Abstract
Reprogramming of somatic cells to induced pluripotent stem (iPS) cells can be achieved by the delivery of a combination of transcription factors, including Oct4, Sox2, Klf4, and c-Myc. Retroviral and lentiviral vectors are commonly used to express these four reprogramming factors separately and obtain reprogrammed iPS cells. Although efficient and reproducible, these approaches involve the time-consuming and labor-intensive production of retroviral or lentiviral particles together with a high risk of working with potentially harmful viruses overexpressing potent oncogenes, such as c-Myc. Here, we describe a simple method to produce bona fide iPS cells from human fibroblasts using poly-β-amino esters as the transfection reagent for the delivery of a single CAG-driven polycistronic plasmid expressing Oct4, Sox2, Klf4, c-Myc, and a GFP reporter gene (OSKMG). We demonstrate for the first time that poly-β-amino esters can be used to deliver a single polycistronic reprogramming vector into human fibroblasts, achieving significantly higher transfection efficiency than with conventional transfection reagents. After a protocol of serial transfections using poly-β-amino esters, we report a simple methodology to generate human iPS cells from human fibroblasts avoiding the use of viral vectors.
Keywords: Human Genetics, Polyamines, Protein Synthesis, Skin, Stem Cell, iPS Cells, Nanoparticle, Poly-β-amino Esters, Reprogamming, Transfection
Introduction
In 2006, Yamanaka and Takahashi (1) showed for the first time that mouse embryonic fibroblast (MEFs)3 can acquire a transcriptional profile and phenotype similar to an embryonic stem (ES) cell by ectopically expressing only four transcription factors. These so-called induced pluripotent stem (iPS) cells were generated by infecting MEFs with four retroviruses constitutively expressing Oct4, Sox2, Klf4, and c-myc (OSKM) (1). Since then several groups have confirmed these results and improved the reprogramming technique, generating iPS cells from mouse and human cells (2–7).
iPS cells share many characteristic of ES cells. They form colonies with similar proliferation and morphological features, express a similar set of pluripotency markers, show comparable promoter methylation levels and telomerase activities and are able to differentiate both in vitro and in vivo into cell derivatives of the three germ layers. Hence, they represent a novel source of pluripotent stem cells with great potential for basic research, drug discovery, or autologous cell therapy.
Retroviruses have been so far the most commonly used vectors to express the OSKM set of transcription factors into fibroblasts (1, 3, 5), neural stem cells, keratinocytes, or cord blood stem cells (8–11) using sometimes different combinations of factors (7). Other viral gene transfer vectors such as lentiviruses have also allowed successful reprogramming (12, 13).
The constitutive retroviral and lentiviral vectors commonly used for reprogramming allow efficient generation of iPS cells from mouse and human somatic cells. However, their permanent integration into the genome limits their use for eventual therapeutic applications due to the risk of both insertional mutagenesis and particularly the reactivation of the reprogramming factors leading to tumor formation (14).
Alternatively, transient expression methods trying to avoid genomic integration have been devised. Such methods include adenovirus (15, 16), plasmid transfection (17–21), and cell penetrating peptides (22, 23).
So far, the simplest protocols for generating transgene-free iPS rely on serial transfection and transient expression of CAG-driven vectors expressing the whole set of reprogramming factors as a single polycistronic transcript, thereafter translated into separate proteins via a ribosomal skipping mechanism allowed by the insertion of the 2A peptide sequence of picornavirus (24) between the cDNAs encoding each transcription factor (17, 19). Following this strategy, Okita et al. (19) obtained mouse iPS lines from MEFs showing no evidence of integration. They performed four serial transfections with FuGENE to transiently express two plasmids (one expressing c-myc and a second construct expressing Oct4, Klf4, and Sox2 as a polycistronic unit). Similarly, we reported that iPS lines without transgene integration can be generated in the mouse by nucleofection of a single plasmid construct expressing OSKM as a single polycistronic unit (17). Nevertheless, the reprogramming protocols described in these works were only applied to MEFs and showed much lower efficiency of iPS generation (<0.001%) when compared with viral induction (∼0.01%). Therefore, the induction of iPS cells from other tissue origins or other species by these methods will eventually require many improvements.
Reprogramming of human fibroblasts is known to require longer expression periods of reprogramming factors than MEFs, and their induction efficiency of iPS generation is reported to be very low (25). In addition, delivery of large DNA constructs, such as the polycistronic constructs used for reprogramming using conventional transfection methods such as liposome or nucleofection, have typically achieved low transfection efficiencies, short periods of transgene expression, and low cell viability.
Therefore, in an attempt to generate transgene-free human iPS cells by transient expression of a single non-viral polycistronic vector in human foreskin fibroblasts, we used poly-β-amino esters as the transfection reagent to transiently express Oct4, Sox2, Klf4, c-myc, and a GFP reporter gene (pCAG-OSKMG). Poly-β-amino esters are a class of polycations that were first developed by Langer and co-workers (26–29) for gene delivery applications. This group has recently described the use of these polymers to successfully deliver a VEGF plasmid into human mesenchymal stem cells and mesenchymal-like stem cells derived from human embryonic stem cells to enhance angiogenesis (30). In this work we successfully delivered a large vector of ∼11 kb into human fibroblasts using poly-β-amino esters, achieving higher efficiencies when compared with conventional commercial transfection reagents, such as Lipofectamine and FuGENE. Moreover, after an optimized protocol of 2 or 3 consecutive transfections, we generated human iPS cells in ∼20–28 days. These iPS cell lines expressed the typical set of pluripotency marker genes, had a stable karyotypes, and were able to differentiate in vitro into derivatives of the three germ layers. Although our approach did not allow the generation of transgene-free iPS cell lines, it represents a simple non-viral alternative to produce iPS cells from human fibroblasts, avoiding the time-consuming and labor-intensive task of producing and working with potentially harmful retroviral or lentiviral particles overexpressing potent oncogenes such as c-Myc.
EXPERIMENTAL PROCEDURES
Synthesis of the Poly-β-amino Esters
Poly-β-aminoesters were synthesized by the addition reaction of primary amines with excess of diol diacrylates. End-capping modification of acrylate-terminated polymers with different kind of diamines improves the ability of the polymers to complex DNA. Three different end-modified poly-β-aminoesters were synthesized after a two-step procedure described elsewhere (28, 29, 31). Chemicals were obtained from Sigma and Panreac and used as received. Phosphate-buffered saline (1×) was purchased from Invitrogen. First, acrylate-terminated poly-5-amino-1-pentanol-co-1,4-butanediol diacrylate was synthesized by the addition of 5-amino-1-pentanol (3.44 g, 33 mmol) to 1,4-butanediol diacrylate (7.93 g, 40 mmol) at a 1.2:1 molar ratio of diacrylate monomer to amine monomer. Polymerization was performed under magnetic stirring at 90 °C for 24 h. Afterward, the poly-5-amino-1-pentanol-co-1,4-butanediol diacrylate obtained was end-modified with three different amine monomers. Freshly synthesized poly-5-amino-1-pentanol-co-1,4-butanediol diacrylate was dissolved in THF (1 g, 2 mmol in 2 ml of THF). Then a solution of each amine monomer, 1) 1,3-diaminopropane (0.148 g, 167 μl, 2 mmol) in THF (8 ml), 2) 1,3-diaminopentane (0.204 g, 239 μl, 2 mmol) in THF (8 ml), or 3) 2-methyl-1,5-pentanediamine (0.232 g, 270 μl, 2 mmol) in THF (8 ml), was added to the poly5-amino-1-pentanol-co-1,4-butanediol diacrylate solution and stirred magnetically overnight at room temperature. Each resulting amino-terminated polymer (B1, B2, and B3) was isolated by precipitation in diethyl ether (100 ml) and dried under vacuum. Synthesized structures were confirmed by 1H NMR and FTIR analysis. NMR spectra were recorded in a 400 MHz Varian (Varian NMR Instruments, Claredon Hills, IL) and methanol-d4 was used as the solvent. IR spectra were obtained using a Nicolet Magna 560(Thermo Fisher Scientific, Waltham, MA) with a KBr beamsplitter using methanol as the solvent in evaporated film.
Nanoparticle Formation and Transfection Procedure Using Poly-β-amino Esters
pmaxGFP plasmid (3486 bp) was purchased from Amaxa and used as a control of transfection. pCAG-OSKMG plasmid (11000 bp), which is a vector expressing the whole set of reprogramming factors as a single polycistronic transcript, was previously developed by our group (17).
Human foreskin fibroblasts were transfected with pmaxGFP plasmid or pCAG-OSKMG plasmid in Opti-MEM medium using the three different poly-β-amino esters. Initial cell density (200,000, 400,000, or 800,000 cells/well of a 6-well plate), amounts of DNA (2.5, 5, or 10 μg/well), DNA to polymer ratio (1:20, 1:40, 1:50, 1:80, or 1:100), and time of transfection (30 min or 1, 2, or 3 h) were optimized to define the optimal conditions for the transfection of the large pCAG-OSKMG plasmid.
Polyplexes were formulated by mixing the poly-β-amino esters (B1, B2, and B3) with the plasmids at different ratios (w/w) of DNA to polymer (between 1:20 to 1:100). Briefly, DNA was first dissolved to a final concentration of 0.1 mg/ml in a 25 mm sodium acetate buffer solution at pH 5.2 (NaAc buffer), which was prepared from a 3 m stock (Sigma). The appropriate amount of B1, B2, or B3 polymers at 20 mg/ml in DMSO was then added to the DNA solution and mixed for some seconds. After 30 min of incubation at room temperature to allow for nanoparticle self-assembly, the particle solution was added to cells and incubated for 1 h. After 1 h of incubation, the medium was replaced by fresh growth medium. Lipofectamine 2000 (Invitrogen) and FuGENE HD, two commercially available transfection reagents, were used according to manufacturer's instructions for comparing the efficiency of transfection.
Particle Size and ζ Potential
Particle size and ζ potential measurements were recorded using a Zetasizer Nano series ZS dynamic Light scattering detector (Malvern Instruments Ltd., 4-milliwatt laser, incident beam 633 nm). Correlation functions were collected at an scattering angle of 90°, and particle sizes were calculated using the Malvern particle sizing software (DTS Version 5.03) using the viscosity and refractive index of water at 25 °C.
Efficiency of Transfection Using Poly-β-amino Esters
The efficiency of transfection of pmaxGFP and pCAG-OSKMG plasmids into human fibroblasts was determined by means of GFP-expressing cells 24 h after the transfection. Green fluorescent protein expression was measured using fluorescence-activated cell sorting on a MoFlo cell sorter (DakoCytomation) applying Summit software. Propidium iodide staining was used to exclude dead cells from the analysis.
Reprogramming with Poly-β-amino Esters
Human foreskin fibroblasts p2 were seeded in 6-well plates at a defined cell density (100,000, 400,000, or 800,000 cells/well) with DMEM medium containing 10% of FBS (fibroblast medium). The next day cells were transfected with pCAG-OSKMG plasmid using poly-β-amino esters. For an optimal transfection polymer-DNA complexes were prepared at 1/50 ratio with 10 μg of DNA (diluted in AcO buffer, pH 5.5, to 0.1 mg/ml) and 25 μl of the polymer at 20 mg/ml for each well of a 6-well plate. Serial transfections with poly-β-amino esters were performed as shown in the timeline depicted in Fig. 3A. After the last transfection fibroblast medium was changed by human embryonic stem cell growth medium, and cells were maintained for 1 week with daily media changes. After 1 week, cells were trypsinized and transferred onto irradiated human feeders using a 1:6 dilution. Approximately, 20–28 days after the plating onto human feeders, the first iPS cell-like colonies appeared.
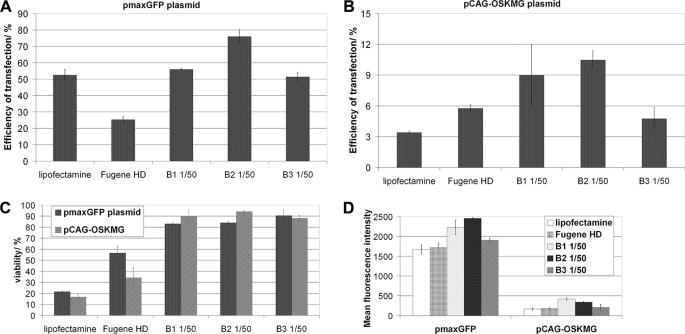
FIGURE 3.
Efficiency of pCAG-OSKMG plasmid delivery to human fibroblasts. The efficiency of transfection using the different polymer formulations for the pmaxGFP plasmid (A) and the pCAG-OSKMG plasmid (B) was quantified by FACS analysis of the GFP-expressing cells 24 h post-transfection. C, cell viability was measured by propidium iodide incorporation 1 day after the transfection. D, mean fluorescence intensity for the different formulations 1 day post-transfection with pmaxGFP and pCAG-OSKMG plasmids is shown.
Integration Analysis
The integration of the transgene was analyzed by PCR. 50 ng of genomic DNA were amplified with Taq polymerase (Roche Applied Science) by using two pairs of primers spanning two different areas of the pCAG-OSKMG vector (OS primers amplify a fragment of the pCAG-OSKMG vector between Oct4 and Sox2: forward (5′-TTCCCTCTGTTCCCGTCACT-3′) and reverse (5′-GCTTGCTGATCTCCGAGTTG-3′), and K1/2 primers amplify a fragment of the pCAG-OSKMG vector in Klf4: forward (5′-AACACACACGACTTCCCCCT-3′) and reverse (5′-GTGGGTGGCTGTTCTTTTCC-3′)).
RT-PCR Analysis
Total RNA from each cell line was isolated using All Prep DNA/RNA columns (Qiagen) following the manufacturer's guidelines. All RNA samples were treated with TURBO DNase inhibitor (Ambion) to remove any residual genomic DNA, and 1 μg of RNA was used to synthesize cDNA using the Invitrogen SuperScript II Reverse Transcriptase kit. 25 ng of cDNA were used to quantify typically pluripotency gene expression markers by using Platinum Syber Green qPCR Super Mix (Invitrogen) in an ABI Prism 7000 thermocycler (Applied Biosystems) and primers as previously described (10). Values were normalized to Gapdh. Similarly, 25 ng of cDNA were used to determine the level of transgene expression using primers that amplify a fragment of the transgene (forward (5′-AAAAGGCCCCCAAGGTAGTG-3′) and reverse (5′-CGGTGGTCCAGATGAACTTC-3′)).
Immunofluorescence Analysis and Alkaline Phosphatase Staining
Cells were grown on plastic cover slide chambers fixed with 4% paraformaldehyde. The following antibodies were used: TRA-1-81 (MAB4381, 1:100) from Chemicon, SSEA-3 (MC-631, 1:2) from the Developmental Studies Hybridoma Bank at the University of Iowa, OCT-3/4 (C-10, Santa Cruz, sc-5279, 1:100), NANOG (Everest Biotech EB06860, 1:100), Tuj1 (Covance, 1:500), GFAP (Dako, 1:1000), α-fetoprotein (Dako, 1:400), FOXA-2 (R&D System, 1:50), smooth muscle actin (Sigma, 1:400), α-sarcomeric actinin (Sigma, 1:200), and anti-GFP (Aves, 1:250). Images were taken using Leica SP5 confocal microscope. Direct AP activity was analyzed using an alkaline phosphatase blue membrane substrate solution kit (Sigma, AB0300) according to the manufacturer's guidelines.
High Resolution, G-banded Karyotype
The analysis was performed on 80% confluent cells growing on Matrigel. Cells were treated with colcemid at 20 ng/ml followed by 45 min of incubation at 37 °C. Then, after trypsinizing the cells into a single cell suspension, they were treated with Carnoy fixative at −20 °C. Finally, the samples were analyzed with the software Cytovision (Applied Imaging).
In Vitro Differentiation of the Generated iPS Cell Lines
After 3–4 days, the embryoid bodies were transferred to 0.1% gelatin-coated glass chamber slides and cultured in differentiation medium (DMEM supplemented with 20% fetal bovine serum, 2 mm l-glutamine, 0.1 mm 2-mercaptoethanol, nonessential amino acids, and penicillin-streptomycin) for 2–3 weeks to allow spontaneous endoderm formation. The medium was changed every other day. For mesoderm/cardiomyocyte differentiation, embryoid bodies were maintained on gelatin-coated plate in differentiated medium supplemented with 100 μm ascorbic acid (Sigma). For ectoderm differentiation, embryoid bodies were cultured on Matrigel-coated plates in N2/B27 medium supplemented with 1 μm retinoic acid for 2–3 weeks.
In Vivo Differentiation
Severe combined immune-deficient (SCID) beige male mice, 8 weeks old approximately, were injected with hiPS cells (1 million for each injection site, approximately) subcutaneously in the testicular parenchyma. All procedures involving animals were approved by the institutional Animal Ethical Board, and the protocols were approved by the Conselleria De Salut of Cataluña. Briefly, the mice were briefly anesthetized with a gaseous mixture of halothane:oxygen:ambient air of 0.5:2:97.5. Injections were performed using a 1-ml syringe with a G25 needle. No more than 150 μl of cell suspension were injected. The mice were sacrificed 8 weeks after the injections or when a tumor was detected by palpation, whichever came first. Tumor samples were collected generally in 2 months and processed for paraffin embedding and hematoxylin and eosin staining following standard procedures.
RESULTS
Preliminary data showed that transfection of human fibroblasts with pCAG-OSKMG vector using Lipofectamine, FuGENE, or nucleofection resulted in very low transfection efficiencies (lower than 8%) and low cell viability (lower than 50%). Thus, we aimed to improve the transfection efficiency of the large DNA vector pCAG-OSKMG to optimize a method of reprogramming of human fibroblasts by transient expression of the reprogramming factors Oct4, Sox2, Klf4, and cMyc. For this purpose we chose poly-β-amino esters as good candidates for transfection of human cells and investigated the ability of these polymers to transfect pCAG-OSKMG into human fibroblasts.
Synthesis and Biophysical Characterization of End-modified Poly-β-amino Esters
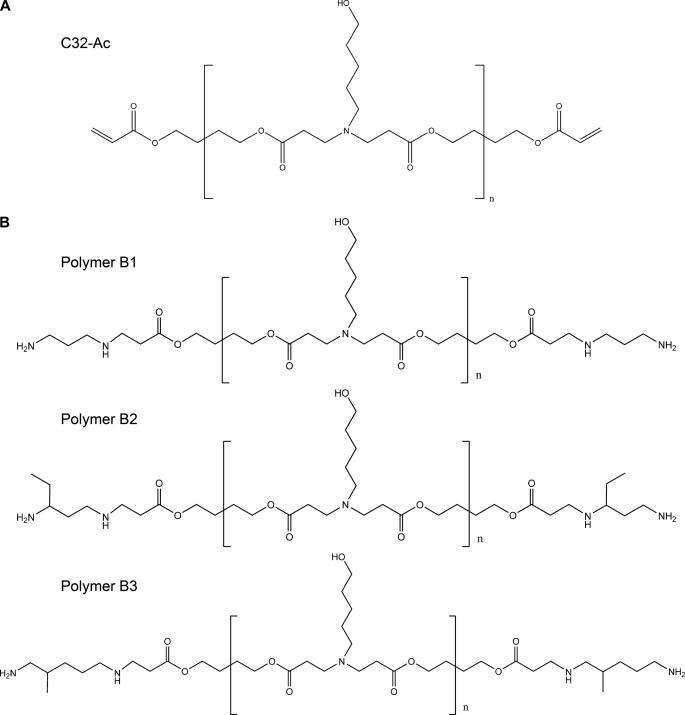
Poly-β-amino esters are a kind of biocompatible polymers that are able to complex DNA by electrostatic interactions forming nanoparticles suitable for gene delivery applications. The presence of ester bonds in their chemical structure confers them the properties of biodegradability and low toxicity. In addition, they are easy to synthesize by a conjugate addition of amine monomers to diacrylates without the production of any byproducts. The development of a large library of 2350 structurally unique poly-β-amino esters using high-throughput combinatorial chemistry was previously reported (26, 27). In that study it was shown that one of the best performing polymers, called C32 polymer, was obtained from an amino alcohol and a hydrophobic diacrylate monomer. The chemical structure of this polymer is shown in Fig. 1A. Moreover, it was reported that chemical end-modification of C32 polymer by the reaction with diamine monomers resulted in improved biomaterials for gene delivery (31, 32).
FIGURE 1.
Chemical structures of poly-β-amino esters. Poly-β-amino esters were synthesized as previously described. First, acrylate-terminated C32 polymer (A) was obtained by the conjugate addition of 5-amino-1-pentanol to 1,4-butanediol diacrylate at a 1.2:1 molar ratio of diacrylate monomer to amine monomer. Next, C32-Ac polymer was end-modified by the reaction with an excess of various diamine monomers to obtain the amine-terminated poly-β-amino esters B1, B2, and B3 (B).
Based on these previous reports, we have synthesized the three different end-modified poly-β-amino esters shown in Fig. 1B. Amine-terminated poly-β-amino esters polymers were obtained using a two-step procedure as previously described (28). B1, B2, and B3 amine-terminated poly-β-amino esters were characterized in terms of molecular structure by 1H NMR and FTIR. 1H NMR spectra were consistent with previously published data (28, 29) and confirmed the synthesized structures.
We next investigated the capacity of B1, B2, and B3 polymers to complex DNA and condense the pCAG-OSKMG vector into nanoparticles with appropriate biophysical properties for transfection. We used the pmaxGFP plasmid (4 kb) as the control plasmid for transfection, because as previously reported, the efficiency of transfection of plasmids around this size using these polymers is between 20 and 50% (33). A polymer complex of the pCAG-OSKMG plasmid was optimized based on visual analysis of GFP-expressing cells under a fluorescent microscope. Initial cell density (100,000, 400,000, or 800,000 cells/well of a 6-well plate), amounts of DNA (2.5, 5, or 10 μg/well), DNA to polymer ratio (1:20, 1:30, 1:40, 1:50, 1:80 or 1:100), and time of transfection (30 min and1, 2, or 3 h) were optimized to define the optimal conditions for the delivery of the large pCAG-OSKMG plasmid. It was found that high cell densities were required to efficiently transfect human fibroblasts with this system. Increased amounts of DNA also had a positive effect on the transfection efficiency. The optimal DNA to polymer ratio (w/w) was 1:50 for the 3 polymers, and incubation of the nanoparticles with the cells for 1 h was enough to allow cellular uptake (Fig. 2). Lower DNA to polymer ratios (1:20, 1:30, 1:40) resulted in insufficient complex of pCAG-OSKMG plasmid and, therefore, no transfection. In contrast, higher ratios (1:60, 1:80, 1:100) resulted in aggregation of the particles, which induced increased toxicity, giving rise to cell death (not shown). In summary, the best performing conditions for pCAG-OSKMG transfection into human fibroblasts were observed at 10 μg of DNA per well, a 1:50 DNA to polymer ratio, and a 1-h incubation time. Therefore, the latter experimental conditions were used in the following experiments.
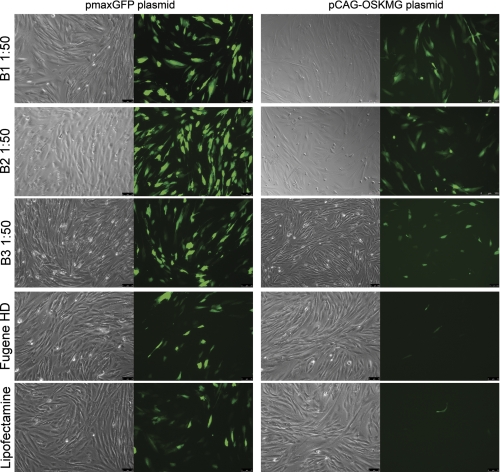
FIGURE 2.
Transfection of pmaxGFP (3.4 kb) and pCAG-OSKMG (11 kb) plasmids into human fibroblasts using amine-terminated poly-β-amino esters. Polyplexes of B1, B2, and B3 polymers with pmaxGFP and pCAG-OSKMG plasmids were formed at an optimal ratio of DNA to polymer of 1:50 (w/w). Conventional transfection reagents Lipofectamine 2000 and FuGENE HD were also used for comparing the efficiency of transfection. Expression of GFP in transfected human fibroblasts 1 day post-transfection with pmaxGFP (3.4 kb) and pCAG-OSKMG (11 kb) plasmids is shown.
In addition, the ability of B1, B2, and B3 polymers to condense DNA into nanoparticles was assessed by particle size and ζ potential analysis. It is well known that nanoparticles with sizes smaller than 300 nm and positive ζ potentials are needed for efficient transfection of cells (32). Complex of pmaxGFP plasmid with B1, B2, and B3 at a 1:50 ratio of DNA to polymer resulted in nanoparticle formation with average particle sizes between 209 and 269 nm and average positive ζ potentials between 12 and 26 mV (Table 1). In the case of pCAG-OSKMG complex, average particle sizes ranged from 189 to 322 nm, with positive ζ potentials between 12 and 16 mV. The nature of the diamine capping moieties in each polymer showed to have a slight influence on these properties. Particles prepared with B3 polymer were the smallest in size. This tendency was more pronounced in the formation of the complex with the large pCAG-OSKMG plasmid. Structurally, B3 has a slightly longer hydrocarbon spacer between the terminal amine and the base C32 polymer when compared with B1 and B2. B1 and B2 polymers showed similar particle sizes; however, B2 was more positively charged than B1, and this tendency was observed for both pmaxGFP and pCAG-OSKMG plasmids. Structurally, these polymers only differed in the presence of a hydrophobic group near the terminal amine. B2 polymer has an ethyl group near the terminal amine, whereas B1 polymer had an unsubstituted alkyl chain. Interestingly, particle size was found to be almost independent from the plasmid length. pCAG-OSKMG-polymer complexes formed slightly bigger particles and were less positively charged than pmaxGFP plasmid. However, the biophysical properties of the nanoparticles obtained for both plasmids accomplished the requirements needed for transfection. Overall, these data demonstrated that B1, B2, and B3 amine-terminated poly-β-amino esters were able to condense DNA vectors up to 11 kb in nanoparticles with appropriate biophysical properties for gene delivery to human fibroblasts.
TABLE 1.
Particle size and ζ potential measurements of the different polymer formulations complexed with pmaxGFP or OSKMG plasmids
The results are expressed as the mean ± S.D. (n = 3).
| Polymer formulation (DNA/polymer ratio) | Particle size |
ζ Potential |
||
|---|---|---|---|---|
| pmaxGFP | pCAG-OSKMG | pmaxGFP | pCAG-OSKMG | |
| nm | mV | |||
| B1 (1:50) | 269 ± 2.2 | 322 ± 16 | 12.7 ± 0.7 | 12.0 ± 1.3 |
| B2 (1:50) | 243 ± 20 | 281 ± 86 | 26.7 ± 0.9 | 14.4 ± 0.7 |
| B3 (1:50) | 209 ± 3.1 | 189 ± 6.1 | 16.3 ± 0.5 | 15.7 ± 0.9 |
Efficiency of pCAG-OSKMG Plasmid Delivery to Human Fibroblasts
The next crucial step in delivery is the cellular uptake of the nanoparticles, and the smaller and more positively charged particles are more favorable for cellular internalization than larger particles and weakly charged particles. However, after cellular uptake, the efficiency of transgene expression may depend on the mechanism of DNA release from the complexes and the intracellular migration of DNA from the cytoplasm to the nucleus. In addition, it has been reported that the transfection efficiency using this kind of polymers is also dependent on the cell type, showing that small structural changes affecting only the ends of the polymer can significantly increase the transfection efficiency to a specific cell type but not to others (28, 31). This demonstrates the difficulty of prediction of the transfection efficiency of these polymers, as not only biophysical parameters but also other parameters, such as the cell type, can influence cellular uptake and transgene expression.
Then we quantified the efficiency of transfection of the synthesized polymers by flow cytometry analysis of the GFP expression 1 day after the transfection with either pmaxGFP or pCAG-OSKMG plasmid. We compared the efficiency of transfection of these polymers with conventional transfection reagents Lipofectamine 2000 and FuGENE HD. Although the nanoparticles obtained with both plasmids showed appropriate biophysical properties (i.e. small particle size and positive charge), transfection efficiency was clearly influenced by the plasmid size. Representative fluorescent micrographs of pmaxGFP and pCAG-OSKMG transfection are shown in Fig. 2.
pmaxGFP plasmid complexed with B1, B2, and B3 polymers showed efficiencies of transfection into human fibroblasts between 50 and 70% and cell viabilities around 90%, whereas Lipofectamine 2000 and FuGENE HD resulted in transfection efficiencies around 50 and 25%, respectively, with significantly lower cell viability (Fig. 3, A and C). In contrast, pCAG-OSKMG plasmid was delivered to human fibroblasts in a less efficient manner than pmaxGFP plasmid, resulting in an average percentage of transfected cells of 10% with B1 and B2 being the best performing polymers (Fig. 3, B and C). These results confirm the behavior observed in the fluorescent microscope images (Fig. 2). Lipofectamine and FuGENE-based protocols achieved only 3 and 5% transfection of cells, respectively, and showed low cell viabilities (Fig. 3, A and C). Importantly, B1, B2, and B3 polymers were able to transfect the large pCAG-OSKMG vector into human fibroblasts with efficiencies up to 3 and 2 times higher than Lipofectamine and FuGENE HD, respectively (Fig. 3, A and B). Moreover, the mean fluorescence intensity derived from the samples transfected with B1, B2, and B3 polymers was higher than the levels of fluorescent intensity showed by Lipofectamine and FuGENE (Fig. 3D), suggesting that the levels of protein produced by cells transfected with poly-β-amino esters were higher.
It is known that cell reprogramming by transient expression methods, such as plasmid transfection, results in much lower induction efficiency of iPS cells when compared with viral methods. In addition, it is widely known that human fibroblasts are shown to have lower reprogramming efficiencies than mouse fibroblasts. Although the complexity of the reprogramming process includes a lot of variables and its mechanism still remains unclear, one of the possible hurdles of the plasmid transfection methodologies could rely on the low transfection efficiency achieved during the delivery of the reprogramming factors. In this sense, increased efficiencies of transgene delivery could eventually improve the reprogramming process. Therefore, in view of the data presented here, poly-β-amino esters represent a good alternative for the delivery of the reprogramming factors into human fibroblasts.
Generation of Human iPS Cells after Transfection of the pCAG-OSKMG Plasmid Using End-modified Poly-β-amino Esters
After testing the potential of B1, B2, and B3 polymers for gene delivery of the pCAG-OSKMG vector, we sought to reprogram human foreskin fibroblasts following a methodology of serial transfections with the polymers. It is known that to induce stable pluripotency to cells, it is necessary to enforce the expression of the pluripotency factors for at least 8–10 days (13). Thus, we designed a panel of reprogramming experiments at different conditions of initial cell density and performing one, two, three, or four consecutive transfections of the pCAG-OSKMG plasmid either with B1 or B2 polymers (Table 2.). At this point, the use of B3 polymer was discontinued because of its higher toxicity and lower transfection efficiency.
TABLE 2.
Summary of the different experimental conditions used on the reprogramming of human fibroblasts (the best conditions are highlighted in bold)
| No. cells/well (6-well plate) | Polymer | OSKMG/polymer ratio | OSKMG | No. of transfections | Total colonies (p0) | GFP positive colonies | GFP negative colonies |
|---|---|---|---|---|---|---|---|
| μg/well | |||||||
| 100,000 (1×) | B1 | 1:50 | 10 | 3 | |||
| 400,000 (1×) | B1 | 1:50 | 10 | 3 | 9 | 8 | 1 |
| 800,000 (1×) | B1 | 1:50 | 10 | 3 | 5 | 5 | 0 |
| 100,000 (1×) | B2 | 1:50 | 10 | 3 | |||
| 400,000 (1×) | B2 | 1:50 | 10 | 3 | 61 | 4 | 57 |
| 800,000 (1×) | B2 | 1:50 | 10 | 3 | 10 | 2 | 8 |
| 800,000 (30×) | B1 | 1:50 | 10 | 4 | |||
| 800,000 (30×) | B2 | 1:50 | 10 | 4 | 12 | 11 | 1 |
| 400,000 (6×) | B1 | 1:50 | 10 | 2 | 200–250 | 20 of 70 | 50 of 70 |
| 400,000 (6×) | B2 | 1:50 | 10 | 2 | |||
| 400,000 (6×) | B1 | 1:50 | 10 | 1 | |||
| 400,000 (6×) | B2 | 1:50 | 10 | 1 |
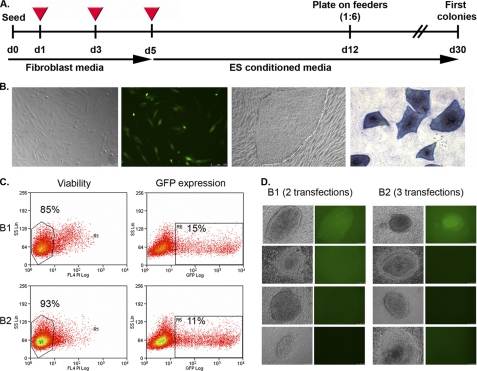
The timeline of the reprogramming protocol is depicted in Fig. 4A. One day after the first transfection with B1 and B2 polymers, 10–15% of cells expressed the pCAG-OSKMG vector, as shown by FACS analysis (Fig. 4C). After the last transfection, fibroblast medium was changed to embryonic stem cell growth medium, and 1 week later cells were trypsinized and plated on irradiated human feeders in a 1:6 dilution. Fig. 4B shows representative images of transfected human fibroblasts with pCAG-OSKMG, a typical hiPS colony reprogrammed with polymer transfection, and a positive alkaline phosphatase staining of these colonies. Two, three, or four consecutive transfections produced iPS-like colonies ∼20–28 days after the seeding on irradiated human feeders. Colonies were left to grow for at least 1 week. After this time, single clones were manually passaged onto fresh irradiated human feeders. Initial colonies did not resemble morphologically a typical embryonic stem cell colony (Fig. 4D). However, after 2 or 3 passages they acquired the characteristic morphology of an embryonic stem cell colony.
FIGURE 4.
Generation of hiPS cells using amine-terminated poly-β-amino esters together with the pCAG-OSKMG vector as the non-viral gene delivery system. A, shown is a timeline of the reprogramming process with two or three consecutive transfections of the pCAG-OSKMG plasmid using B1 and B2 polymers respectively. B, shown are representative microscope images (phase contrast and fluorescent images) of initial human fibroblasts expressing the GFP 1 day after the transfection of pCAG-OSKMG, a typical iPS cell colony obtained with the use of amine-terminated poly-β-amino esters, and the positive staining for alkaline phosphatase of these iPS cell colonies, as a first marker of pluripotency. C, FACS analysis of GFP expression and viability of human fibroblasts 1 day post-transfection with pCAG-OSKMG using B1 or B2 polymers are shown. D, representative microscope images (phase contrast and fluorescent images) of initial human iPS clones (passage zero) obtained by transfection with B1 and B2 polymers are shown. After 2 or 3 passages these iPS clones acquired the characteristic morphology of an embryonic stem cell colony.
We observed that a percentage of the generated colonies maintained the expression of GFP, suggesting that the transgene was still active in these colonies. Thus, the GFP reporter gene allowed the screening of GFP negative colonies, where the plasmid was either not integrated into the cell genome or was integrated and subsequently silenced. Then, we only selected colonies that were GFP negative by visual inspection under the fluorescent microscope to screen for possible transgene-free human iPS clones. Experiments performed with three transfections using B2 polymer and two transfections using B1 polymer resulted in a higher numbers of colonies, whereas one transfection was not enough to give rise to reprogrammed colonies (Table 2).
We selected colonies from the best-performing experimental conditions. A total of 61 colonies at passage 0 from initially seeded 400,000 human fibroblasts were obtained using a protocol of 3 serial transfections with pCAG-OSKMG and B2 polymer (B2-three transfections) (Table 2). We picked a total of 57 colonies (GFP negative) from which we successfully established 15 hiPS cell lines. When a protocol of two serial transfections with pCAG-OSKMG and B1 polymer was followed (B1-two transfections), a total of 200–250 colonies at passage 0 from initially seeded 2,400,000 human fibroblasts were obtained (Table 2). We picked a total of 50 colonies (GFP negative) from which we established 13 hiPS cell lines.
We characterized 6 hiPS cell lines, 3 clones from B2-three transfections protocol (clones #40, #56, and #26) and three from B1-two transfections protocol (clones #61, #37, and #27). All of them showed strong alkaline phosphatase activity (Fig. 5).
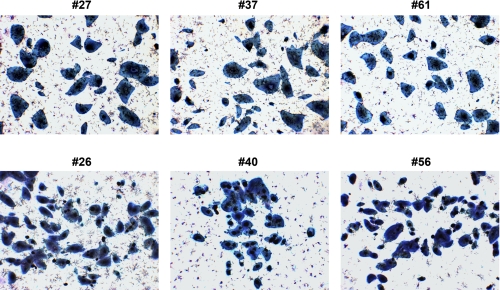
FIGURE 5.
Alkaline phosphatase staining of iPS cell lines #27, #37, #61, #26, #40, and #56.
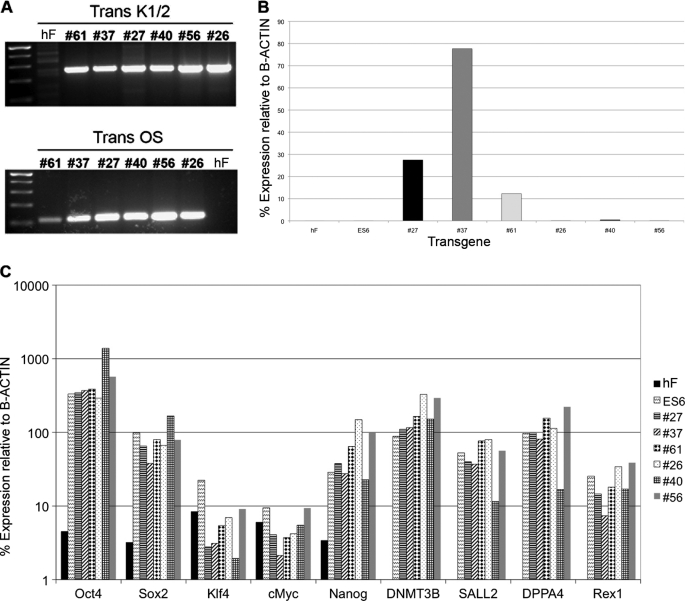
Then we subjected the established iPS cell lines to a PCR of genomic DNA by using two pair of primers (K1/2 and OS primers) specially designed to amplify specific regions of the pCAG-OSKMG vector and found that all the six clones analyzed had integrated the transgene (Fig. 6A). It is reported that the probability of having a transgene-free iPS clone by using non-viral methods is extremely low. Although we did not have any transgene-free human iPS cell line among the selected clones, we proceeded to further characterize them in terms of transgene silencing, pluripotency, and differentiation potential.
FIGURE 6.
Characterization of the hiPS cell lines reprogrammed using amine-terminated poly-β-amino esters. A, shown is a genomic DNA PCR analysis of the transgene integration in hiPS clones #27, #37, #61, #26, #40, and #56, confirming the insertion of the transgene. B, quantitative RT-PCR analysis of the transgene silencing in hiPS clones #27, #37, #61, #26, #40, and #56 is shown. C, quantitative RT-PCR analysis shows up-regulation of endogenous expression of the pluripotency markers Oct4, Sox2, Klf4, cMyc, Nanog, DNMT3B, SALL2, DPPA4, and Rex1 in hiPS clones #27, #37, #61, #26, #40, and #56.
Transgene expression at the RNA level was analyzed by quantitative RT-PCR. We observed that iPS clones at very early passages still had some expression of the polycistronic transcript (data not shown). However, after several passages we found that clones #26, #40, and #56 had successfully silenced it (Fig. 6B). In contrast, clones #27, #37, and #61 still maintained some transgene expression (Fig. 6B), suggesting that these clones were not able to silence the transgene. These results are in accordance with previous published data showing that a percentage of the integrated clones did not silence the transgene even after several passages (17).
Quantitative RT-PCR revealed that all the tested clones had reactivated a set of endogenous pluripotency markers including Oct4, Sox2, Nanog, and Rex1 among others (Fig. 6C), comparable with a human embryonic stem cell line (ES6) (35), showing that the obtained iPS cell lines had pluripotency properties similar to human embryonic stem cells.
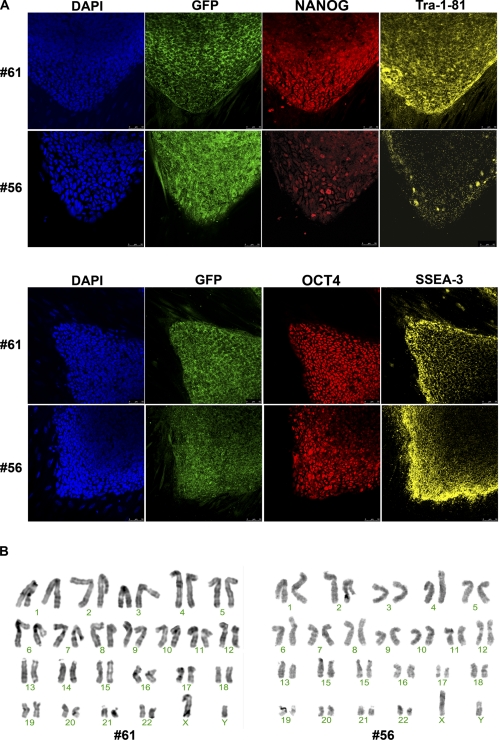
Then we proceeded to analyze the clones by immunofluorescence for Nanog, Tra-1–81, Oct4, and SSEA-3 expression and revealed the expression of these pluripotency markers in all (Fig. 7A), confirming their pluripotency. In addition, we also analyzed the expression of GFP by immunofluorescence (Fig. 7A). Even the clones that had silenced the transgene showed some GFP expression at the level of protein. This is explained by the fact that GFP is known to be a very stable protein and, therefore, still detectable by immunofluorescence by the time of transgene silencing. In addition, high resolution G-banded karyotype of the six clones revealed a normal, diploid, male chromosomal content (Fig. 7C).
FIGURE 7.
Expression of pluripotency markers and karyotype analysis of hiPS cell lines reprogrammed using amine-terminated poly-β-amino esters. A, immunofluorescence analysis for pluripotency markers of hiPS cell lines #61 and #56 is shown. The colonies express the embryonic markers SSEA-3 and TRA-1–81 and the transcription factors OCT4 and NANOG. Underlying fibroblasts provide a negative control. Scale bars, 50 μm. B, high resolution, G-banded karyotype indicating a normal, diploid, male chromosomal content in hiPS cell lines #61 and #56 is shown.
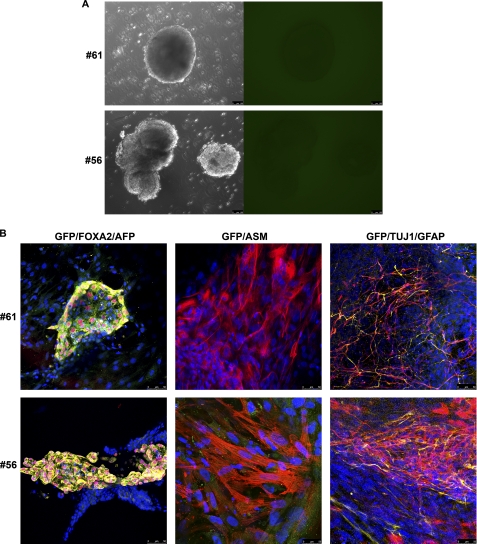
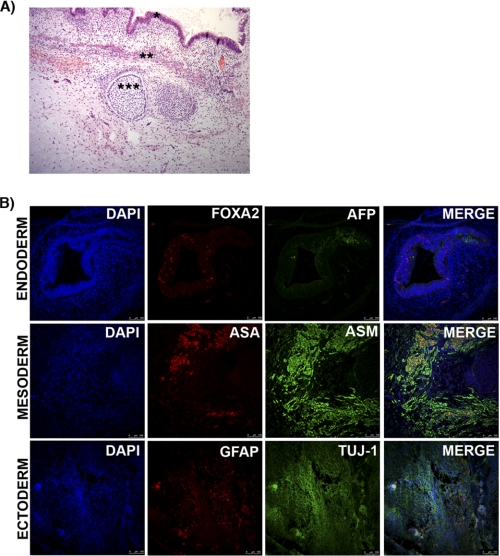
We also tested the differentiation potential of these clones by generating embryoid bodies and differentiating them into the three germ layers (Fig. 8A). All were able to differentiate into derivatives of the three germ layers, showing positive immunofluorescence staining for markers of endoderm, mesoderm, and ectoderm (Fig. 8B). Interestingly, we also observed that differentiating cells from the iPS clones that had not silenced the transgene gradually decreased the expression of GFP, suggesting complete transgene silencing during the differentiation process. Finally, teratoma formation assay was conducted by injecting hiPS cells in the testis of SCID mouse. After 8 weeks, all the lines were able to form complex teratoma, indicating that our iPS lines were able to differentiate in vivo into derivatives of the three germ layers. In agreement with our observation by in vitro differentiation tests, the six iPS clones showed no expression of GFP, suggesting complete silence of transgene expression (Fig. 9).
FIGURE 8.
In vitro differentiation of hiPS cell lines reprogrammed using amine-terminated poly-β-amino esters. hiPS cell lines generated with amine-terminated poly-β-amino esters transfection of pCAG-OSKMG were differentiated in vitro through the formation of embryoid bodies (A), which did not express GFP as observed by fluorescent microscopy, indicating the silencing of the transgene. B, after generation of embryoid bodies, hiPS cells were specifically directed to differentiate into the three primary germ cell layers: endoderm-FOXA2 (red) and AFP (yellow), mesoderm-α-smooth muscle (ASM; red), and ectoderm-Tuj1 (red) and glial fibrillary acidic protein (GFAP; yellow). GFP (green) was also analyzed.
FIGURE 9.
Representative images of in vivo differentiation of hiPS line #61 into the three primary germ cell layers. A, after 8 weeks teratomas were collected and analyzed by conventional histology. hiPS cells were differentiated in vivo to endoderm (*), mesoderm (**), or ectoderm fates (***). B, shown is the immunofluorescence analysis for the three primary germ cell layers: endoderm (nuclear FOXA2 in red and α1-fetoprotein (AFP) in green); mesoderm (α-smooth muscle (ASM) in red and α-sarcomeric actinin (ASMA); ectoderm (glial fibrillary acidic protein (GFAP) in red and neuron-specific class III β-tubulin (Tuj-1) in green. Scale bar, 100 μm.
In summary, although the six iPS clones presented evidence of transgene integration, the reprogramming methodology developed in this work represents a simple method to reprogram human fibroblasts without the need of using viral vectors. The use of poly-β-amino esters in this methodology as a transfection reagent for the delivery of the reprogramming DNA vector into human fibroblasts has been crucial, as Lipofectamine and FuGENE showed very low transfection efficiencies.
DISCUSSION
Here we have described for the first time the successful delivery of the large pCAG-OSKMG plasmid (11 kb) to human fibroblasts by using three different end-modified poly-β-amino esters as non-viral delivery system. These kinds of polymers were previously described to be good candidates for gene delivery applications in human cells (30). Therefore, we chose them as a promising non-viral delivery system to develop an optimized plasmid transfection strategy to generate human iPS cells from human fibroblasts.
We first demonstrated that poly-β-amino esters could condense the pCAG-OSKMG plasmid-forming nanoparticles with appropriate biophysical properties to allow cellular uptake. Interestingly, particle size was found to be almost independent from the plasmid length. pCAG-OSKMG-polymer complexes formed particles of similar size and ζ potential than pmaxGFP-polymer complexes. Nevertheless, cellular uptake and transgene expression 24 h after transfection were clearly influenced by the plasmid size, as the pCAG-OSKMG plasmid was less efficiently transfected than the pmaxGFP plasmid. This could be explained by the fact that the efficiency of transgene expression may depend on other parameters such as the mechanism of DNA release from the complexes and the intracellular migration of DNA from the cytoplasm to the nucleus. Remarkably, these polymers could improve the delivery of the pCAG-OSKMG plasmid into human fibroblasts with a 3-fold increase in the transfection efficiency when compared with conventional transfection reagents, including Lipofectamine and FuGENE.
Next, we developed a simple non-viral protocol to generate hiPS cells by transient expression of the single non-viral polycistronic vector (pCAG-OSKMG) in human foreskin fibroblasts. We used the synthesized polymers to sequentially deliver the pCAG-OSKMG plasmid containing the pluripotency associated transcription factors Oct4, Sox2, Klf4, and cMyc and a GFP reporter gene. The use of these polymers for the delivery of the reprogramming vector pCAG-OSKMG into human fibroblasts has been essential for the generation of human iPS-like colonies after an optimized plasmid-transfection strategy. Our group previously reported the reprogramming of mouse embryonic fibroblasts by transient expression of the pCAG-OSKM vector (17). However, human fibroblasts are harder to reprogram than mouse embryonic fibroblasts, and commercial transfection reagents have shown very poor transfection efficiencies of large plasmids (such as the one used here) into human fibroblasts. In this work we report a significant improvement of the transfection efficiency of the pCAG-OSKMG plasmid by the use of poly-β-amino esters, which allowed us to develop an optimized and simple methodology for the reprogramming of human fibroblasts. These polymers could deliver the factors into human fibroblasts and maintain their expression during a period of time enough to generate human iPS cell colonies. We characterized six hiPS cell lines that showed expression of the typical set of pluripotency marker genes, had a stable karyotype, and were able to differentiate in vitro and in vivo into derivatives of the three germ layers. However, we found that all the six clones analyzed had integrated the transgene. Although this strategy was not able to produce transgene-free hiPS cells, it constitutes an alternative and a simple approach to generate hiPS cells without the need of working with time-consuming and harmful viral systems.
Moreover, recent investigations have shown that the reprogramming process could be enhanced by modification of the cell signaling (36), treatment with chemical drugs (34), or using adult stem cells as a starting cell population (8, 9, 11). Therefore, the combination of these findings with the methodology presented here could represent a promising approach to further improve the reprogramming of human cells in future studies.
Acknowledgments
We are grateful to Meritxell Carrió and Vanesa Tobajas for expert assistance with cell culture techniques, Mercè Gaudes Martí, Lola Mulero Pérez, and Cristina Pardo for bioimaging assistance, and Montserrat Barragan and Gustavo Tiscornia for helpful discussions.
This work was supported in part by a fellowship from the Sara Borrell program (to E. G.), by the Juan de La Cierva program (N. M.), the Swiss National Science Foundation (to F. G.), CIBER, MICINN, Fundacion Cellex, Sanofi-Aventis, and the G. Harold and Leila Y. Mathers Charitable Foundation (to J. C. I. B.).
- MEF
- mouse embryonic fibroblast
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- hiPS
- human iPS
- OSKM
- Oct4, Sox2, Klf4, and c-myc.
REFERENCES
- 1. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Plath K., Hochedlinger K. (2007) Cell Stem Cell 1, 55–70 [DOI] [PubMed] [Google Scholar]
- 3. Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 4. Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 5. Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008) Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 6. Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H., Jiang W., Cai J., Liu M., Cui K., Qu X., Xiang T., Lu D., Chi X., Gao G., Ji W., Ding M., Deng H. (2008) Cell Stem Cell 3, 587–590 [DOI] [PubMed] [Google Scholar]
- 7. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 8. Eminli S., Utikal J., Arnold K., Jaenisch R., Hochedlinger K. (2008) Stem Cells 26, 2467–2474 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M. J., Ruau D., Han D. W., Zenke M., Schöler H. R. (2008) Nature 454, 646–650 [DOI] [PubMed] [Google Scholar]
- 10. Aasen T., Raya A., Barrero M. J., Garreta E., Consiglio A., Gonzalez F., Vassena R., Biliæ J., Pekarik V., Tiscornia G., Edel M., Boué S., Izpisúa Belmonte J. C. (2008) Nat. Biotechnol. 26, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 11. Giorgetti A., Montserrat N., Aasen T., Gonzalez F., Rodríguez-Pizà I., Vassena R., Raya A., Boué S., Barrero M. J., Corbella B. A., Torrabadella M., Veiga A., Izpisua Belmonte J. C. (2009) Cell Stem Cell 5, 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., Jaenisch R. (2008) Cell Stem Cell 2, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wernig M., Lengner C. J., Hanna J., Lodato M. A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R. (2008) Nat. Biotechnol. 26, 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 15. Zhou W., Freed C. R. (2009) Stem Cells 27, 2667–2674 [DOI] [PubMed] [Google Scholar]
- 16. Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. (2008) Science 322, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez F., Barragan Monasterio M., Tiscornia G., Montserrat, Pulido N., Vassena R., Batlle Morera L., Rodriguez Piza I., Izpisua Belmonte J. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8918–8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. (2009) Nature 458, 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008) Science 322, 949–953 [DOI] [PubMed] [Google Scholar]
- 20. Woltjen K., Michael I. P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M., Kaji K., Sung H. K., Nagy A. (2009) Nature 458, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin II., Thomson J. A. (2009) Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., Siuzdak G., Schöler H. R., Duan L., Ding S. (2009) Cell Stem Cell 4, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R., Kim K. S. (2009) Cell Stem Cell 4, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryan M. D., Drew J. (1994) EMBO J. 13, 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. (2008) Cell Stem Cell 3, 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson D. G., Lynn D. M., Langer R. (2003) Angew. Chem. Int. Ed. Engl. 42, 3153–3158 [DOI] [PubMed] [Google Scholar]
- 27. Anderson D. G., Akinc A., Hossain N., Langer R. (2005) Mol. Ther. 11, 426–434 [DOI] [PubMed] [Google Scholar]
- 28. Green J., Zugates G., Tedford N., Huang Y., Griffith L., Lauffenburger D., Sawicki J. A., Langer R., Anderson D. G. (2007) Advanced Materials 19, 2836–2842 [Google Scholar]
- 29. Zugates G. T., Peng W., Zumbuehl A., Jhunjhunwala S., Huang Y. H., Langer R., Sawicki J. A., Anderson D. G. (2007) Mol. Ther. 15, 1306–1312 [DOI] [PubMed] [Google Scholar]
- 30. Yang F., Cho S. W., Son S. M., Bogatyrev S. R., Singh D., Green J. J., Mei Y., Park S., Bhang S. H., Kim B. S., Langer R., Anderson D. G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3317–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Green J. J., Zhou B. Y., Mitalipova M. M., Beard C., Langer R., Jaenisch R., Anderson D. G. (2008) Nano. Lett. 8, 3126–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Green J. J., Langer R., Anderson D. G. (2008) Acc. Chem. Res. 41, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang F., Green J. J., Dinio T., Keung L., Cho S. W., Park H., Langer R., Anderson D. G. (2009) Gene Ther. 16, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raya A., Rodríguez-Pizà I., Arán B., Consiglio A., Barri P. N., Veiga A., Izpisúa Belmonte J. C. (2008) Cold Spring Harb. Symp. Quant. Biol. 73, 127–135 [DOI] [PubMed] [Google Scholar]
- 36. Marson A., Foreman R., Chevalier B., Bilodeau S., Kahn M., Young R. A., Jaenisch R. (2008) Cell Stem Cell 3, 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]