Abstract
Plectin belongs to the plakin family of cytoskeletal crosslinkers, which is part of the spectrin superfamily. Plakins contain an N-terminal conserved region, the plakin domain, which is formed by an array of spectrin repeats (SR) and a Src-homology 3 (SH3), and harbors binding sites for junctional proteins. We have combined x-ray crystallography and small angle x-ray scattering (SAXS) to elucidate the structure of the central region of the plakin domain of plectin, which corresponds to the SR3, SR4, SR5, and SH3 domains. The crystal structures of the SR3-SR4 and SR4-SR5-SH3 fragments were determined to 2.2 and 2.95 Å resolution, respectively. The SH3 of plectin presents major alterations as compared with canonical Pro-rich binding SH3 domains, suggesting that plectin does not recognize Pro-rich motifs. In addition, the SH3 binding site is partially occluded by an intramolecular contact with the SR4. Residues of this pseudo-binding site and the SR4/SH3 interface are conserved within the plakin family, suggesting that the structure of this part of the plectin molecule is similar to that of other plakins. We have created a model for the SR3-SR4-SR5-SH3 region, which agrees well with SAXS data in solution. The three SRs form a semi-flexible rod that is not altered by the presence of the SH3 domain, and it is similar to those found in spectrins. The flexibility of the plakin domain, in analogy with spectrins, might contribute to the role of plakins in maintaining the stability of tissues subject to mechanical stress.
Keywords: Cytoskeleton, Intermediate Filaments, Protein Structure, SH3 Domains, X-ray Crystallography, X-ray Scattering, Plakin, Spectrin Repeat
Introduction
Plakins are a family of high molecular weight proteins that interconnect elements of the cytoskeleton and tether them to membrane-associated structures; hence, they are also known as cytolinkers (1–2). Mammalian plakins include desmoplakin, plectin, the bullous pemphigoid antigen 1 (BPAG1),4 the microtubule actin crosslinking factor 1 (MACF1, also known as ACF7, trabeculin, or macrophin), envoplakin, periplakin, and epiplakin. Plakins are also present in invertebrates; Drosophila melanogaster has a single plakin gene named shortstop (also know as kakapo) that encodes at least two protein forms, Shot I and Shot II. Similarly, the only plakin gene in Caenorhabditis elegans, vab-10, encodes two variants VAB-10A and VAB-10B.
Plectin is a highly versatile plakin that associates with intermediate filaments (3), microtubules (4), and actin fibers (5), and crosslinks these cytoskeletal networks (5–6). Plectin also connects intermediate filaments to membrane-associated complexes. In stratified epithelia such as the skin, plectin is localized at the hemidesmosomes, which are junctional complexes that link the intermediate filaments to the basement membrane, and links the integrin α6β4 and the bullous pemphigoid antigen 2 (BPAG2, also known as type XVII collagen or BP180) to the cytokeratins (7–8). Plectin is also localized at the desmosomes (9), which mediate cell-cell contacts, and connects the nuclear envelope to the intermediate filaments by binding to the outer nuclear membrane protein nesprin-3α (10–11). In striated muscle, plectin is localized at the Z-line and the costameres, where it associates with the intermediate filament protein desmin and with components of the dystrophin glycoprotein complex (12–15). The contribution of plectin to preserve the integrity of tissues that are exposed to mechanical stress is illustrated by the effect of mutations in the PLEC gene, which lead to a severe skin blistering disease called epidermolysis bullosa simplex that is characterized by defects at the level of the hemidesmosomal cell-basal membrane junction and is frequently associated with late-onset muscular dystrophy (16–17).
Plectin (∼500 kDa) has a tripartite structure consisting of N- and C-terminal regions separated by a central rod domain (Fig. 1A). Other epithelial plakins such as BPAG1e/n, desmoplakin, periplakin, and envoplakin have the same structure. Near the N terminus plectin contains an actin binding domain (ABD) built up of a tandem pair of calponin homology domains (CH1 and CH2), similar to the ABDs present in dystrophin and other members of the spectrin superfamily. The ABD binds to the first pair of fibronectin type III repeats of the integrin β4 subunit via the CH1 domain (18–20); the ABD also binds to F-actin (21–22), nesprin-3α (11), and to nonfilamentous vimentin (23). Adjacent to the ABD extends a ∼1000 residue long region named the plakin domain that is conserved in all plakins, except in epiplakin. The plakin domain of plectin and other plakins contain protein-protein interaction sites and they are important for the localization of plakins at junctional complexes. In plectin this region harbors binding sites for the integrin β4 subunit (24), the cytoplasmic domain of BPAG2 (25), β-dystroglycan (13), β-synemin (15), and the tyrosine kinase Fer (26). The central region consists of a coiled-coil rod domain that acts as a structural spacer of the protein-protein binding sites located in the N- and C-terminal regions and mediates homo-dimerization of plectin. Finally, the C-terminal region contains six plakin repeat domains and harbors binding sites for intermediate filaments.
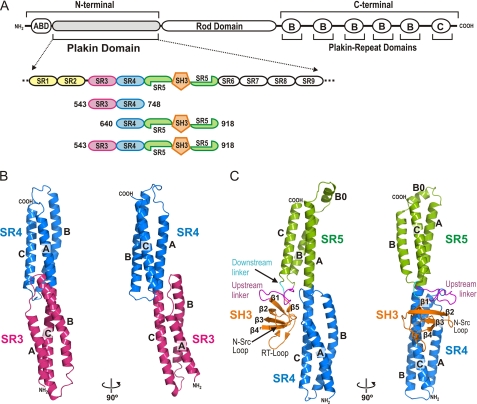
FIGURE 1.
Crystal structure of the SR3-SR4 and SR4-SR5-SH3 regions of the plakin domain of plectin. A, schematic representation of the domain structure of plectin and a close up view on the structure of the plakin domain. The three proteins characterized in this study are aligned underneath. B and C, two orthogonal views of ribbon representations of the crystal structures of the SR3-SR4 (B) and the SR4-SR5-SH3 (C) regions. The SR4 is shown in equivalent orientations in both panels.
Initial analysis of the sequence of the plakin domain suggested that this region contains multiple α-helical bundles (27), some of which were later identified as spectrin repeats (SR) (28). The presence of SRs in the plakin domain was confirmed by the crystal structures of two di-repeat fragments of BPAG1 (29) and plectin (30). The SR-fold (∼100-residues long) consists of three α-helices connected by short loops that pack in a left-handed helical bundle with up-down-up topology (31–32). The α-helices show a heptad pattern in which positions a and d are occupied by hydrophobic residues that pack at the core of the bundle. Adjacent SRs in the structures of BPAG1, plectin, and those of other pairs of SRs are connected by a helical linker in which the third α-helix (C) of the N-terminal repeat and the first α-helix (A) of the C-terminal SR are fused into a single helix. Thus, arrays of multiple SRs form rod-like structures, as it was illustrated by the crystal structure of a tetra-repeat rod of α-actinin (33). We have previously identified eight canonical SRs in the sequence of the plakin domain of plectin (SR1 to SR5 and SR7 to SR9), and an additional shorter SR-like domain (SR6) (Fig. 1A) (30). The available crystal structures of plectin and BPAG1 correspond to the SR1-SR2 and SR3-SR4, respectively. The SR2 and SR3 of plectin are connected by a ∼20-residues long linker predicted to be non-helical, while repeats SR3 to SR9 occur contiguous in the plectin sequence as observed in the structure of the SR3-SR4 pair of BPAG1 (29). In addition to the SRs, there is a Src-homology 3 (SH3) domain toward the middle of the plakin domain (29); this is of interest because SH3 domains mediate protein-protein interactions, frequently binding to Pro-rich sequences (34). The SH3 domain is inserted in the central repeat, SR5, and this arrangement is almost identical to that of α-spectrin in which there is an SH3 within the central SR; for which the relative organization of the SH3 and the SRs is unknown. Overall, the structural organization of the plakin domain is highly similar to that of α- and β-spectrins and other members of the spectrin superfamily, both regarding the repertoire of modules (ABD, SR, and SH3) and their arrangement.
Despite recent advances in the characterization of the structure and function of the plakin domain, important issues remained unanswered. Does the plakin domain adopt a rod-like structure formed by juxtaposed SRs similar to that of spectrins and related proteins? If so, does the insertion of the SH3 domain alter the SR array? Is the structure of the SH3 domain compatible with ligand binding and is its binding site accessible? To address these questions, we have combined x-ray crystallography and small angle x-ray scattering (SAXS) to elucidate the structure of the central region of the plakin domain of plectin, which includes the SR3, SR4, SR5, and the SH3; a region that is involved in protein-protein interactions. Our results have implications for the contribution of the plakin domain of plectin and other plakins to the stability of junctional complexes subject to mechanical forces.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA sequences coding for residues 543–748, 640–918, and 543–918 of human plectin (numbering correspond to the plectin 1c variant, UniprotKB accession number Q15149–2) were cloned into a modified version of the pET15b vector (35). Proteins were expressed in Escherichia coli strain BL21(DE3) and were purified by nickel-chelating affinity chromatography as described (22). The His tag present at the N terminus of the fusion proteins was cleaved by digestion with tobacco etch virus protease, and was removed by a second nickel-affinity chromatography.
Crystallization and Structure Determination of the SR3-SR4 Region
Crystals of SR3-SR4 (residues 543–748) were grown at 4 °C using vapor diffusion methods by mixing a protein solution at 31 mg/ml in 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 0.1 mm DTT with an equal volume of mother liquor consisting of 0.1 m HEPES (pH 7.5), 17% (w/v) polyethylene glycol (PEG) 4000, and 8% (v/v) isopropanol. Prior to data collection, crystals were transferred into a cryoprotectant solution consisting of 0.1 m HEPES (pH 7.5), 18% (w/v) PEG 4000, and 6% (v/v) isopropanol, and 20% (v/v) glycerol that was vitrified by direct immersion in liquid nitrogen. Data were collected at 100 K using a Microstar-H rotating anode x-ray generator (Bruker AXS) and a mar345dtb detector (Marresearch GmbH). Diffraction intensities were indexed and integrated with XDS and reduced with XSCALE (36).
Crystals belong to space group P21212 and contain two molecules in the asymmetric unit (∼54% solvent content) (Table 1). A mixed search model for molecular replacement (37) was built by homology modeling using as template the crystal structure of the equivalent region of BPAG1 (PDB code 2IAK)(29) that corresponds to ∼85% of the residues of the plectin fragment and have ∼65% sequence identity. Our data were then phased by molecular replacement using the program PHASER (38) within the CCP4 suite (39). Refinement was done against data to 2.22 Å resolution using phenix.refine (40), alternated with manual model building using COOT (41). This partial structure was completed based on additional electron density observed in the 2mFo−DFc maps. Simulated annealing was used in the initial stages of refinement, while gradient-driven positional refinement, individual isotropic B-factor restrained refinement and TLS refinement (42) were used at later stages. Three TLS groups identified by using the TLS Motion Determination server (43) were refined in each molecule. Non-crystallographic symmetry restraints were not included during refinement. Solvent molecules were built in peaks over 3σ of mfobs−Dfcalc maps and 1σ of 2mfobs−Dfcalc maps when reasonable H-bonding pattern was observed. Two elongated electron densities not belonging to the protein chains were modeled as two PEG fragments, ethylene glycol and di(hydroxyethyl)ether, respectively. The final model contains residues 545–746 of molecule A, residues 543–745 of the molecule B, 210 water molecules and two PEG fragments. The model has excellent geometry with 99.5% of the main chain torsion angles located in the favored regions of the Ramachandran plot.
TABLE 1.
Summary of crystallographic analysis
| Data Collection | |||
| Protein | SR3-SR4 (543–748) | SR4-SR5-SH3 (640–918) | |
| Data set | Native | Native | 1 mm EMTS |
| Space group | P21212 | P212121 | P212121 |
| Cell dimensions | a = 97.4 Å | a = 72.7 Å | a = 72.2 Å |
| b = 119.4 Å | b = 108.5 Å | b = 107.9 Å | |
| c = 44.6 Å | c = 112.1 Å | c = 113.0 Å | |
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418 |
| Resolution (Å) | 2.22 (2.30-2.22)a | 2.95 (3.05-2.95)a | 3.40 (3.53-3.40)a |
| Unique reflections | 26066 | 18051 | 22352b |
| Redundancy | 7.0 (5.9)a | 14.3 (14.5)a | 7.8 (7.7)a |
| Completeness (%) | 98.4 (92.1)a | 93.6 (85.5)a | 95.1 (98.6)a |
| Rmeasc (%) | 6.0 (44.4)a | 5.9 (43.9)a | 8.9 (62.4)a |
| Mean I/σ(I) | 23.1 (4.9)a | 29.4 (6.9)a | 16.5 (3.9)a |
| Risod (%) | 32.1 | ||
| Phasing power (iso acent/iso cent/ano) | 0.84/0.92/0.64 | ||
| FOM SHARP (acentric/centric) | 0.16/0.17 | ||
| Refinement statistics | |||
| Resolution range (Å) | 20–2.22 | 20–2.95 | |
| Unique reflections, work/free | 24757/1315 | 17102/925 | |
| Rwork (%) | 20.4 | 24.1 | |
| Rfreee (%) | 24.8 | 26.5 | |
| Number of | |||
| Residues | 407 | 542 | |
| Waters | 210 | 0 | |
| Ca2+ | 0 | 2 | |
| Other hetero-compounds | 2 | 0 | |
| Average B value (Å2) | |||
| Wilson plot | 34.0 | 75.3 | |
| Protein | 40.1/41.7 | 91.2/88.3 | |
| Solvent | 44.1 | - | |
| Ca2+ | - | 84.2 | |
| Other hetero-compounds | 52.9 | - | |
| Rmsd bond lengths (Å) | 0.004 | 0.013 | |
| Rmsd angles (°) | 0.679 | 1.031 | |
| Ramachandran plotf, residues in | |||
| Favored regions | 401 (99.5%) | 517 (96.8%) | |
| Additionally allowed | 2 (0.5%) | 17 (3.2%) | |
| Outliers | 0 (0.0%) | 0 (0.0%) | |
| PDB accession code | 3PDY | 3PE0 | |
a Numbers in parenthesis correspond to the outer resolution shell.
b Keeping Bijvoet pairs separate.
c Rmeas is the multiplicity independent R factor as described by Diederichs and Karplus (66).
d Riso = Σ||Fder| − |Fnat‖Σ/|Fnat|, where Fder is the heavy-atom derivative structure factor and Fnat is the protein structure factor.
e Calculated using 5% of reflections that were not included in the refinement.
f As defined in the program MOLPROBITY (67).
Crystallization and Structure Determination of the SR4-SR5-SH3 Region
Crystals of the SR4-SR5-SH3 protein (residues 640–918) were grown at 4 °C using vapor diffusion methods by mixing a protein solution at 15 mg/ml in 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 1 mm DTT with an equal volume of mother liquor consisting of 0.1 m imidazole (pH 7.2), 0.2 m Ca acetate, 9% (w/v) PEG 8000, and 2 mm DTT. After crystals were grown, the drop containing the crystals was equilibrated for ∼36 h against a solution with twice the concentration of all the components in the initial crystallization solution. Before data collection, the crystal was transferred to 0.2 m imidazole (pH 7.2), 0.4 m calcium acetate, 18% (w/v) PEG 8000, 4 mm DTT, and increasing concentrations of glycerol up to 20%.
Crystals for heavy atom derivatization were grown as for native crystals, but using 0.1 m imidazole (pH 7.6), 0.2 m calcium acetate, 10% (w/v) PEG 8000, and 2 mm DTT as crystallization solution. Crystals were soaked for 3.5 h in 0.1 m imidazole (pH 7.6), 0.2 m calcium acetate, 12% (w/v) PEG 8000, 1 mm ethylmercurithiosalicylate (EMTS). Excess of EMTS was removed during the equilibration in cryoprotectant solutions consisting of 0.1 m imidazole (pH 7.6), 0.2 m calcium acetate, 12% (w/v) PEG 8000, and 5% to 20% glycerol. Data of the native and EMTS-soaked crystals were collected and processed as for the SR3-SR4 crystals.
SR4-SR5-SH3 crystals belong to the space group P212121. The asymmetric unit contains two plectin molecules (∼64% solvent content) that are related by a non-crystallographic translation about 1/2 along axis c (0.00 0.00 0.46), which was evident as a peak in the native Patterson map. Phases were obtained by single isomorphous replacement with anomalous scattering (SIRAS) using the native and mercurial datasets. Calculation of approximate substructure amplitudes, heavy atom substructure determination, and initial phase calculations were done with ShelxC/D/E (44) and the HKL2MAP graphical interface (45). Phase probability distributions from the top four Hg-sites found by Shelx, were further refined with SHARP (46), and allowed for the identification of one additional minor Hg site. Phase improvement and extension with Solomon (47) and DM (48) allowed the calculation of an interpretable electron density map (supplemental Fig. S1). Two copies of the SR4 from the SR3-SR4 structure were located by phased-molecular replacement using MOLREP (49); in addition, α-helices corresponding to the SR5 were built and two copies of a mixed model (37) of the SH3 domain based on the crystal structure of the SH3 of α-spectrin (PDB code 2PQH) were docked into the SIRAS map. Refinement was done with phenix.refine against native data to 2.95 Å. At early stages, refinement included simulated annealing, while individual positional and grouped B-factor (two groups per residue) refinement combined with the refinement of five TLS groups in each plectin molecule was done at later stages. Non-crystallographic symmetry restraints and main-chain H-bond distance restraints were used during refinement. A map calculated using the anomalous differences of the native dataset and the SIRAS phases has the two strongest peaks near Gln-792 in each of the molecules in the asymmetric unit (supplemental Fig. S2A) and the anomalous signal at theses positions was higher than at any of the sulfur atoms of the protein. Based on the environment (near the carbonyls of Asp-789 and Gln-792) these peaks were modeled as Ca2+ ions whose B-factors were refined anisotropically. The final model has good geometry (Table 1) and contains residues 642–915 in both chains, with the exception of residues 871–873 in molecule A and 830–832 in molecule B, for which no clear electron density was observed.
Analysis of Structures
Identification of residue pairs that could form disulfide bonds was done with the program SSBOND (50). Analysis of protein motions by pair-wise comparison of conformers was done with the program DynDom (51). Molecular figures were prepared using PyMOL (52).
Titration of Thiol Groups
Sulfhydryl groups were titrated under denaturing conditions with 5,5′-dithio-nis(2-nitrobenzoic acid) as described (22).
SAXS Measurements and Analysis
SAXS measurements were performed at the cSAXS beamline at the Swiss Light Source (Villigen, Switzerland). The energy of the x-ray beam was adjusted to 12 KeV (λ = 1.0 Å) and the distance from the sample to the detector (PILATUS 2 m, Dectris Ltd.) was 2.15 meters, covering a scattering vector range (q = 4πsinθ/λ) from 0.015 to 0.5 Å−1, as determined by the silver behenate scattering profile. The buffers used for the last gel filtration step of the protein purifications, containing 5% (v/v) glycerol and 2.5 mm DTT, and protein samples were measured consecutively in the same borosilicate capillary (Ø 1 mm, Hilgenberg GmbH), using an in house designed thermostatized copper holder. Sample temperature was set to 10 ± 1 °C for all the measurements. Data frames were recorded every 0.5 s, for a total exposure time of 30 s, at 10 different positions (0.5 mm spacing) in the capillary, to reduce radiation damage. Data sets were collected from SR3-SR4-SR5-SH3 protein at concentrations of 1.2, 2.4, 4.8, and 9.6 mg/ml. Data reduction and analysis was performed using the ATSAS package (53), according to standard procedures.
The theoretical scattering profiles of atomic models were calculated and fitted to the experimental data with the program CRYSOL (54). The pair-distance distribution, P(r), of atomic models was calculated with the program HYDROPRO (55).
Construction of an Atomic Model of the SR3-SR4-SR5-SH3 Region
The structure of the SR3-SR4 region was superposed onto that of SR4-SR5-SH3 by fitting the positions of the Cα atoms of the central region of the helical bundle of the SR4, residues 647–661, 686–706, and 721–742. Models were built by combining the regions 543–660 and 692–727 of the moved SR3-SR4 structure with the regions 661–691, 728–915 of the structure of SR4-SR5-SH3.
Protein Data Bank Accession Numbers
The atomic coordinates and structure factors of the SR3-SR4 and SR4-SR5-SH3 structures have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under ID codes 3PDY and 3PE0. The coordinates of the atomic model of the SR3-SR4-SR5-SH3 are available from the authors.
RESULTS
To study the structure of the central region of the plakin domain, the following three fragments of human plectin were expressed as soluble proteins (Fig. 1A). A first construct contains the third and fourth SR (SR3-SR4), residues 543–748; a second fragment corresponds to the fourth and fifth SR and the SH3 (SR4-SR5-SH3), residues 640–918. A third construct, SR3-SR4-SR5-SH3, contains the whole region covered by the previous fragments, residues 543–918.
Structure of the SR3-SR4 Region of Plectin
The SR3-SR4 protein yielded crystals that diffracted to 2.22 Å. The structure was refined to a final Rfree of 24.8% (Table 1) (Fig. 1B). The asymmetric unit contains two SR3-SR4 polypeptides that have a similar overall structure with minor differences (see below). Each repeat has the canonical SR fold, consisting of three α-helices (A, B, and C). The structures of each individual SR in the two molecules present in the asymmetric unit of the crystal are almost identical; the rmsd for all equivalent main chain atoms is 1.19 Å and 0.54 Å for the SR3 and SR4, respectively. The only significant differences between the two structures are located at the first two turns of helix A and the B/C loop of the SR3, which pack slightly tighter in one of the molecules than in the other, due to different crystal contacts.
The structure of the SR3-SR4 of plectin is very similar to that of the equivalent repeats of BPAG1 (PDB code 2IAK) (29), as expected from the 52% sequence identity between plectin and BPAG1 in this region. After superposition of each individual repeat of the BPAG1 structure onto the equivalent SR of plectin the rmsd for all main chain atoms is 1.00–1.32 Å for the SR3 (values from the comparison with each of the two copies of plectin in the asymmetric unit) and 0.75–0.79 Å for the SR4. Our structure of the SR3-SR4 of plectin was refined against significantly higher resolution data than that of BPAG1, 2.22 Å versus 3.0 Å, and reveals details that were not visible in the BPAG1 structure. These include the A/B loop, the first two turns of helix B and the side chains of residues in the helices A and B of the SR4 (supplemental Fig. S3A). The helix A is about two turns shorter than the adjacent helices B and C; thus, the A/B loop covers the hydrophobic core. In addition to the structural similarity of the individual repeats, the arrangement of the SR3-SR4 tandem is also very similar in plectin and BPAG1, and it is different from that observed in other structures of tandem SRs.
Comparison of the two molecules of plectin present in the asymmetric unit reveals small differences in the relative orientation of the SR3 and SR4. After superposition of the SR3 of both molecules there is a ∼4.5 Å difference in the position of the Cα atoms of residues in the A/B loop of SR4. The two conformations are related by a closure motion with the hinge located at the SR3/SR4 boundary (supplemental Fig. S4). Superposition of the BPAG1 structure with those of plectin reveals variations in the interdomain orientation of similar amplitude as observed between the plectin molecules and further illustrates the moderate plasticity of the SR3-SR4 tandem.
Structure of the SR4-SR5-SH3 Region of Plectin
The crystal structure of the SR4-SR5-SH3 region of plectin was solved by SIRAS using a mercurial derivative and was refined against data to 2.95 Å resolution (Table 1) (Fig. 1C). The asymmetric unit contains two molecules of SR4-SR5-SH3 which are almost identical (the rmsd for all equivalent main-chain atoms is 0.33 Å) as expected from the required use of non-crystallographic symmetry restraints throughout refinement.
The structure of the SR4 in the SR4-SR5-SH3 molecule is very similar to that observed in the SR3-SR4 polypeptide. The only noticeable difference in the SR4 between the two structures resides in the A/B loop (supplemental Fig. S3). In SR4-SR5-SH3 the A/B loop contacts the SH3 and the SR5 (see below). The sites for this contact are absent in the SR3-SR4 structure. Thus, the native conformation of the A/B loop in the full-length protein is better represented by the SR4-SR5-SH3 structure.
The SR5 exhibits the characteristic SR fold. In addition, it has a unique 10-residues long helix-B0 upstream of helix B, which protrudes at the end of the longitudinal axis of the three-helix bundle. Helix B0 has a patch of hydrophobic residues (Val-784, Leu-787, Leu-790, and Leu-791) that in our crystal structure binds in a coiled-coil fashion to the SR4 of a neighboring molecule (supplemental Fig. S2B). It is reasonable that, in the full-length plectin, the hydrophobic-rich surface of helix B0 might establish analogous interactions with the region downstream of SR5, which is predicted to consist of a SR-like domain (SR6) (30). The structure contains a Ca2+ ion near the C terminus of helix B0. Calcium was present in high concentration in the crystallization solution (0.2 m calcium acetate). In addition, Ca2+ concentrations up to 0.1 m had no effect on the thermal stability of the protein in solution (data not shown). Thus, the observed bound Ca2+ does not represent a functional Ca2+ binding site at physiological concentrations.
The SR4-SR5 tandem forms an elongated structure. Helices A and B of SR4 and helices B and C of SR5 lie approximately on opposite sides of the A-C interdomain helix. Thus, there is no direct contact between the SR4 and the SR5 other than those that occur at the helical linker. In summary, the SH3 domain does not alter the overall canonical fold of the SR5 or the rod-like structure of the tandem array of SRs.
The SH3 of plectin exhibits the characteristic overall fold observed in other SH3 domains. It consists of five β-strands (β1 to β5) that adopt a β-barrel structure (56). The N terminus of the SH3 domain is connected to helix B of SR5 by a 16-residue long segment, whereas the C terminus of the SH3 is linked to helix C by a short sequence. We called these segments the upstream and downstream linkers, respectively.
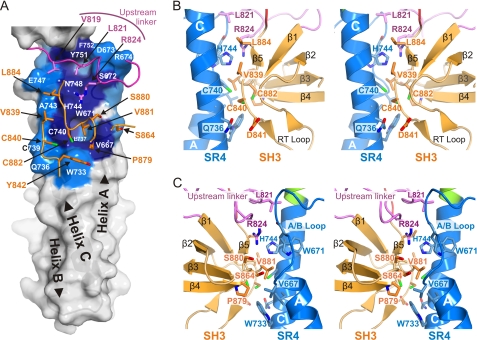
Despite being inserted within the sequence of the SR5, the SH3 domain does not contact the SR5, but it makes an extensive interaction with the SR4, which occludes ∼560 Å2 of the solvent accessible area of the SH3 and a similar area of the SR4. The inter-domain contacts are mainly hydrophobic (Fig. 2). The SR4 binding surface in the SH3 domain is centered on Val-881 in the β4/β5 loop; its side chain docks into a hydrophobic pocket in the SR4 formed by residues Val-667, Trp-671, Trp-733, Leu-737, Cys-740, and His-744, which are located in the A/B loop and the helix C. The N-terminal part of the β1/β2 loop (known as the RT-loop), also contributes to the interaction. Cys-740 in SR4 and Cys-840 and Cys-882 in the SH3 form a Cys-cluster buried at the SR4-SH3 interface. Analysis of a native anomalous difference map calculated using the SIRAS-derived phases revealed three independent peaks that can be assigned to the sulfur atoms of these Cys. The distances between the anomalous difference peaks are larger than the expected S-S distance in a disulfide bridge (supplemental Fig. S5). Titration of the sulfhydryl groups of the SR4-SR5-SH3 protein in the absence of reducing agents yields 7.0 ± 0.1 free sulfhydryls per molecule, which reveals that all Cys are reduced. Thus, the thiol groups of Cys-740, Cys-840, and Cys-882 were modeled reduced in our structure.
FIGURE 2.
The intramolecular interface between the SR4 and the SH3 domain of plectin. A, footprint of the SH3 domain and the upstream linker on the surface of the SR4. Residues in SR4 that participate in the intramolecular contact are labeled on the surface and are colored according to the percentage of the accessible surface buried as defined by the protein interfaces, surfaces, and assemblies service (PISA) (65): dark blue for residues mostly occluded (≥70% buried area) and light blue for residues partially occluded (≥20% and <70% buried area). The backbone of the upstream linker (purple) and the SH3 domain (orange) is partially shown as a wire, and the side chains of the main residues that contact the SR4 are shown as sticks and are labeled with arrows. The positions of the three α-helices of the SR4 are labeled on top of the surface and the amino-to-carboxyl direction indicated with arrowheads. B and C, stereo representations of the intramolecular contacts between the SR4 (blue), the SH3 (orange), and the upstream linker (purple). The two lateral views in panels B and C are related by a 180° rotation around a vertical axis.
The SR4-SH3 interaction is further stabilized by the upstream linker (residues 817–833), which is inserted as a wedge between the SR4 and the SH3. Leu-821 docks into a pocket on the SR4. Arg-824 is buried in between the SR4 and SH3 and probably makes H-bonds with both domains. The downstream linker (residues 886–889) makes minor contacts with the SH3 and the N terminus of the upstream linker, but it does not contact the SR4 directly. This short linker consists mostly of a Pro cluster (886PPP888), suggesting that it provides a well defined anchorage of the SH3 C terminus to the SR5. Thus, it is likely that the downstream linker favors the interaction of the SH3 with the SR4 by limiting the conformational freedom of the SH3 domain.
The SR4/SH3 Interface Is Conserved in Plakins but Not in α-Spectrins
The residues of the SR4, the SH3, and the linkers that form the intramolecular interface are conserved in other plakins (supplemental Fig. S6), suggesting that the arrangement of the SR4 and SH3 observed in plectin is conserved throughout the plakin family.
Similar to plakins, α-spectrins contain an SH3 domain inserted in the B/C loop of its ninth SR (SRs numbered according to Ref. 57). Nonetheless, α-spectrins show significant differences with plakins at the residues involved in the SR4-SH3 interaction in plectin. The positions of the SR8 of α-spectrin, equivalent to the residues that form the hydrophobic-rich interaction surface in the SR4 of plectin, are occupied by polar residues. In the SH3 domain, the positions of two Cys residues of plectin (Cys-840 and Cys-882) that face the SR4 are occupied by tyrosines in α-spectrins. The sequences of the upstream and downstream linkers in α-spectrins do not show similarities with those of plakins. In summary, all the structural elements of the SR4-SH3 interaction present in plakins are different in α-spectrins.
The Putative Binding Site of the SH3 Domain of Plectin Is Distorted and Occluded
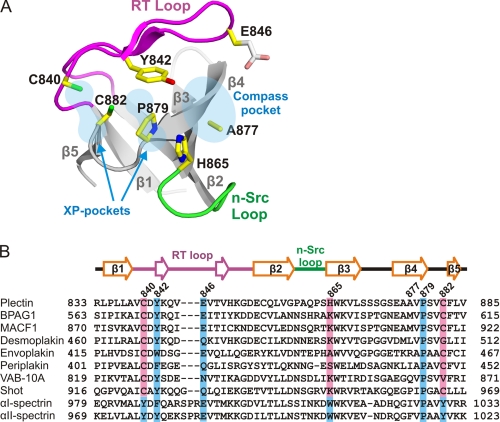
SH3 domains are generally involved in protein-protein interactions. A majority of SH3 domains recognize ligands with Pro-rich sequences containing the core motif XPXXP that, when bound, adopts a poly-Pro type II helix conformation (34, 56, 58). The poly-Pro binding site in canonical SH3 domains is located on the surface between the RT and n-Src loops. It has two parallel pockets, each of which recognizes an XP dipeptide on the ligand; hence, they are known as the XP-pockets. These pockets are formed by conserved aromatic residues in the RT-loop, the β4 strand, and the β4/β5 loop. Adjacent to one of the XP-pockets, the RT and n-Src loops form the “specificity” or “compass” pocket that typically recognizes a basic residue that either precedes or follows the XPXXP motif. The putative binding site of the SH3 domain of plectin shows significant differences with canonical poly-Pro binding sites (Fig. 3A). First, three well-conserved aromatic residues that create the XP-pockets in canonical SH3 domains are substituted by amino acids with shorter side chains in plectin, namely Cys-840, His-865, and Cys-882. The presence of these three non-aromatic residues creates a rather flat surface in plectin instead of the two XP-pockets. Second, the RT-loop of plectin is three-residues shorter than that of typical SH3 domains and the compass pocket of plectin is wider than that of other SH3 domains.
FIGURE 3.
Structure of the putative binding site of the SH3 domain. A, ribbon representation of the SH3 domain of plectin. The side chains of residues in positions equivalent to those that form the binding pockets in canonical SH3 domains are shown as sticks. The side chain of Glu-846 was not modeled beyond its Cβ atom due to lack of well-defined electron density; an arbitrary conformation is shown here. B, multiple sequence alignment of the SH3 domain of human plectin (Uniprot Q15149–2) with those of other human plakins: BPAG1e (Q03001-3), MACF1 (Q9UPN3-2), desmoplakin (P15924), envoplakin (Q92817), and periplakin (O60437); plakins from invertebrates: VAB-10A from Caenorhabditis elegans (Q86NF8) and Shot from Drosophila melanogaster (A1Z9J1); and two human α-spectrins: αI-spectrin (erythrocyte, P02549), and αII-spectrin (brain, Q13813). The secondary elements of the plectin structure are shown above the alignment. Conserved residues that form the binding pockets in α-spectrin are shown with blue boxes, whereas residues of plakins unlikely to sustain binding in a canonical manner are shown with light red boxes.
The aforementioned differences in the putative binding site of plectin are present in the other members of the plakin family (Fig. 3B). Cys-840 and Cys-882 are highly conserved in plakins. On the other hand, His-865 and Ala-877, which contribute to the compass pocket, are poorly conserved within the family; His-865 is most frequently substituted by Lys, while there is higher variability at the position equivalent to Ala-877.
Finally, the pseudo poly-Pro binding site of plectin is partially occluded by the intramolecular interaction between the SR4 and the SH3 domains. Cys-840, Tyr-842, Pro-879, and Cys-882 contribute to the interface (see above) suggesting that in plectin and other plakins the area of the XP-pockets has evolved to engage in an intramolecular interaction instead of ligand recognition. On the other hand, the compass pocket is accessible on the surface.
Structure of the SR3-SR4-SR5-SH3 Region
Attempts to crystallize the SR3-SR4-SR5-SH3 region of the plakin domain of plectin were unsuccessful. Nonetheless, we have built an atomic model of this region using as templates the crystal structures of the SR3-SR4 and SR4-SR5-SH3 fragments, which share the SR4.
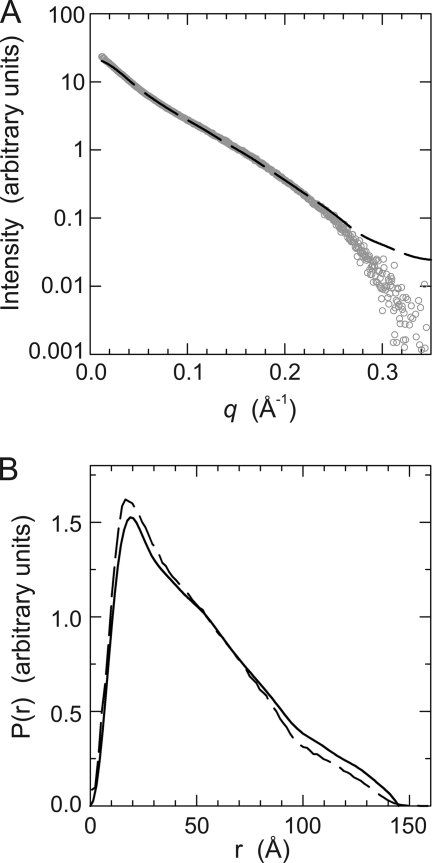
To experimentally evaluate the composite atomic model we have analyzed the structure in solution of the SR3-SR4-SR5-SH3 region, residues 543–918, by SAXS, which can provide accurate shape information. The experimental scattering profile is shown in Fig. 4A. The apparent molecular weight of SR3-SR4-SR5-SH3 calculated from the estimated excluded volume was 45.2 ± 0.5 kDa, which is in agreement with the molecular mass calculated from its sequence (43310 Da) and confirms that this fragment of plectin is a monomer in solution. Guinier analysis of the scattering data revealed that SR3-SR4-SR5-SH3 has a radius of gyration (RG) of 43.9 ± 4.5 Å (supplemental Fig. S7). This value lies within the experimental error with the RG of 42.9 ± 2.3 Å determined in the calculation of the pair-distribution probability function (P(r)) using data covering a range of the scattering vector from 0.017 to 0.35 Å−1. The P(r) has a maximum dimension (Dmax) of 145 Å (Fig. 4B).
FIGURE 4.
SAXS analysis of the SR3-SR4-SR5-SH3 region. A, scattering profile of SR3-SR4-SR5-SH3 (open circles). The theoretical scattering curve of the atomic model (dashed line) shows a good fit to the experimental data. B, pair-distance distribution curves estimated from the SAXS data using the program GNOM (solid line) and calculated for a hydrated model of the atomic structure (dashed line).
The SAXS scattering curve and the distance distribution calculated for the atomic model were almost identical to the experimental scattering profile and the SAXS-derived P(r) (Fig. 4). The RG and the Dmax of the model are 40.3 Å and 155 Å, respectively; yet 99.99% of the distances occur at D<147 Å, in very close agreement with the RG and the Dmax values determined from the SAXS measurements. Moreover, there is a good agreement between the atomic model and low resolution envelopes derived from the SAXS data (supplemental Fig. S8). In summary, the atomic model of the SR3-SR4-SR5-SH3 region provides a realistic representation of the structure of this region in solution.
The model of the SR3-SR4-SR5-SH3 region reveals that helices A and B of the SR3 lie on the same side of the SR4 as the SH3 domain creating a C-shaped groove (supplemental Fig. S9). At the center of this groove, residues in helices A and C of the SR4 form a hydrophobic surface that is flanked by polar and acidic residues.
DISCUSSION
The plakin domain of plectin and other plakins harbors protein-protein interaction sites. The central region of the plakin domain of plectin characterized herein participates in the interaction with junctional proteins such as the integrin β4 subunit and the cytoplasmic domain of BPAG2 in epithelia and with β-dystroglycan and β-synemin in skeletal muscle. The precise binding sites in the plakin domain are not known. The presence of an SH3 domain in this region is of interest because SH3 domains often mediate protein-protein interactions. SH3 domains frequently recognize Pro-rich ligands. In the putative binding site of the SH3 domain of plectin there are substantial structural differences with that of canonical domains. Substitutions of residues that form the PXXP-binding site of archetypical SH3 domains compromise their recognition of these ligands. Thus, the structure of the SH3 domain of plectin is not compatible with it binding to Pro-rich motifs. The residues that form the putative binding site of the SH3 of plectin are also present in other plakins, suggesting that none of them recognize Pro-rich sequences in a canonical manner. In fact, to date no Pro-rich ligand has been identified for the SH3 domain of plectin or other plakins. Thus, in plakins we refer to this area as a poly-Pro pseudo-binding site. On the other hand, this pseudo-binding site is involved in the interaction with the SR4, suggesting that it has evolved to stabilize this intramolecular contact.
Some SH3 domains engage in protein-protein interactions via alternative mechanisms including the binding to short non-Pro-rich sequence motifs or to globular domains via tertiary contacts; and some of these interactions involve regions of the SH3 domain other than the canonical PXXP-binding groove (34). Thus, it is still possible that the SH3 domain of plectin might mediate or contribute to the association with junctional proteins via a non-PXXP binding mechanism.
In addition to the SH3 domain, the SRs of the plakin domain of plectin may also contain protein-protein binding sites, similar to those found in α- and β-spectrins (59). It is possible that in plectin the binding-sites for some ligands include both the SH3 domain and one or more SRs. The domain arrangement of the SR3-SR4-SR5-SH3 region, as observed in the crystal structures and the SAXS-validated atomic model, defines multi-domain surfaces that may be relevant for ligand binding. For example, the groove created by the SR3, SR4, and SH3 domains might be a potential protein interaction site.
The SR3-SR4-SR5 region forms a rod-like structure ∼145 Å in length. Most likely the rod-like structure continues throughout the additional repeats, SR6 to SR9, to reach an estimated total extension over 400 Å. Thus, in addition to harbor protein-protein interaction sites the plakin domain sustains two structural roles. First, it acts as a structural spacer that in combination with the central rod domain separates the binding sites in the N and C terminus of plectin. Thus, it facilitates the crosslinking role of plakins. Second, the SR3-SR4-SR5 array, and by extension the rest of the plakin domain, resembles a semi-flexible rod. Analysis of the SR3-SR4 structures reveals a putative hinge movement of small amplitude centered at the SR3/SR4 linker. Similar bending movements have been described in multi-repeat fragments of α- and β-spectrin (60–61) and they are likely to occur at other inter-repeat linkers of the plakin domain. The SH3 domain, which is connected to the SR5 by a short and Pro-rich downstream linker and to the SR4 via a relatively large interface, is likely to reduce the bending at the SR4/SR5 junction, suggesting that there are preferential points of flexibility. Finally, the plakin domain is likely to straighten or stretch under tensile forces; thus, binding to junctional proteins might be altered by force-driven conformational changes. Vice versa, association to other proteins could modify the mechanical properties of the plakin domain. In summary, the plakin domain might contribute to the stability of the cytoskeleton by allowing reversible deformation and the storage of energy during events of mechanical stress.
An interesting feature of the SR4/SH3 interface is the presence of a Cys-cluster formed by Cys-740, Cys-840, and Cys-882 at the center of the contact surfaces, and the nearby Cys-739 located at the edge of the interaction area. The pair Cys-739/Cys-840 is at an optimal distance for the formation of a disulfide bond, alternatively Cys-840 might form a disulfide bond with Cys-740. In both cases the disulfide bond would link the SR4 to the SH3. The reducing environment of the cytosol would maintain the thiols free, as observed in the crystal structure; but in states of oxidative stress the reactive oxygen species might induce the formation of at least one disulfide bond between the SR4 and the SH3, which could have two effects. First, it might affect the affinity of plectin for some ligands due to a reduction in the conformational adaptability of this region. Similarly, oxidation of a Cys near the intermediate filament-binding site located in the C-terminal region of plectin reduces its affinity for vimentin (62). Second, disulfide crosslinking might reduce the flexibility of this segment of the plakin domain, increasing the stability of the plakin domain. Finally, Cys-740 and Cys-840 are highly conserved in the plakin family (supplemental Fig. S6), suggesting that other plakins might also be subject to reversible redox regulation. The role of plakins during oxidative stress is not known; it is possible that they contribute to the proposed cytoprotective role against stress of intermediate filament proteins (63).
Overall, the structures of the plakin domain presented here extend the similarities between plakins and other members of the spectrin superfamily. Nonetheless, there are significant differences in the organization of the SH3 domain and the SR-array between plakins and α-spectrins. Thus, plakins constitute a distinct group within the spectrin superfamily. The pseudo-binding site of the SH3 domain and the SR4/SH3 interface are widely conserved among plakins including those of nematodes and insects, which contain a single plakin gene. Thus, the plakin-specific role of the SH3 was acquired early after the spectrin-plakin branching and the SR4/SH3 association has been maintained ever since, suggesting that it may be required for the architecture and function of all plakins.
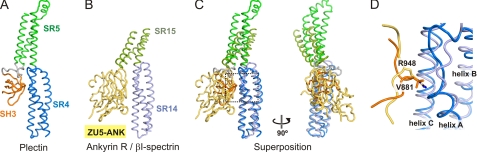
The SR4-SR5-SH3 of plectin has striking similarities with the recently determined structure of the SR13-SR14-SR15 of βI-spectrin in complex with the ZU5-ANK domain of ankyrin (64). The orientation of the ZU5-ANK, the SR14 and the SR15 in the complex resembles the arrangement of the SH3, SR4, and SR5 domains of plectin, respectively. After superposition of the SR14 of βI-spectrin on the SR4 of plectin, the ZU5-ANK and the SH3 domains, despite having unrelated tertiary structures, occupy an equivalent position (Fig. 5). Moreover, the B/C loop of the SR15 of βI-spectrin coincides with the insertion points of the SH3 domain in the same loop of the SR5 of plectin. Finally, Arg-948 of ankyrin occupies the equivalent position to Val-841 of the SH3 of plectin, and its guanidinium group docks in a polar pocket in the SR14 located in the equivalent position of the hydrophobic cavity in the SR4 of plectin that accommodates the side chain of Val-841. The similarities between the ankyrin-binding site in the SR14-SR15 fragment of βI-spectrin and the SH3-interaction site in the SR4-SR5 of plectin suggest that they might reveal a protein-protein interaction area (inter- or intramolecular), which is also present in arrays of other SRs and that involves the A/B loop and the helix C of an SR and the B/C loop of the following SR. Nonetheless, further structural characterization of SR interactions will be necessary to understand whether the plectin and βI-spectrin/ankyrin structures illustrate a preferential binding mode or that they share a serendipitous resemblance.
FIGURE 5.
Similar domain arrangement in the SR4-SR5-SH3 of plectin and the complex formed by βI-spectrin and ankyrin R. A, worm representation of the structure of the SR4-SR5-SH3 of plectin; the orientation is the same as in Fig. 1C. B, structure of the ZU5-ANK domain of ankyrin R bound to the SR14-SR15 of βI-spectrin (PDB code 3KBT); the SR13, which is present in the crystal structure of the complex but does not contact the ZU5-ANK domain, has been omitted. C, orthogonal views of the ankyrin/βI-spectrin and the plectin structures after superposition of the SR14 of βI-spectrin onto the SR4 of plectin. D, close-up view of the SH3/SR4 and ZU5-ANK/SR14 interfaces in plectin and the ankyrin/βI-spectrin complex, respectively. The side chain of Val-881 in the SH3 of plectin and Arg-948 in the ZU5-ANK domain occupy equivalent positions and are shown as sticks. For clarity, only a segment of the backbones around Val-881 and Arg-948 are shown.
In summary, in this study we have progressed from the elucidation of structures of di-repeat fragments of the plakin domain to that of a multi-repeat region. The emerging rod-like structure suggests that in addition to the scaffolding role, the plakin domain also contributes to the mechanical stability of the cytoskeleton and its anchorage to junctional complexes. Our work paves the way to the elucidation of the complete structure of the plakin domain and to assess its role in maintaining the integrity of healthy tissues and in disease linked to defects of plakins.
Acknowledgments
We thank Jaime Pascual and Tassos Perrakis for critical comments on the manuscript. We acknowledge the Swiss Light Source for provision of the synchrotron radiation facilities.
This work was supported by the Spanish Ministry of Science and Innovation and the European Regional Development Fund (Grants BFU2006-01929 and BFU2009-08389, to J. M. dP.). This work was also supported by grants from the Netherlands Science Foundation and the Dutch Cancer Society (to A. S.).
The atomic coordinates and structure factors (codes 3PDY and 3PE0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
- BPAG
- bullous pemphigoid antigen
- ABD
- actin binding domain
- CH
- calponin homology
- Dmax
- maximum dimension
- EMTS
- ethylmercurithiosalicytate
- MACF
- microtubule actin crosslinking factor
- PDB
- Protein Data Bank
- rmsd
- root mean square deviation
- RG
- radius of gyration
- SAXS
- small angle X-ray scattering
- SIRAS
- single isomorphous replacement with anomalous scattering
- SH3
- Src-homology 3
- SR
- spectrin repeat.
REFERENCES
- 1. Jefferson J. J., Leung C. L., Liem R. K. (2004) Nat. Rev. Mol. Cell Biol. 5, 542–553 [DOI] [PubMed] [Google Scholar]
- 2. Sonnenberg A., Liem R. K. (2007) Exp. Cell Res. 313, 2189–2203 [DOI] [PubMed] [Google Scholar]
- 3. Foisner R., Leichtfried F. E., Herrmann H., Small J. V., Lawson D., Wiche G. (1988) J. Cell Biol. 106, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herrmann H., Wiche G. (1987) J. Biol. Chem. 262, 1320–1325 [PubMed] [Google Scholar]
- 5. Seifert G. J., Lawson D., Wiche G. (1992) Eur J. Cell Biol. 59, 138–147 [PubMed] [Google Scholar]
- 6. Svitkina T. M., Verkhovsky A. B., Borisy G. G. (1996) J. Cell Biol. 135, 991–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Pereda J. M., Ortega E., Alonso-García N., Gómez-Hernández M., Sonnenberg A. (2009) Cell Adh. Migr. 3, 361–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Litjens S. H., de Pereda J. M., Sonnenberg A. (2006) Trends Cell Biol. 16, 376–383 [DOI] [PubMed] [Google Scholar]
- 9. Eger A., Stockinger A., Wiche G., Foisner R. (1997) J. Cell Sci. 110, 1307–1316 [DOI] [PubMed] [Google Scholar]
- 10. Ketema M., Wilhelmsen K., Kuikman I., Janssen H., Hodzic D., Sonnenberg A. (2007) J. Cell Sci. 120, 3384–3394 [DOI] [PubMed] [Google Scholar]
- 11. Wilhelmsen K., Litjens S. H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. (2005) J. Cell Biol. 171, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reipert S., Steinböck F., Fischer I., Bittner R. E., Zeöld A., Wiche G. (1999) Exp. Cell Res. 252, 479–491 [DOI] [PubMed] [Google Scholar]
- 13. Rezniczek G. A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K. E., Winder S. J., Wiche G. (2007) J. Cell Biol. 176, 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schröder R., Warlo I., Herrmann H., van der Ven P. F., Klasen C., Blümcke I., Mundegar R. R., Fürst D. O., Goebel H. H., Magin T. M. (1999) Eur. J. Cell Biol. 78, 288–295 [DOI] [PubMed] [Google Scholar]
- 15. Hijikata T., Nakamura A., Isokawa K., Imamura M., Yuasa K., Ishikawa R., Kohama K., Takeda S., Yorifuji H. (2008) J. Cell Sci. 121, 2062–2074 [DOI] [PubMed] [Google Scholar]
- 16. Rezniczek G. A., Walko G., Wiche G. (2010) Dermatol. Clin. 28, 33–41 [DOI] [PubMed] [Google Scholar]
- 17. Pfendner E., Rouan F., Uitto J. (2005) Exp. Dermatol. 14, 241–249 [DOI] [PubMed] [Google Scholar]
- 18. de Pereda J. M., Lillo M. P., Sonnenberg A. (2009) EMBO J. 28, 1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Litjens S. H., Koster J., Kuikman I., van Wilpe S., de Pereda J. M., Sonnenberg A. (2003) Mol. Biol. Cell 14, 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Litjens S. H., Wilhelmsen K., de Pereda J. M., Perrakis A., Sonnenberg A. (2005) J. Biol. Chem. 280, 22270–22277 [DOI] [PubMed] [Google Scholar]
- 21. Fontao L., Geerts D., Kuikman I., Koster J., Kramer D., Sonnenberg A. (2001) J. Cell Sci. 114, 2065–2076 [DOI] [PubMed] [Google Scholar]
- 22. García-Alvarez B., Bobkov A., Sonnenberg A., de Pereda J. M. (2003) Structure 11, 615–625 [DOI] [PubMed] [Google Scholar]
- 23. Sevcík J., Urbániková L., Kost'an J., Janda L., Wiche G. (2004) Eur. J. Biochem. 271, 1873–1884 [DOI] [PubMed] [Google Scholar]
- 24. Koster J., van Wilpe S., Kuikman I., Litjens S. H., Sonnenberg A. (2004) Mol. Biol. Cell 15, 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koster J., Geerts D., Favre B., Borradori L., Sonnenberg A. (2003) J. Cell Sci. 116, 387–399 [DOI] [PubMed] [Google Scholar]
- 26. Lunter P. C., Wiche G. (2002) Biochem. Biophys. Res. Commun. 296, 904–910 [DOI] [PubMed] [Google Scholar]
- 27. Virata M. L., Wagner R. M., Parry D. A., Green K. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baines A. J. (2003) Cell Mol. Biol. Lett. 8, 195–214 [PubMed] [Google Scholar]
- 29. Jefferson J. J., Ciatto C., Shapiro L., Liem R. K. (2007) J. Mol. Biol. 366, 244–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sonnenberg A., Rojas A. M., de Pereda J. M. (2007) J. Mol. Biol. 368, 1379–1391 [DOI] [PubMed] [Google Scholar]
- 31. Pascual J., Pfuhl M., Rivas G., Pastore A., Saraste M. (1996) FEBS Lett. 383, 201–207 [DOI] [PubMed] [Google Scholar]
- 32. Pascual J., Pfuhl M., Walther D., Saraste M., Nilges M. (1997) J. Mol. Biol. 273, 740–751 [DOI] [PubMed] [Google Scholar]
- 33. Ylänne J., Scheffzek K., Young P., Saraste M. (2001) Structure 9, 597–604 [DOI] [PubMed] [Google Scholar]
- 34. Kaneko T., Li L., Li S. S. (2008) Front Biosci. 13, 4938–4952 [DOI] [PubMed] [Google Scholar]
- 35. Alonso-García N., Inglés-Prieto A., Sonnenberg A., De Pereda J. M. (2009) Acta. Crystallogr. D. Biol. Crystallogr. 65, 858–871 [DOI] [PubMed] [Google Scholar]
- 36. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 37. Schwarzenbacher R., Godzik A., Grzechnik S. K., Jaroszewski L. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 1229–1236 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D. Biol. Crystallogr 50, 760–76315299374 [Google Scholar]
- 40. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsl. 42, contribution 8 [Google Scholar]
- 41. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 43. Painter J., Merritt E. A. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 44. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 45. Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 46. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Acta Crystallogr. D. Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 47. Abrahams J. P., Leslie A. G. (1996) Acta Crystallogr D Biol Crystallogr 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 48. Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 [Google Scholar]
- 49. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 50. Hazes B., Dijkstra B. W. (1988) Protein Eng. 2, 119–125 [DOI] [PubMed] [Google Scholar]
- 51. Hayward S., Berendsen H. J. (1998) Proteins 30, 144–154 [PubMed] [Google Scholar]
- 52. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, U.S.A [Google Scholar]
- 53. Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Svergun D., Barberato C., Koch M. H. J. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 55. García De La Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Musacchio A. (2002) Adv Protein Chem. 61, 211–268 [DOI] [PubMed] [Google Scholar]
- 57. Pascual J., Castresana J., Saraste M. (1997) Bioessays 19, 811–817 [DOI] [PubMed] [Google Scholar]
- 58. Zarrinpar A., Bhattacharyya R. P., Lim W. A. (2003) Sci. STKE 2003, RE8. [DOI] [PubMed] [Google Scholar]
- 59. Baines A. J. (2009) Biochem. Soc. Trans. 37, 796–803 [DOI] [PubMed] [Google Scholar]
- 60. Grum V. L., Li D., MacDonald R. I., Mondragón A. (1999) Cell 98, 523–535 [DOI] [PubMed] [Google Scholar]
- 61. Kusunoki H., MacDonald R. I., Mondragón A. (2004) Structure 12, 645–656 [DOI] [PubMed] [Google Scholar]
- 62. Spurny R., Abdoulrahman K., Janda L., Rünzler D., Köhler G., Castañón M. J., Wiche G. (2007) J. Biol. Chem. 282, 8175–8187 [DOI] [PubMed] [Google Scholar]
- 63. Toivola D. M., Strnad P., Habtezion A., Omary M. B. (2010) Trends Cell Biol. 20, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ipsaro J. J., Mondragón A. (2010) Blood 115, 4093–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 66. Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 67. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]