein allowing it to dock within the cell membrane, in close proximity to substrate, thus leading to increased catalytic activity. See “Experimental Procedures” for details regarding generation of model.
RESULTS
Activity of Human PLCβ3 in Phospholipid Detergent-Mixed Micelles
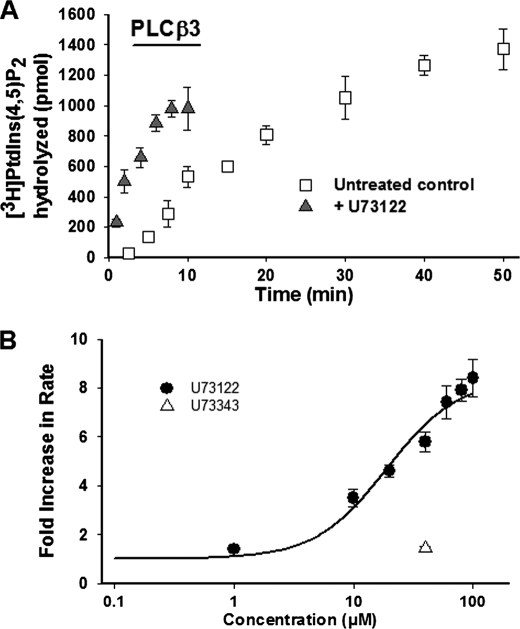
hPLCβ3 activity with respect to time in DDM/PIP2-mixed micelles at a substrate mole fraction of 0.1 is presented in Fig. 2A. Consistent with previous studies (38, 39), two distinct phases of activity were observed as follows: a short lag time and an initial burst of activity that was linear when ≤15% of PIP2 was utilized, followed by a second, slower phase of hydrolysis. Additional studies were performed at fixed calcium and PIP2 concentrations, and were designed such that ≤15% of total substrate was utilized in all cases or when substrate consumption was linear with respect to time.
FIGURE 2.
A, time course of PIP2 hydrolysis by hPLCβ3 in DDM-mixed micelles in the presence and absence of U73122 (40 μm). PIP2, reconstituted in DDM, was combined with the assay buffer and hPLCβ3 in a final volume of 100 μl. Assays were initiated by moving samples to a 37 °C water bath and incubating for the indicated times. Incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Enzyme hydrolysis rates were determined based on initial time points within the linear phase of activity. Data represent mean ± S.D. from triplicate determinations from one representative experiment. B, concentration-dependent activation of hPLCβ3 by U73122 in DDM-mixed micelles. PIP2, reconstituted in DDM, was combined with assay buffer and hPLCβ3 in a final volume of 100 μl. U73122, at indicated concentrations, was added to the assay mixture prior to the addition of the enzyme. Assays were initiated by moving samples to a 37 °C water bath. Incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Data represent mean ± S.D. from triplicate determinations from one representative experiment. EC50(PLCβ3) was determined as described under “Experimental Procedures” and was found to be 19.1 μm in this experiment. The estimated EC50 from these data should be interpreted within the context of the current set of results, as solubility limitations of U73122 in DMSO stock solutions prevented assessment of activation at concentrations greater than 100 μm.
U73122, but Not U73343, Increases Human PLCβ3 Activity in a Concentration-dependent Manner
In DDM mixed micelles, U73122 unexpectedly increased the activity of hPLCβ3. The activation was rapid and eliminated the observed lag time in control experiments (Fig. 2A). The effect of U73122 was concentration-dependent with a maximum increase in hPLCβ3 activity of ∼8-fold over control. Application of a modified Hill equation yielded an estimated EC50 value of 13.6 ± 5 μm (n = 4) for the observed activation (Fig. 2B). The estimated EC50 from these data should be interpreted within the context of the current set of results, as solubility limitations of U73122 in DMSO stock solutions prevented assessment of activation at concentrations greater than 100 μm. U73343, a close structural analog of U73122 containing N-alkylsuccinimide instead of N-alkylmaleimide moiety (Fig. 1), had no effect on activity at 40 μm (Fig. 2B). Because U73343 is incapable of covalently modifying the enzyme due to the absence of a maleimide moiety, this result provides evidence that the observed activation of hPLCβ3 by U73122 occurs via covalent modification of the enzyme.
U73122 Increases the Activity of Other Human PLC Isozymes
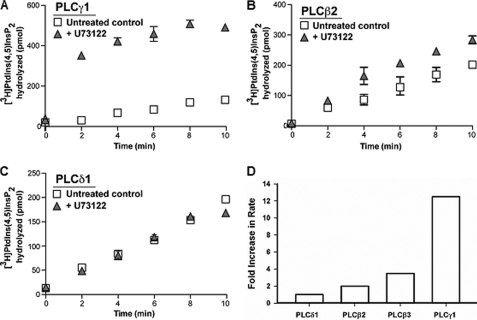
Incubation of other PLC isozymes with 10 μm U73122 for 10 min in cholate-mixed micelles increased the activity of hPLCγ1 and hPLCβ2 but not hPLCδ1 (Fig. 3, A–C). The fold increase in rate was much greater for hPLCγ1 (10–15-fold) than for hPLCβ2 (2-fold) (Fig. 3D). These data demonstrate that the enzyme activation by U73122 is not unique to hPLCβ3 and that it is applicable to other hPLCs. Importantly, U73122 neither activates nor inhibits the activity of hPLCδ1.
FIGURE 3.
Time course of PIP2 hydrolysis by hPLCγ1 (A), hPLCβ2 (B), and hPLCδ1 (C) in cholate-mixed micelles in the presence and absence of U73122 (10 μm). Purified PLC isozymes were incubated with U73122 in the presence of 0.03 mm cholate for 10 min at 32 °C prior to the addition of cholate-solubilized micelles containing 50 μm PIP2. Data represent mean ± S.E. and are representative of two or more separate experiments. D, relative fold increase in rate caused by incubation with 10 μm U73122 for the four PLC isozymes evaluated in mixed micellar systems.
U73122-mediated Activation of Human PLCβ3 Is via Time-dependent and Irreversible Modification of Protein Sulfhydryl Groups
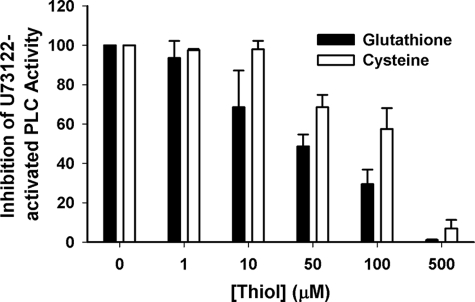
Preincubation of hPLCβ3 with U73122 increased activity compared with control in a time-dependent manner, and maximal activation was achieved within 30 s (Fig. 4). Thiol-containing compounds, glutathione and cysteine, which react readily with electrophiles such as a maleimide, attenuated the U73122-mediated activation of hPLCβ3 in a concentration-dependent manner (Fig. 5). The ability of sulfhydryl reagents to prevent the U73122-mediated activation suggests that U73122 alkylates hPLCβ3 by reacting with nucleophiles, likely protein sulfhydryl groups (i.e. cysteine residues) leading to the activation. To confirm that the activation was irreversible, U73122-modified enzyme was subjected to ultrafiltration to remove excess and reversibly bound U73122, and following centrifugation, the enzyme activity was measured again. Reconstituted hPLCβ3 that had been subject to ultrafiltration maintained increased activity as compared with untreated enzyme. Taken together, these results suggest that activation of hPLCβ3 was caused by direct and irreversible covalent modification of one or more sulfhydryl groups on the enzyme.
FIGURE 4.
Effect of preincubation time (hPLCβ3 with U73122) on the U73122-mediated activation of hPLCβ3 in DDM-mixed micelles. U73122 was preincubated with hPLCβ3 at 40 μm in assay buffer (50 μl total volume) for the indicated times in the absence of PIP2 and DDM. Preincubation was quenched with the addition of 1 mm glutathione. Assays were initiated by the addition of preincubated enzyme to mixed micelles in a final volume of 100 μl, and incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Data represent mean ± S.D. from triplicate determinations from one representative experiment.
FIGURE 5.
Effect of glutathione and cysteine on the U73122-mediated activation of hPLCβ3 in DDM-mixed micelles. PIP2, reconstituted in DDM, was combined with assay buffer, the thiol compound at the indicated concentration, and hPLCβ3 in a final volume of 100 μl. U73122 (40 μm) was added to the assay mixture prior to the addition of the enzyme. Assays were initiated by moving samples to a 37 °C water bath. Incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Zero concentration of the thiol compound represents fully activated hPLCβ3. Data represent mean ± S.D. for triplicate determinations from one representative experiment.
Mass Spectral Evidence for Alkylation of Human PLCβ3 by U73122
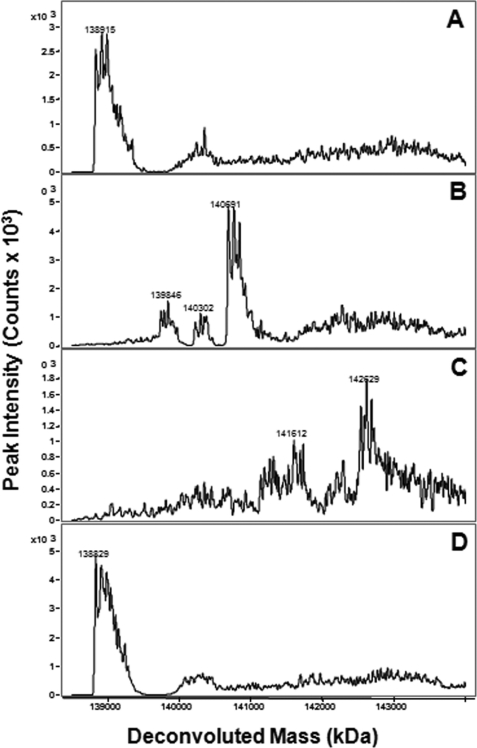
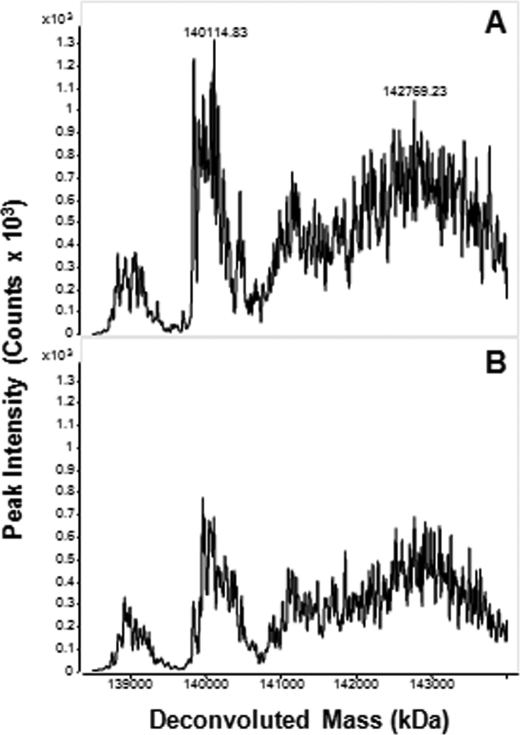
To unequivocally establish that alkylation of hPLCβ3 by U73122 is a requisite step for its activation, mass spectra were obtained of intact hPLCβ3 alone and after incubation with either 40 μm U73122 or 40 μm U73343 for various time intervals. Mass spectra of hPLCβ3 revealed a complex spectral pattern for a protein of ∼138 kDa (Fig. 6A). When incubated with 40 μm U73122, spectra revealed the presence of protein species with discrete increases in mass, consistent with the covalent addition of up to eight molecules of U73122 per molecule of hPLCβ3 (Fig. 6, B and C). Extended incubation times did not lead to more than eight U73122 additions, although shorter incubation times yielded hPLCβ3 species modified with two to seven molecules of U73122. Even at the shortest incubation time (5 s), protein species with only a single addition of U73122 were not detected. As expected, mass spectra of hPLCβ3 incubated with U73343 for extended time periods remained unchanged (Fig. 6D).
FIGURE 6.
Mass spectra of intact hPLCβ3 following incubation with either U73122 or U73343. Both compounds were incubated with hPLCβ3 at 40 μm for various time intervals in assay buffer at 37 °C in the absence of the substrate and detergent. Incubations were terminated with the addition of 1 mm glutathione, and an aliquot was analyzed via LC/MS. A, hPLCβ3 alone; B, hPLCβ3 following incubation with U73122 for 1 min; C, hPLCβ3 following incubation with U73122 for 15 min; D, hPLCβ3 following incubation with U73343 for 15 min. The complexity of each spectra reflects the heterogeneity of the protein likely including phosphorylated and acetylated forms. See “Experimental Procedures” for details.
Interestingly, the reaction of U73122 with hPLCβ3 led to a shift in chromatographic retention time of the intact protein (data not shown). With increasing incubation times, the observed peak (4.2 min) began to split, and as time progressed, the latter half of the observed split peak (4.3 min) became the dominant peak. This shift to longer chromatographic retention is likely due to increased lipophilicity of intact hPLCβ3 as a result of the addition of the highly lipophilic U73122.
Mass spectral data indicated that up to four molecules of U73122 were added to each molecule of hPLCβ3 in 1 min (Fig. 6B), by which time maximal activation had been achieved (Fig. 4). This observation implies that not all eight additions of U73122 were required to achieve maximal activation of the enzyme.
U73122 Alkylates Specific Cysteine Residues of Human PLCβ3
hPLCβ3 contains 14 cysteine residues (Fig. 7). LC/MS/MS-based peptide mapping of protease-digested hPLCβ3 identified peptides containing 12 of the 14 cysteines. Peptide mapping revealed that incubation of hPLCβ3 with U73122 for 30 min leads to the modification of 8 of the 12 detectable cysteines; this result is consistent with the observed mass changes of hPLCβ3 when incubated with U73122 for extended time periods (Fig. 6, B and C). The following cysteines were alkylated by U73122: Cys-193, Cys-221, Cys-360, Cys-516, Cys-614, Cys-892, Cys-1176, and Cys-1207.
FIGURE 7.
Primary sequence of hPLCβ3 (NP000923). Cysteine residues are in boldface type. The following cysteines were found alkylated by U73122 and are underlined: Cys-193, Cys-221, Cys-360, Cys-516, Cys-614, Cys-892, Cys-1176, and Cys-1207.
Effect of NEM on Human PLCβ3 Activity and on U73122-mediated Activation and Alkylation of Human PLCβ3
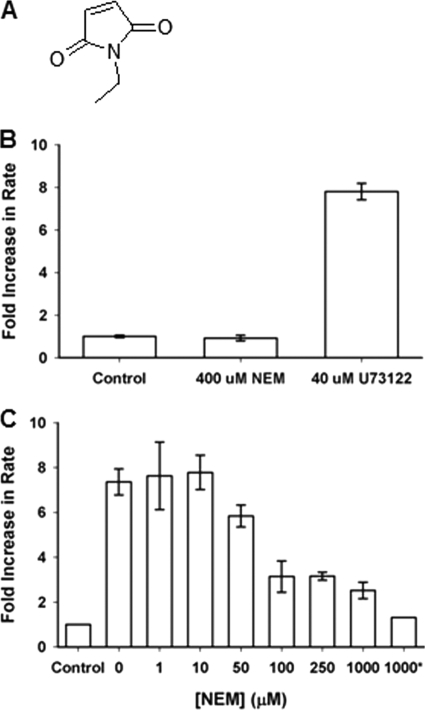
NEM is a small water-soluble compound containing the maleimide functional group but lacking the long alkyl chain and steroid skeleton present in U73122 (Fig. 8A). At concentrations up to 400 μm, NEM had no effect on the activity of hPLCβ3 in DDM-mixed micelles (Fig. 8B). Despite this lack of activation by NEM, mass spectra of intact hPLCβ3 that had been incubated with NEM revealed protein species of increasing mass, consistent with the addition of eight NEM molecules (Fig. 9A). Together, these data demonstrate that although NEM alkylates hPLCβ3, it has no effect on enzyme activity.
FIGURE 8.
Effects of NEM on hPLCβ3 activity and U73122-mediated activation of the enzyme in DDM-mixed micelles. A, structure of NEM. B, effect of NEM on hPLCβ3 activity in DDM-mixed micelles. PIP2, reconstituted in DDM, was combined with assay buffer and hPLCβ3 in a final volume of 100 μl. NEM or U73122, at indicated concentrations, was added to the assay mixture prior to the addition of enzyme. Assays were initiated by moving samples to a 37 °C water bath. Incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Data represent mean ± S.D. from triplicate determinations. C, effect of NEM on the U73122-mediated activation of hPLCβ3 in DDM-mixed micelles. PIP2, reconstituted in DDM, was combined with assay buffer and hPLCβ3 in a final volume of 100 μl. NEM, at indicated concentrations, and U73122 (40 μm) were added to the assay mixture prior to the addition of enzyme. * indicates that NEM was preincubated with enzyme prior to adding the enzyme to the assay mixture. Assays were initiated by moving samples to a 37 °C water bath. Incubation times were adjusted to ensure that less than 15% PIP2 was hydrolyzed in all cases. Data represent mean ± S.D. for triplicate determinations from one representative experiment.
FIGURE 9.
Covalent modification of hPLCβ3 by NEM in DDM-mixed micelles. A, mass spectra of intact hPLCβ3 following incubation with NEM. NEM was incubated with hPLCβ3 at 400 μm for 15 min in assay buffer at 37 °C in the absence of the substrate and detergent. Incubations were terminated with the addition of 1 mm glutathione, and an aliquot was analyzed via LC/MS. B, mass spectra of intact hPLCβ3 following sequential incubation with NEM and U73122. NEM was incubated with hPLCβ3 at 400 μm for 15 min in assay buffer at 37 °C in the absence of the substrate and detergent, followed by the addition of U73122 at 40 μm and incubation for an additional 15 min. Incubations were terminated with addition of 1 mm glutathione, and an aliquot was analyzed via LC/MS. Treatment of hPLCβ3 with NEM (400 μm) followed by treatment with U73122 (40 μm) does not further modify hPLCβ3, consistent with almost complete inhibition of U73122-mediated activation by NEM at concentrations greater than 250 μm.
On the other hand, NEM was able to attenuate the ability of U73122 to activate hPLCβ3 in a concentration-dependent manner (Fig. 8C). Therefore, it follows that NEM and U73122 react with the same cysteine residues on hPLCβ3 but that NEM is unable to activate the protein. Interestingly, NEM, at concentrations as high as 1 mm, was unable to completely inhibit the U73122-mediated activation, unless hPLCβ3 was incubated with NEM prior to the addition of U73122 (Fig. 8C). When NEM-modified hPLCβ3 was subsequently incubated with U73122, mass spectra revealed no further additions of U73122 to hPLCβ3 (Fig. 9B). Together, these data provide unequivocal evidence that NEM modifies the same cysteine residues as U73122 but that the NEM modification does not cause activation of the enzyme.
DISCUSSION
U73122 has long been regarded as a prototypical inhibitor of PLCs and almost exclusively reported as a potent and selective inhibitor of PLC activity in cellular systems. In fact, modulation of a cellular event by U73122 is often purported as evidence of the involvement of PLCs in that event. Recent studies have questioned the use of U73122 as a specific inhibitor of PLCs by uncovering a number of “off-target” interactions, likely due to covalent modification of nucleophiles by the maleimide moiety in U73122 (27–29). These findings point to the complication of using U73122 as a tool to study PLC function in complex cellular systems and suggest the need for a clearer understanding of how U73122 interacts with PLCs at the molecular level.
In this study, interaction of U73122 with hPLCβ3 was investigated in a cell-free system to elucidate the mechanism by which U73122 inhibits PLC enzymes in cellular systems. To limit the presence of confounding components that may act as nucleophilic targets for U73122, the simplest system available, i.e. mixed micelles, was employed. The expectation was that U73122 would irreversibly inhibit the lipase activity via a mechanism involving covalent modification of nucleophilic residues on hPLCβ3 (i.e. cysteines). Surprisingly, U73122 caused an increase, rather than a decrease, of the lipase activity in a concentration-dependent manner (Fig. 2B). Interestingly, U73122 also caused activation of hPLCβ2 and hPLCγ1 enzymes in mixed micelles, indicating that the enzyme activation mechanism is applicable to other members of the PLC enzyme family (Fig. 3, A–C). This is the first study demonstrating activation of PLC enzymes by U73122 in a well defined cell-free system. Interestingly, a recent study in whole cells observed weak and transient activation effects in addition to potent inhibition (28), providing both a precedent and potential relevance to cellular systems for the current observation.
The activation observed in mixed micelles reached maximal effect rapidly and was sustained upon longer incubation (Fig. 4). Furthermore, the activation was attenuated with competing nucleophiles, glutathione and cysteine, in solution (Fig. 5). These results strongly suggested that the activation of hPLCβ3 occurs via covalent modification of one or more nucleophiles, presumably sulfhydryl groups, by the reactive maleimide moiety of U73122. A simple replacement of the maleimide moiety in U73122 with a succinimide moiety rendered the resulting compound (U73343) completely ineffective as a PLC activator. This result provided strong support to the hypothesis that covalent modification of hPLCβ3 by U73122 was a requisite step for enzyme activation.
Mass spectral analysis of the hPLCβ3 protein that was allowed to react with U73122 as a function of time provided direct evidence for covalent modification of the protein by U73122. With increasing incubation times, reaction of up to eight U73122 molecules per molecule of the hPLCβ3 protein could easily be detected based on increases in mass-to-charge of intact hPLCβ3 by multiples of the molecular weight of U73122 (∼0.5 kDa) (Fig. 6). These data unequivocally demonstrate that U73122 reacts covalently with as many as eight distinct sites on hPLCβ3 and suggest an underlying molecular mechanism for modulation of PLC activity in cell-free experimental systems.
There are 14 cysteine residues in hPLCβ3 (Fig. 7). To unambiguously confirm cysteines as the sites of alkylation by U73122, as well as to identify specific residues modified by U73122, LC/MS/MS was used for peptide mapping of U73122-modified hPLCβ3. Peptides containing 12 of the 14 cysteines were identified from protease-digested hPLCβ3, excluding Cys-669 and Cys-834. When hPLCβ3 was incubated with U73122 under conditions similar to those that led to its activation, 8 of the 12 identified cysteine residues were found to be alkylated, including Cys-360 and Cys-614 located in the so-called “X” and “Y” boxes that make up the highly conserved catalytic domain of all PLCs. Other modified residues include Cys-516 in the less conserved linker region between the X and Y boxes, as well as Cys-193, Cys-221, Cys-892, Cys-1176, and Cys-1207 outside of the catalytic domain. Interestingly, Cys-193 and Cys-221 are located within the pleckstrin homology domain, a region thought to be critical for the membrane recruitment of PLCβ isozymes, and recently implicated in the interaction of PLCβ2 with Rac1 during substrate catalysis (41). Time-dependent activation studies (Fig. 4) demonstrated that maximal activation was observed within 30 s of preincubation of U73122 with hPLCβ3; however, after 1 min of incubation, mass spectra of intact hPLCβ3 revealed only four additions of U73122 (Fig. 6). These results suggest that alkylation of four or fewer residues are primarily responsible for the activation. Site-directed mutagenesis of each alkylated cysteine residue would be useful to explore the role of individual cysteines in the observed activation by U73122.
Maleimides are inherently reactive and readily interact with thiols, such as cysteine residues, on cellular proteins; therefore, it was important to determine whether protein alkylation at cysteine residues on hPLCβ3 was the sole mechanism for the U73122-mediated activation. NEM (Fig. 8) is a small water-soluble maleimide, which contains an ethyl group in place of the lipophilic alkylamino steroid functionality of U73122; and it is capable of modifying sulfhydryl groups on proteins. Interestingly, NEM had no effect on the activity of hPLCβ3 in DDM-mixed micelles at concentrations as high as 400 μm, despite the fact that mass spectra of hPLCβ3 incubated with NEM identified protein species consistent with reaction of up to eight NEM molecules (Fig. 9A). This result indicates that simply alkylating the sulfhydryl groups on PLCs does not cause activation of the enzyme. In addition, U73343, which is nearly identical to U73122 with respect to chemical structure except that it does not contain the reactive maleimide moiety (but contains succinimide moiety instead), does not cause activation of the enzyme. Together, these results suggest that the activation mechanism involves covalent modification of several sulfhydryl groups on hPLCβ3 by a hydrophobic steroid moiety. As expected based on the above observations, the combined treatment of NEM and the succinimide derivative, U73343, had no effect on activity (data not shown). Importantly, the lack of any effect of NEM on the activity of hPLCβ3, activation or inhibition, also suggests that the eight cysteine residues alkylated by NEM do not play a direct role in the catalytic activity of hPLCβ3 in this system. Although NEM was unable to activate hPLCβ3, excess NEM attenuated the U73122-mediated activation in a concentration-dependent manner. In addition, mass spectral data demonstrated that NEM pretreatment prevented covalent modification of hPLCβ3 by U73122 (Fig. 9B). Together, these studies imply that alkylation of several sulfhydryl groups on hPLCβ3, none of which is playing a direct role in the catalytic function, by a lipophilic steroid causes activation of the enzymatic activity.
The following hypothesis for activation of hPLCβ3 is consistent with the results presented herein. U73122 irreversibly binds to multiple cysteine residues on hPLCβ3 and serves as either a lipid anchor or interfacial recognition site for the enzyme, facilitating adsorption of the enzyme to the substrate interface (i.e. the micelle surface). The protein-linked U73122 increases the rate of lipase activity by keeping the enzyme in close proximity to the membrane where the substrate resides (Fig. 10). Because the proposed mechanism of enzyme activation involves U73122 chaperoning the enzyme molecules to the lipid micelles where the substrate is located, it is not surprising that PLC isozymes other than hPLCβ3 were also activated by this agent. The proposed mechanism for activation is consistent with the recent observation that U73122 enhances in vitro yeast vacuole membrane fusion mediated by the soluble NSF attachment protein receptor domain of Vam7p through enhanced membrane binding (31).
Implications for PLC Biochemistry
Because all PLC isozymes work on primarily two substrates, PIP or PIP2, their selectivity in cellular functions must derive from their differential activation by receptors to which they are linked. The molecular mechanisms underlying cellular PLC activation and subsequent substrate hydrolysis at cellular membranes are not completely understood. Recruitment of the cytosolic PLC enzymes to the cell membrane and conformational changes upon membrane binding that expose the active site and increase enzyme activity are believed to be an activation mechanism for a number of lipases (i.e. the opening of the lid providing access to the active site) (42–49) and have recently been hypothesized for PLCβ isozymes (41), although the triggers for these structural changes have not yet been elucidated.
The importance of cysteine residues in protein function is well established. For example, specific cysteine residues are required for activation of the ion channel, TRPA1 (50, 51), are important for enzymatic activity of caspases (52, 53) as well as other lipases (54), are crucial for regulating the activity of cellular phosphatases such as PTEN (55), and are used as palmitoylation sites for integral membrane proteins such as claudin (56) and signaling proteins like H-ras (57, 58). Post-translational modifications of proteins, such as phosphorylation, have clearly been shown to trigger rapid conformational changes in many proteins leading to significant alterations in protein function (59–62); in fact, recent studies have implicated reversible cysteine modifications (e.g. S-nitrosation) in changes to protein function (63–65). For example, nitrosation of caspase 3 by nitric oxide at a specific cysteine residue was reported to be critical for promotion of apoptosis (52, 53). S-Nitrosothiols have also been shown to modify phosphatases such as PTEN, a phosphatidylinositol 1,4,5-trisphosphate phosphatase, leading to a reversible inhibition of its phosphatase activity (55) and supporting a hypothesis for cysteine-dependent changes in protein function. Interestingly, a recent report suggests that low concentrations of the reactive oxygen species, H2O2, generates intracellular calcium oscillations in rat astrocytes by activating PLCγ1 via a sulfhydryl-dependent oxidation mechanism (66). Based on the proposed U73122-mediated activation mechanism for PLC enzymes in cell-free systems, it is tempting to speculate that activation of PLC enzymes inside the cell occurs via recruitment of cytosolic PLCs to the cell membrane through transient modification of sulfhydryl groups on the enzyme by lipophilic moieties, triggered as a result of receptor activation.
Implications for the Use of U73122 as a Prototypical PLC Inhibitor
The present results provide unequivocal evidence that in cell-free systems U73122 activates PLC enzymes. This contrasts with the “established” role of U73122 as a specific inhibitor of this family of enzymes. It is conceivable that U73122 has opposite effects on PLC enzymes depending on the environment in which it finds the enzyme molecules. For example, it could activate the cytosolic PLC enzymes by a mechanism similar to the one proposed for PLC activation in cell-free systems but inhibit PLCs that are already associated with the cell membrane (the site of PLC catalysis) due to receptor-mediated activation. Alternatively, the observed inhibition of PLCs by U73122 in cellular systems may result indirectly by covalent modification of other cellular proteins (e.g. receptors). The results presented in this study, as well as other recent evidence (29), suggest that U73122, a highly reactive molecule, is capable of rapidly alkylating nucleophiles in experimental systems. Thus, it is reasonable to assume that the effects of U73122, particularly in cell-based assays, will be complex and dependent on the presence of free nucleophiles in the media, in the cell membrane, and in the cytosol. For example, this study uncovered evidence for covalent modification of bovine serum albumin, a component of the mixed micellar assay, by U73122 (data not shown). Therefore, care should be taken when making conclusions based on the observed effects of U73122 in cellular systems. Studies intended to implicate PLC enzymes in cellular phenotypes should avoid using this compound as a probe molecule, as its reactivity with other cellular nucleophiles may lead to off-target effects that could be mistakenly attributed to PLCs involvement. This conclusion highlights the risk associated with using U73122 as a probe for cellular functions involving PLC enzymes and a need for the development of small molecules that directly and selectively modulate PLC enzymes.
Acknowledgments
We thank Drs. T. Kendall Harden, Jason Snyder, and Aurelie Gresset (Department of Pharmacology, School of Medicine, University of North Carolina, Chapel Hill) for providing hPLCβ3 and hPLCγ1 and for helpful discussions regarding cell-free PLC assays.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM057391 (to J. S.).
- PLC
- phospholipase C
- DDM
- dodecyl maltoside
- h
- human
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP
- phosphatidylinositol 4-phosphate
- U73122
- 1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione
- PI
- phosphatidylinositol
- NEM
- N-ethylmaleimide.
REFERENCES
- 1. Harden T. K., Sondek J. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 355–379 [DOI] [PubMed] [Google Scholar]
- 2. Rebecchi M. J., Pentyala S. N. (2000) Physiol. Rev. 80, 1291–1335 [DOI] [PubMed] [Google Scholar]
- 3. Rhee S. G. (2001) Annu. Rev. Biochem. 70, 281–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rhee S. G., Bae Y. S. (1997) J. Biol. Chem. 272, 15045–15048 [DOI] [PubMed] [Google Scholar]
- 5. Rhee S. G., Suh P. G., Ryu S. H., Lee S. Y. (1989) Science 244, 546–550 [DOI] [PubMed] [Google Scholar]
- 6. Rhee S. G., Choi K. D. (1992) J. Biol. Chem. 267, 12393–12396 [PubMed] [Google Scholar]
- 7. Jones N. P., Peak J., Brader S., Eccles S. A., Katan M. (2005) J. Cell Sci. 118, 2695–2706 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Tomar A., George S. P., Khurana S. (2007) Am. J. Physiol. Cell. Physiol. 292, C1775–C1786 [DOI] [PubMed] [Google Scholar]
- 9. Ji Q. S., Winnier G. E., Niswender K. D., Horstman D., Wisdom R., Magnuson M. A., Carpenter G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nebigil C. G. (1997) Biochemistry 36, 15949–15958 [DOI] [PubMed] [Google Scholar]
- 11. Balda M. S., González-Mariscal L., Contreras R. G., Macias-Silva M., Torres-Marquez M. E., García-Sáinz J. A., Cereijido M. (1991) J. Membr. Biol. 122, 193–202 [DOI] [PubMed] [Google Scholar]
- 12. Cereijido M., González-Mariscal L., Contreras R. G., Gallardo J. M., García-Villegas R., Valdés J. (1993) J. Cell Sci. Suppl. 17, 127–132 [DOI] [PubMed] [Google Scholar]
- 13. Ward P. D., Klein R. R., Troutman M. D., Desai S., Thakker D. R. (2002) J. Biol. Chem. 277, 35760–35765 [DOI] [PubMed] [Google Scholar]
- 14. Bleasdale J. E., Bundy G. L., Bunting S., Fitzpatrick F. A., Huff R. M., Sun F. F., Pike J. E. (1989) Adv. Prostaglandin Thromboxane Leukot. Res. 19, 590–593 [PubMed] [Google Scholar]
- 15. Carvou N., Norden A. G., Unwin R. J., Cockcroft S. (2007) Cell. Signal. 19, 42–51 [DOI] [PubMed] [Google Scholar]
- 16. Hou C., Kirchner T., Singer M., Matheis M., Argentieri D., Cavender D. (2004) J. Pharmacol. Exp. Ther. 309, 697–704 [DOI] [PubMed] [Google Scholar]
- 17. Smith R. J., Sam L. M., Justen J. M., Bundy G. L., Bala G. A., Bleasdale J. E. (1990) J. Pharmacol. Exp. Ther. 253, 688–697 [PubMed] [Google Scholar]
- 18. Thompson A. K., Mostafapour S. P., Denlinger L. C., Bleasdale J. E., Fisher S. K. (1991) J. Biol. Chem. 266, 23856–23862 [PubMed] [Google Scholar]
- 19. Ward P. D., Ouyang H., Thakker D. R. (2003) J. Pharmacol. Exp. Ther. 304, 689–698 [DOI] [PubMed] [Google Scholar]
- 20. Chen Y. J., Sheng W. Y., Huang P. R., Wang T. C. (2006) J. Biomed. Sci. 13, 667–674 [DOI] [PubMed] [Google Scholar]
- 21. Feisst C., Albert D., Steinhilber D., Werz O. (2005) Mol. Pharmacol. 67, 1751–1757 [DOI] [PubMed] [Google Scholar]
- 22. Hughes S. A., Gibson W. J., Young J. M. (2000) Naunyn Schmiedebergs Arch. Pharmacol. 362, 555–558 [DOI] [PubMed] [Google Scholar]
- 23. Pulcinelli F. M., Gresele P., Bonuglia M., Gazzaniga P. P. (1998) Biochem. Pharmacol. 56, 1481–1484 [DOI] [PubMed] [Google Scholar]
- 24. Klose A., Huth T., Alzheimer C. (2008) Mol. Pharmacol. 74, 1203–1214 [DOI] [PubMed] [Google Scholar]
- 25. Macmillan D., McCarron J. G. (2010) Br. J. Pharmacol. 160, 1295–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burgdorf C., Schäfer U., Richardt G., Kurz T. (2010) J. Cardiovasc. Pharmacol. 55, 555–559 [DOI] [PubMed] [Google Scholar]
- 27. Gloyna W., Schmitz F., Seebeck J. (2005) Regul. Pept. 125, 179–184 [DOI] [PubMed] [Google Scholar]
- 28. Horowitz L. F., Hirdes W., Suh B. C., Hilgemann D. W., Mackie K., Hille B. (2005) J. Gen. Physiol. 126, 243–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilsher N. E., Court W. J., Ruddle R., Newbatt Y. M., Aherne W., Sheldrake P. W., Jones N. P., Katan M., Eccles S. A., Raynaud F. I. (2007) Drug Metab. Dispos. 35, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 30. Enyeart J. J., Danthi S. J., Liu H., Enyeart J. A. (2005) J. Biol. Chem. 280, 30814–30828 [DOI] [PubMed] [Google Scholar]
- 31. Fratti R. A., Collins K. M., Hickey C. M., Wickner W. (2007) J. Biol. Chem. 282, 14861–14867 [DOI] [PubMed] [Google Scholar]
- 32. Han S., Kim T. D., Ha D. C., Kim K. T. (2005) J. Biol. Chem. 280, 38228–38234 [DOI] [PubMed] [Google Scholar]
- 33. Mariappan M. M., Senthil D., Natarajan K. S., Choudhury G. G., Kasinath B. S. (2005) J. Biol. Chem. 280, 28402–28411 [DOI] [PubMed] [Google Scholar]
- 34. Suzuki Y., Zhang H., Saito N., Kojima I., Urano T., Mogami H. (2006) J. Biol. Chem. 281, 28499–28507 [DOI] [PubMed] [Google Scholar]
- 35. Wei Y., Zavilowitz B., Satlin L. M., Wang W. H. (2007) J. Biol. Chem. 282, 6455–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gresset A., Hicks S. N., Harden T. K., Sondek J. (2010) J. Biol. Chem. 285, 35836–35847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hicks S. N., Jezyk M. R., Gershburg S., Seifert J. P., Harden T. K., Sondek J. (2008) Mol. Cell 31, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. James S. R., Paterson A., Harden T. K., Downes C. P. (1995) J. Biol. Chem. 270, 11872–11881 [DOI] [PubMed] [Google Scholar]
- 39. James S. R., Smith S., Paterson A., Harden T. K., Downes C. P. (1996) Biochem. J. 314, 917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wagner C. D., Hall J. T., White W. L., Miller L. A., Williams J. D. (2007) J. Mass Spectrom. 42, 139–149 [DOI] [PubMed] [Google Scholar]
- 41. Jezyk M. R., Snyder J. T., Gershberg S., Worthylake D. K., Harden T. K., Sondek J. (2006) Nat. Struct. Mol. Biol. 13, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 42. Aloulou A., Rodriguez J. A., Fernandez S., van Oosterhout D., Puccinelli D., Carrière F. (2006) Biochim. Biophys. Acta 1761, 995–1013 [DOI] [PubMed] [Google Scholar]
- 43. Berg O. G., Cajal Y., Butterfoss G. L., Grey R. L., Alsina M. A., Yu B. Z., Jain M. K. (1998) Biochemistry 37, 6615–6627 [DOI] [PubMed] [Google Scholar]
- 44. Egloff M. P., Sarda L., Verger R., Cambillau C., van Tilbeurgh H. (1995) Protein Sci. 4, 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roussel A., Canaan S., Egloff M. P., Rivière M., Dupuis L., Verger R., Cambillau C. (1999) J. Biol. Chem. 274, 16995–17002 [DOI] [PubMed] [Google Scholar]
- 46. Roussel A., Miled N., Berti-Dupuis L., Rivière M., Spinelli S., Berna P., Gruber V., Verger R., Cambillau C. (2002) J. Biol. Chem. 277, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 47. Thomas A., Allouche M., Basyn F., Brasseur R., Kerfelec B. (2005) J. Biol. Chem. 280, 40074–40083 [DOI] [PubMed] [Google Scholar]
- 48. Winkler F. K., D'Arcy A., Hunziker W. (1990) Nature 343, 771–774 [DOI] [PubMed] [Google Scholar]
- 49. Yang Y., Lowe M. E. (2000) J. Lipid Res. 41, 48–57 [PubMed] [Google Scholar]
- 50. Hinman A., Chuang H. H., Bautista D. M., Julius D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19564–19568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Macpherson L. J., Dubin A. E., Evans M. J., Marr F., Schultz P. G., Cravatt B. F., Patapoutian A. (2007) Nature 445, 541–545 [DOI] [PubMed] [Google Scholar]
- 52. Mitchell D. A., Marletta M. A. (2005) Nat. Chem. Biol. 1, 154–158 [DOI] [PubMed] [Google Scholar]
- 53. Tannenbaum S. R., Kim J. E. (2005) Nat. Chem. Biol. 1, 126–127 [DOI] [PubMed] [Google Scholar]
- 54. Saario S. M., Salo O. M., Nevalainen T., Poso A., Laitinen J. T., Järvinen T., Niemi R. (2005) Chem. Biol. 12, 649–656 [DOI] [PubMed] [Google Scholar]
- 55. Yu C. X., Li S., Whorton A. R. (2005) Mol. Pharmacol. 68, 847–854 [DOI] [PubMed] [Google Scholar]
- 56. Van Itallie C. M., Gambling T. M., Carson J. L., Anderson J. M. (2005) J. Cell Sci. 118, 1427–1436 [DOI] [PubMed] [Google Scholar]
- 57. Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. (1989) Cell 57, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 58. Hancock J. F., Parton R. G. (2005) Biochem. J. 389, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Canagarajah B. J., Khokhlatchev A., Cobb M. H., Goldsmith E. J. (1997) Cell 90, 859–869 [DOI] [PubMed] [Google Scholar]
- 60. Goldsmith E. J., Sprang S. R., Hamlin R., Xuong N. H., Fletterick R. J. (1989) Science 245, 528–532 [DOI] [PubMed] [Google Scholar]
- 61. Ohki S., Eto M., Kariya E., Hayano T., Hayashi Y., Yazawa M., Brautigan D., Kainosho M. (2001) J. Mol. Biol. 314, 839–849 [DOI] [PubMed] [Google Scholar]
- 62. Ohki S., Eto M., Shimizu M., Takada R., Brautigan D. L., Kainosho M. (2003) J. Mol. Biol. 326, 1539–1547 [DOI] [PubMed] [Google Scholar]
- 63. Handy D. E., Loscalzo J. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 64. Konorev E. A., Kalyanaraman B., Hogg N. (2000) Free Radic. Biol. Med. 28, 1671–1678 [DOI] [PubMed] [Google Scholar]
- 65. Miersch S., Mutus B. (2005) Clin. Biochem. 38, 777–791 [DOI] [PubMed] [Google Scholar]
- 66. Hong J. H., Moon S. J., Byun H. M., Kim M. S., Jo H., Bae Y. S., Lee S. I., Bootman M. D., Roderick H. L., Shin D. M., Seo J. T. (2006) J. Biol. Chem. 281, 13057–13067 [DOI] [PubMed] [Google Scholar]