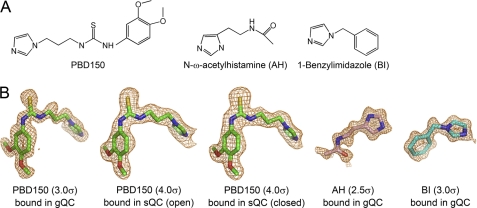
Abstract
Aberrant pyroglutamate formation at the N terminus of certain peptides and proteins, catalyzed by glutaminyl cyclases (QCs), is linked to some pathological conditions, such as Alzheimer disease. Recently, a glutaminyl cyclase (QC) inhibitor, PBD150, was shown to be able to reduce the deposition of pyroglutamate-modified amyloid-β peptides in brain of transgenic mouse models of Alzheimer disease, leading to a significant improvement of learning and memory in those transgenic animals. Here, we report the 1.05–1.40 Å resolution structures, solved by the sulfur single-wavelength anomalous dispersion phasing method, of the Golgi-luminal catalytic domain of the recently identified Golgi-resident QC (gQC) and its complex with PBD150. We also describe the high-resolution structures of secretory QC (sQC)-PBD150 complex and two other gQC-inhibitor complexes. gQC structure has a scaffold similar to that of sQC but with a relatively wider and negatively charged active site, suggesting a distinct substrate specificity from sQC. Upon binding to PBD150, a large loop movement in gQC allows the inhibitor to be tightly held in its active site primarily by hydrophobic interactions. Further comparisons of the inhibitor-bound structures revealed distinct interactions of the inhibitors with gQC and sQC, which are consistent with the results from our inhibitor assays reported here. Because gQC and sQC may play different biological roles in vivo, the different inhibitor binding modes allow the design of specific inhibitors toward gQC and sQC.
Keywords: Crystal Structure, Drug Design, Enzyme Inhibitors, Enzyme Structure, Metalloenzymes, Post-translational Modification, X-ray Crystallography, Glutaminyl Cyclase, Golgi Apparatus, Pyroglutamate
Introduction
The formation of pyroglutamate (pGlu)3 at the N terminus of numerous bioactive peptides, hormones, and chemokines, such as thyrotropin-releasing hormone and monocyte chemoattractant proteins, is believed to protect the peptides or proteins from exopeptidase degradation and/or to endow them with a proper conformation for binding to their receptors (1, 2). Glutaminyl cyclases (QCs, EC 2.3.2.5) catalyze the formation of N-terminal pGlu residue from a glutaminyl or glutamyl residue (3–6). The enzymes are abundant in the neuroendocrine tissues and peripheral blood lymphocytes of mammals (known as type II QCs) (7, 8) and have also been identified in plants, parasites, and bacteria (type I QCs) (9–11). These two types of QCs are quite different in their molecular structure and protein stability (3, 4, 9, 12).
In humans, QCs are involved in several pathological conditions, such as amyloidotic diseases, osteoporosis, rheumatoid arthritis, malignant pheochromocytoma, and melanoma (13–17). QC is capable of catalyzing the pGlu formation on N-terminally truncated amyloid-β peptides (18, 19), in particular, Aβ3(pGlu)-40/42, a major constituent of amyloid-β deposits in sporadic and familial Alzheimer disease (20). Because of their abundance, resistance to proteolysis, rapid aggregation, and neurotoxicity, the pGlu-modified amyloid-β peptides were suggested to play a crucial role in the initiation of pathological cascades, resulting in the development of Alzheimer disease (21). Based on this hypothesis, QC inhibitors may be useful for the treatment of Alzheimer disease. Recently, Schilling et al. (13) have shown that oral application of a QC inhibitor, PBD150, in transgenic mouse models and Drosophila model of Alzheimer disease resulted in significantly reduced depositions of Aβ3(pGlu)-40/42 in brain, which led to a significant improvement of learning and memory in these transgenic animals. PBD150 inhibits human QC with a Ki value in the low nanomolar range (22). This inhibitor was developed by applying a ligand-based optimization approach starting from imidazole. More recently, the potency of the inhibitor was further improved by an order of magnitude by the addition of a methyl group to its imidazole ring (23). However, although the crystal structure of human QC is now available (Protein Data Bank code 2AFM) (4), the detailed interaction mechanism between human QC and PBD150 remains to be elucidated to optimize the enzyme-inhibitor interactions.
In addition to the pathological role in brain tissues, a significantly increased gene (QPCT) expression of QC was reported in the cancer cells of melanoma (24). The elevated QC level was believed to interfere with the vaccine treatments of melanoma because vaccines using peptides that have glutaminyl or glutamyl residue at their N terminus might be interrupted by QCs in the tumor-destructive process. Moreover, the increased gene expression of QC was also reported in the peripheral blood monocytes from patients with rheumatoid arthritis (15), a disorder characterized by chronic inflammation and destruction of bone and cartilage in diarthrodial joints. The increased QC activity in monocytes presumably promotes the maturation of some pGlu-modified chemokines, such as monocyte chemoattractant proteins, which in turn enhance the activation and migration of monocytes, thus accelerating the progression of the inflammatory disease. Therefore, QC inhibitors may also be used for suppressing inflammation as well as increasing the effectiveness of melanoma vaccines.
Apart from the currently well studied human QC that is encoded by the gene QPCT located at chromosome 2p22.2, an isoform of the enzyme was recently identified, encoded by the QPCTL gene that maps to chromosome 19q13.32 (25, 26). The first one possesses an N-terminal secretion signal and is thus believed to be a secretory QC (sQC); in contrast, the latter one carries an N-terminal signal anchor and has been demonstrated to be a Golgi-resident QC (gQC). Except for the different N-terminal signal peptides, these two QCs have similarly sized (∼330 residues) catalytic domains with a sequence identity of ∼45% between them. A tissue distribution analysis in a mouse model revealed that both QCs are ubiquitously expressed (25). However, the expression of gQC showed no significant difference between different organs, whereas the expression of sQC was higher in neuronal tissues. Another notable difference between these two QCs is that gQC has 2–15-fold weaker QC activities on several synthetic substrates when compared with the activities of sQC (25). This finding suggests that these two QCs have distinct active site structures and different sensitivities toward QC inhibitors.
To gain insights into the molecular properties of the Golgi-resident QC, we describe here the atomic resolution (1.13 and 1.05 Å) crystal structures of the Golgi-luminal catalytic domain of human gQC. The structures reveal a relatively wide open and negatively charged active site when compared with the reported structure of sQC. We also determined the structures of gQC-PBD150 and sQC-PBD150, revealing a large loop movement in the active site of gQC upon inhibitor binding. To further compare the inhibitor binding modes between gQC and sQC, we also solved the high-resolution structures of gQC in complex with the inhibitors N-ω-acetylhistamine and 1-benzylimidazole because the structures of sQC in complex with these two inhibitors are available (4). Lastly, we have analyzed the inhibition effects of these inhibitors on gQC and sQC to confirm our observations on the inhibitor-bound QC structures.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were purchased as follows: l-glutaminyl 2-naphthylamide from Bachem (Weil am Rhein, Germany); 2-naphthylamide, N-ω-acetylhistamine, and cacodylate from Sigma-Aldrich; and 1-benzylimidazole from Fluka (St. Louis, MO). Human pyroglutamyl aminopeptidase I was prepared as described previously (5). The QC inhibitor PBD150 was chemically synthesized according to a previous report (22). All other chemicals were of analytical or HPLC grade.
Protein Expression and Purification
The DNA with sequence coding for the Golgi-luminal region (Ser53–Leu382) of human gQC (nucleotide entry: NP_060129) was synthesized and then inserted into a thioredoxin fusion expression vector as described previously via HindIII and XhoI cloning sites (27). The resulting construct contained a thioredoxin fusion protein, two His6 tags, and a Factor Xa cleavage site upstream of the gQC coding region. The vector was transformed into Escherichia coli BL21 (DE3) CodonPlus-RIL cells (Stratagene, La Jolla, CA). The bacteria were grown in Terrific Broth containing ampicillin (70 μg/ml) and chloramphenicol (34 μg/ml) at 37 °C until the cell density reached an A600 of 0.8–0.9. The cultures were induced with 1 mm isopropyl β-d-thiogalactopyranoside for 16–20 h at 20 °C. The cells were then harvested by centrifugation (8983 × g for 30 min at 4 °C) followed by freezing at −80 °C.
Frozen bacterial pellets were resuspended in the lysis buffer (50 mm Tris-HCl, pH 7.8, containing 150 mm NaCl), and the cells were lysed using a cell disruptor (Constant Systems, Kennesaw, GA). The cell lysate was clarified by centrifugation (104,630 × g for 60 min at 4 °C), and the supernatant was loaded onto a nickel-nitrilotriacetic acid (Amersham Biosciences) column preequilibrated with buffer A (50 mm Tris-HCl, 150 mm NaCl, 10 mm imidazole, and 5% glycerol, pH 7.8). The column was washed with the same buffer, and the bound materials were eluted by a linear gradient of 0–100% buffer B (50 mm Tris-HCl, 150 mm NaCl, 300 mm imidazole, and 5% glycerol, pH 7.8). The fractions for thioredoxin fusion gQC were pooled and then digested with Factor Xa (0.3 units/ml) (Novagen, Darmstadt, Germany). To reduce the interference from imidazole in the protein solution during Factor Xa digestion, the digestion reaction was carried out in a dialysis bag, and the imidazole was removed by dialysis against buffer C (50 mm Tris-HCl, 150 mm NaCl, and 5% glycerol, pH 7.8) at 25 °C for 16–20 h. Subsequently, because gQC has relatively high histidine content (4.5%), the cleaved gQC products were isolated from the digest by a second nickel-nitrilotriacetic acid column preequilibrated with buffer C. The bound gQCs were eluted by a linear gradient of 0–100% buffer D (50 mm Tris-HCl, 150 mm NaCl, 40 mm imidazole, and 5% glycerol, pH 7.8). The proteins were concentrated and further desalted by a Sephacryl S-100 column (Amersham Biosciences) eluted with buffer E (50 mm Tris-HCl, pH 7.8, containing 5% glycerol). The purified gQCs were shown to be homogeneous as judged by SDS-PAGE with Coomassie Blue staining.
The expression and purification of sQC was according to a previous protocol (27). The mutant W207F of sQC was constructed on the basis of the protocol of the QuikChange® site-directed mutagenesis Kit (Stratagene).
Enzyme Activity Assay
The QC activity of recombinant gQC was evaluated at 25 °C using the fluorescent substrate l-glutaminyl 2-naphthylamide (Gln-βNA). The 100-μl reaction mixture contained 250 μm fluorogenic substrate, ∼0.2 units of human pyroglutamyl aminopeptidase I (1 unit is defined as the amount hydrolyzing 1 μmol of pGlu-βNA per minute under the same condition), and an appropriately diluted aliquot of recombinant gQC in 50 mm Tris-HCl and 5% glycerol, pH 8.0. The excitation and emission wavelengths were set at 320 and 410 nm, respectively. The reaction was initiated by the addition of gQC. Enzymatic activity was determined by the amount of released βNA, calculated using a standard curve for βNA under the same assay condition. The measurements were made using a FluoroLog®-3 spectrofluorometer (HORIBA Jobin Yvon, Edison, NJ).
Inhibitor Assay
For testing the inhibition activities of imidazole derivatives and cacodylate on gQC and sQC, the reaction mixture was the same as described above, except for the addition of the inhibitor compound. The enzyme was first incubated with each of the inhibitors at 25 °C for 5 min, and then the enzyme-inhibitor mixture was added to the reaction mixture to initiate the cyclization reaction. The IC50 values were obtained by fitting the initial reaction rate versus the inhibitor concentrations using the software KaleidaGraph (Synergy Software, Reading, PA). Ki values were calculated according to the equation IC50 = Ki (1 + S/Km) (28), where S is the substrate concentration and Km is the Michaelis-Menten constant. The Km values of the substrate Gln-βNA on gQC and sQC were referenced from previous reports (3, 25). The concentrations of gQC and sQC used were ∼0.1 μm.
Crystallization and X-ray Data Collection
The purified gQC was concentrated to ∼7 mg/ml using the centrifugal filter Ultracel® 10 K (Millipore, Billerica, MA). The initial crystallization screening of ∼900 conditions was achieved by the Core Facility for Protein Production and X-Ray Structural Analysis in Academia Sinica (Taipei, Taiwan). The crystals were grown at 20 °C by the sitting-drop vapor-diffusion method. Finally, two crystallization conditions were selected, i.e. (i) 30% (v/v) PEG 200, 5% (w/v) PEG 3000, and 0.1 m Mes, pH 6.0, and (ii) 50% (v/v) PEG 200, 0.2 m MgCl2, and 0.1 m cacodylate, pH 6.5, because the pH values are close to the physiological condition of Golgi apparatus. The purified gQC was mixed with an equal volume of the crystallization solution, and rod-like crystals with dimensions reaching 0.1 × 0.08 × 0.3 mm appeared within 6–10 days (supplemental Fig. S1C). X-ray-diffraction experiments were performed at the beamlines 13B1 and 13C1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan). Because of the presence of high concentration PEGs in the crystallization solutions, additional cryoprotectants were not required for the crystals. All diffraction data were processed and scaled with the HKL2000 package (29). The data collection statistics are listed in Table 1. The space group of the crystals is P212121, with typical unit cells of a = 53 Å, b = 69 Å, and c = 77 Å and a solvent content of ∼35%, in which an asymmetric unit comprises one gQC molecule. For the structures of gQC-inhibitor complexes, the crystals obtained from the pH 6.0 condition were soaked in the mother liquor added with 5 mm inhibitor at 20 °C for 3–5 days.
TABLE 1.
Data collection and refinement statistics
Abbreviations used are: Acet., N-ω-acetylhistamine; Benz., 1-benzylimidazole; ASU, asymmetric unit.
| Data collection | |||||||
| Crystal | gQC-pH 6.0 | gQC-pH 6.5 | gQC-PBD150 | gQC-Acet. | gQC-Benz. | sQC-PBD150 | sQC-W207F |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | R3 | R32 |
| Resolution (Å) | 30-1.13 | 30-1.05 | 30-1.40 | 30-1.13 | 30-1.12 | 50-1.95 | 50-1.95 |
| Cell dimensions (Å) | |||||||
| a | 53.2 | 53.1 | 53.4 | 53.3 | 53.6 | 155.8 | 118.8 |
| b | 68.6 | 68.5 | 69.9 | 68.6 | 69.2 | 155.8 | 118.8 |
| c | 77.4 | 77.3 | 77.4 | 77.4 | 77.4 | 80.5 | 332.1 |
| Total observations | 1,412,060 | 905,192 | 406,637 | 1,380,721 | 574,803 | 169,715 | 349,893 |
| Unique reflections | 104,166 | 127,164 | 53,439 | 105,259 | 107,676 | 53,126 | 65,863 |
| Redundancy | 13.6 (9.7)a | 7.1 (7.1) | 7.6 (7.6) | 13.1 (7.4) | 5.3 (2.9) | 3.2 (3.1) | 5.3 (5.4) |
| Completeness (%) | 97.9 (91.0) | 96.4 (96.7) | 92.3 (100.0) | 98.7 (92.2) | 96.9 (91.0) | 99.1 (95.1) | 99.7 (99.8) |
| I/σ(I) | 78.1 (14.5) | 37.4 (3.5) | 36.9 (3.3) | 62.1 (4.4) | 35.2 (2.2) | 17.6 (2.3) | 40.3 (3.1) |
| Rmerge (%) | 5.4 (18.5) | 8.3 (59.4) | 6.3 (52.8) | 6.3 (29.2) | 5.3 (41.6) | 7.5 (49.2) | 4.7 (49.1) |
| No. of molecules in an ASU | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| Refinement | |||||||
| Resolution (Å) | 30-1.13 | 30-1.05 | 30-1.40 | 30-1.13 | 30-1.12 | 30-1.95 | 30-1.95 |
| Reflections (>0 σ(F)), working/test | 93,667/5203 | 114,172/6403 | 47,869/2678 | 94,677/5244 | 96,642/5393 | 47,256/2679 | 59,162/3335 |
| Rfactor/Rfree | 0.126/0.153 | 0.138/0.158 | 0.157/0.207 | 0.131/0.161 | 0.150/0.181 | 0.181/0.239 | 0.143/0.190 |
| r.m.s. deviation bond lengths (Å)/ angles (°) | 0.027/2.14 | 0.027/2.13 | 0.024/2.04 | 0.026/2.16 | 0.028/2.27 | 0.021/1.86 | 0.029/1.90 |
| Average B-factor (Å2)/no. of atoms | |||||||
| Protein | 10.8/2460 | 12.6/2474 | 14.0/2444 | 13.1/2474 | 13.0/2444 | 40.2/5222 | 35.5/5216 |
| Inhibitor/ligand | 9.1/5 | 15.4/22 | 14.6/11 | 11.8/12 | 29.1/44 | ||
| Zinc ion | 5.8/1 | 6.7/1 | 8.6/1 | 7.7/1 | 6.9/1 | 25.8/2 | 28.5/2 |
| Water | 26.6/374 | 27.9/362 | 30.1/404 | 29.3/448 | 28.5/463 | 48.2/284 | 45.3/507 |
| Ramachandran plot (%) | |||||||
| Most favored | 91.5 | 91.5 | 92.6 | 91.2 | 91.9 | 88.2 | 90.3 |
| Additionally allowed | 8.5 | 8.5 | 7.0 | 8.8 | 7.7 | 11.8 | 9.5 |
| Generously allowed | 0 | 0 | 0.4 | 0 | 0.4 | 0 | 0.2 |
| Disallowed | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PDB ID code | 3PB4 | 3PB6 | 3PB7 | 3PB8 | 3PB9 | 3PBB | 3PBE |
a Values in parentheses correspond to highest resolution shell.
Regarding the sQC mutant W207F, the purified protein (∼8 mg/ml) was mixed with an equal volume of the crystallization buffer (1.7 m (NH4)2SO4, 4% (v/v) dioxane, and 100 mm Mes, pH 6.5) and crystallized at 25 °C by the hanging-drop vapor-diffusion method. For the structure of the sQC-PBD150 complex, 2 μl of wild-type sQC (∼10 mg/ml) was mixed with 0.5 μl of PBD150 (5 mm) and 2 μl crystallization buffer (the same as for W207F) and then crystallized as described above. Subsequent cryoprotectant preparation and x-ray data collection were referenced from a previous report (4).
Structure Determination and Refinement
The crystal structure of gQC was solved by the sulfur single-wavelength anomalous dispersion phasing method using 1.13 Å resolution data collected at the wavelength of 1.0 Å (see Table 1). The positions and occupancies of the six sulfur atoms in gQC were determined with SHELX C/D/E (30). The initial phases were calculated with Phaser (31), and the phases were improved by density modification with DM (32). An initial ∼86% model was automatically traced into the electron density map with ARP/wARP (33), and the remaining model was manually built with O and Coot (34, 35). The resulting model was subjected to rigid-body refinement, simulated annealing, energy minimization, and B-factor refinement with CNS (36). Throughout the refinements, a random selection for 5% of the data was set aside as a free data set, and the model was refined against the remaining data with F > 0 as a working data set. The parameters for ideal protein geometry of Engh and Huber (37) were used during refinements. Subsequently, several rounds of model building with Coot and refinement with Refmac5 (38) were performed to improve the quality and completeness of the structure. The well ordered water molecules and one zinc ion were located with Coot. The refinement converged on a final Rfactor/Rfree of 0.126/0.153 at 30–1.13 Å resolution range. The structures of gQC at pH 6.5 and gQC-inhibitor complexes were determined by the molecular replacement method with Molrep (39) using the refined gQC structure as the search model. The structures of sQC mutant and sQC-PBD150 complex were also solved by the molecular replacement method using the wild-type sQC (Protein Data Bank code 2AFM) as the search model. These structures were refined as described above, and the stereochemical quality of the refined structures was checked with Procheck (40). The final refinement statistics are listed in Table 1. The molecular figures were produced with PyMOL (46).
RESULTS
Protein Purification, Crystallization, and Structure Determination
The Golgi-luminal region (Ser53–Leu382) of human gQC, which corresponds to the catalytic domain of the enzyme (Fig. 1), was overexpressed in E. coli cells and purified to near homogeneity. Purified gQC showed an ∼3-fold weaker QC activity on the substrate l-glutaminyl 2-naphthylamide when compared with the activity of sQC (supplemental Fig. S1A), consistent with the previous results (25). The purified enzyme revealed CD spectra typical of an α/β-protein (supplemental Fig. S1B), similar to the CD curve for sQC, confirming that the recombinant gQC was correctly folded. The gQC crystals grown at the physiological pH conditions of Golgi apparatus were selected for structure determination (supplemental Fig. S1C).
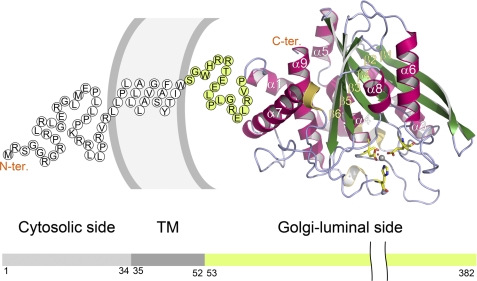
FIGURE 1.
Overall structure and domain organization of gQC. The structure of the Golgi-luminal catalytic domain of gQC is shown as a ribbon diagram. The amino acid sequences corresponding to the nine α-helices (α1–α9), six β-strands (β1–β6), and three 310-helices can also be seen in Fig. 2. The catalytic zinc ion is shown as a gray ball. The zinc-coordinated amino acid residues (Asp186, Glu226, and His351) and water molecule are drawn with the yellow stick models and a red ball, respectively. The single-letter code sequence with green background (Ser53–Pro71) represents the N-terminal flexible region of our structure. The remaining sequence with white background depicts the transmembrane (TM) (Leu35–Trp52) and cytosolic (Met1–Arg34) domains of an entire gQC molecule, as predicted with the program HMMTOP (45).
The crystal structure of human gQC was solved by the sulfur single-wavelength anomalous dispersion phasing method due to the six sulfur atoms of the enzyme. Using x-ray data from the crystals grown at pH 6.0 (Table 1), the six sulfur atom sites were successfully located with an average occupancy of 0.33. The structure was solved and refined to 1.13 Å resolution with the Rfactor/Rfree values being 0.126/0.153. Other gQC structures have also been determined and refined to high resolutions on the basis of the refined structure. In general, the gQC structures comprise 309–313 residues, one zinc ion, ∼400 water molecules, and/or one of the inhibitor molecules, with the N-terminal 17–21 residues being excluded from the models due to their high flexibility. Ramachandran plot analysis showed that >91% of the residues are in the most favored region and that no residue is in the disallowed region (Table 1). The crystals for the sQC-PBD150 complex have a space group R3, different from the R32 space group of the free-form sQC (4).
Overall Structure
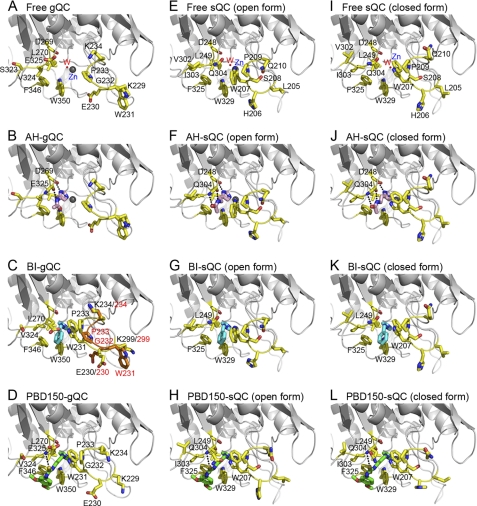
The structure of gQC, at pH 6.0, reveals a globular and α/β-mixed scaffold, which comprises a central six-stranded β-sheet surrounded by two (α6 and α8) and six (α1, α2, α3, α4, α5, and α9) α-helices on opposite sides, with one α-helix (α7) located at one edge of the β-sheet (Fig. 1), thus displaying an open sandwich topology. The central β-sheet is twisted, consisting of two antiparallel (β1 and β2) and four parallel (β3, β4, β5, and β6) strands. In addition, there are three short 310-helices adjacent to α2, α7, or α9 (Fig. 2). The remaining portion of the protein mainly consists of unstructured loops, where the major components of the active site are located. From the long distance (∼36 Å) and opposite sides between the first residue and the zinc catalytic center (Fig. 1), it is reasonable to assume that the active site of gQC is away from the Golgi membrane. The structure of gQC at pH 6.5 showed no significant conformational changes (r.m.s. deviation = 0.229 Å for all Cα atoms) from that at pH 6.0, except for a loop near the catalytic center (supplemental Fig. S2).
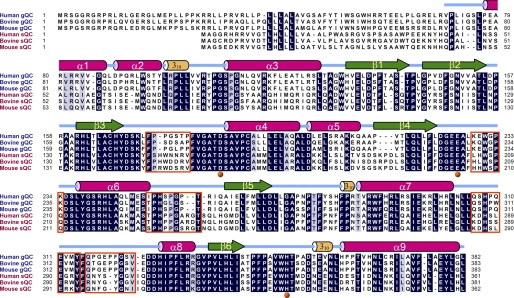
FIGURE 2.
Sequence alignment of human gQC with several representative gQCs and sQCs. The secondary structural elements, e.g. α-helices, β-strands, and 310-helices, according to our refined structures of gQC, are illustrated. The residues that are identical in five out of the six sequences are shaded in gray, and the completely conserved residues are depicted in black. The zinc-coordinated residues are marked with orange balls under the alignment. The residues at the loops that have different conformations between gQC and sQC, as described under “Results,” are boxed in orange. The GenBank accession numbers of these sequences are as follows: NP_060129 (human gQC), NP_001069408 (bovine gQC), NP_080387 (mouse gQC), NP_036545 (human sQC), NP_803472 (bovine sQC), and AAI51028 (mouse sQC).
Comparison with the sQC Structure
The gQC structure shares a highly conserved scaffold to the structure of sQC (Fig. 3A), with the r.m.s. deviations being 0.983 and 0.963 Å (for all the Cα atoms) when compared with the structures of the open form (or conf-A in Ref. 4) and closed form (or conf-B) sQCs, respectively. The notable conformational changes were found in several loops, i.e. Phe174–Pro180 (referring to the gQC sequence), Lys229–Lys234, Ile250–Thr257, Gln306–Tyr314, and Gln316–Val324 (Figs. 2 and 3A). Among these loops with distinct conformations, three of them create a large portion of the active site structure in gQC and sQC. As a consequence, the active site pocket of gQC is relatively wide open in contrast to the narrow active site structure of sQC (Fig. 3B). Moreover, the globular structure of gQC appears smaller (57 × 48 × 44 Å) than that of sQC (63 × 58 × 41 Å) (Fig. 3B) despite their almost identical residue numbers. This is due to the different conformations in the loops Phe174–Pro180 and Ile250–Thr257 between gQC and sQC and an additional α-helix located at the N terminus of sQC. Another notable difference between gQC and sQC is the negatively charged surface around the active site pocket of gQC when compared with that of sQC (Fig. 3B). This is attributed to some residue substitutions around the active site between these two QCs, as shown in Fig. 3B.
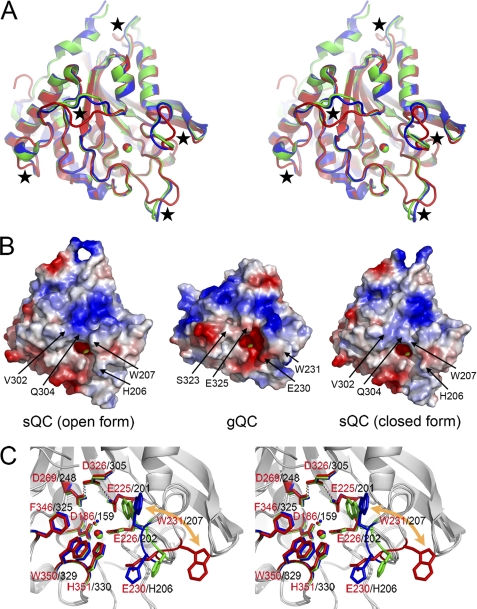
FIGURE 3.
Comparison of the gQC and sQC structures. A, superimposition of the structures of gQC (red), open form sQC (blue), and closed form sQC (green) in stereo view. The loops with different conformations between gQC and sQC are marked with black stars. B, surface charge potential of the gQC and sQC structures. The charge potentials were calculated by using the software PyMOL (46) with the values ranging from −67.258 to 67.258, colored from red to blue. Note that gQC has a more negatively charged surface around its active site. The notable residue substitutions that contribute to the distinct charge potentials between these two QCs are labeled. The tryptophan residue with a large positional change is also labeled. C, superimposition of the active site structures of gQC (red), open form sQC (blue), and closed form sQC (green) in stereo view. Note that the tryptophan residue, i.e. Trp231 in gQC and Trp207 in sQC, moves ∼15 Å in between these two QCs. By contrast, the catalytic glutamate residue (Glu225 in gQC) and the catalytically essential hydrogen-bond network (Glu225···Asp326···Asp269 in gQC) in these structures are superimposed very well.
Comparison of the Active Site Structures of gQC and sQC
The active site of gQC is located near the C-terminal edge of the central parallel strands β3, β4, and β5 (Fig. 1), forming a relatively open active site pocket accessible to the solvent. The single zinc ion (26) at the bottom of the active site pocket is tetrahedrally coordinated to the two carboxylic oxygen atoms from Asp186 and Glu226, the imidazole nitrogen atom from His351, and a water molecule (Fig. 4, A and B). The three zinc-coordinating residues are completely conserved among animal gQCs and sQCs (Fig. 2). A cis-peptide bond between Asp186 and Ser187 was observed, stabilized by a number of hydrogen bonds. This cis-peptide bond was also found in the structures of sQC and some binuclear aminopeptidases (4, 41, 42). In addition, several other residues surrounding the zinc catalytic center (Fig. 4A) are also highly conserved in animal gQCs (supplemental Fig. S3), especially the hydrophobic and completely conserved Trp231, Leu270, Val324, Phe346, and Trp350. This may reflect that gQCs prefer substrates with hydrophobic residues at their second or third positions (25, 26). Unexpectedly, the structure of gQC at pH 6.5 showed a cacodylate molecule bound to the zinc ion in the replacement of the zinc-coordinated water molecule (Fig. 4C), likely due to the presence of 0.1 m cacodylate in the crystallization buffer. Our inhibition assays demonstrated that cacodylate possesses weak inhibition activities toward gQC and sQC, with the Ki values in the low millimolar range (Table 2).
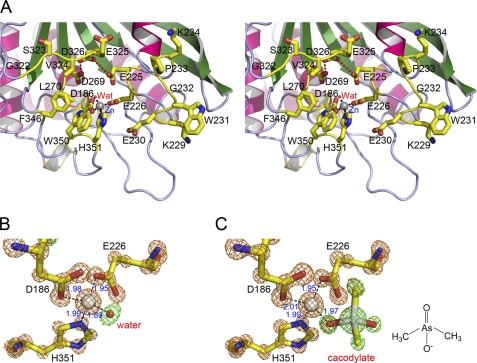
FIGURE 4.
The active site structures of gQC. A, a close-up and stereo view of the active site structure of gQC at pH 6.0. The model representations are the same as in Fig. 1. Wat, water molecule. B, a close-up view of the tetrahedral zinc coordination environment of gQC at pH 6.0. The 7.0σ Fo − Fc stimulated annealing omit densities for the zinc ion (gold) and the zinc-coordinated residues (gold) and water molecule (green) are overlaid with the refined models. The coordination bonds are drawn with distances indicated in angstroms. C, a close-up view of the zinc coordination environment of gQC at pH 6.5. Note that the zinc-coordinated water is replaced by a cacodylate molecule. The chemical structure of cacodylate is shown on the left. The Fo − Fc omit densities are also shown, with that for the cacodylate molecule being colored in green.
TABLE 2.
Inhibition effects of cacodylate and imidazole derivatives on gQC and sQC
Assays were performed at 25 °C in 50 mm Tris-HCl, pH 8.0, using l-glutaminyl 2-naphthylamide as substrate. Results are means ± S.E. from three experiments.
| Compound |
Ki value |
|
|---|---|---|
| gQC | sQC | |
| μm | ||
| Cacodylate | 6696 ± 1274 | 1824 ± 68 |
| N-ω-Acetylhistamine | 5.748 ± 0.366 | 1.698 ± 0.493 |
| 1-Benzylimidazole | 0.262 ± 0.062 | 0.607 ± 0.089 |
| PBD150 | 1.817 ± 0.154 | 0.095 ± 0.003 |
The active site pocket of gQC has a dimension of ∼18 × 15 × 8 Å, broader than that of sQC (∼13 × 11 × 7 Å). The different orientations in two loops, particularly the loop Lys229–Lys234, contribute to the distinct active site conformations (Fig. 3A). Trp231 in this loop of gQC shows an outward positioning in its indole ring (Fig. 3C), whereas the corresponding Trp201 in sQC has an inward orientation. Notably, sQC itself has two distinct active site conformations due to the opposite indole orientations of Trp207 (Fig. 3C), whereas only one conformation was found in gQC. A previous study indicated that mutating Trp207 to phenylalanine in sQC reduced the turnover rate (kcat) of the enzyme by ∼3.7-fold (4). Here, the structure of this sQC mutant (W207F) shows that Phe207 has similar orientations in the two sQC molecules of the asymmetric unit (supplemental Fig. S4), suggesting that flipping in the Trp207 indole ring of sQC plays a catalytic role. This may explain the ∼3-fold weaker activity of gQC when compared with sQC.
From superimposition of the active site structures of gQC and sQC (Fig. 3C), the hydrophobic substrate-binding pocket in sQC is primarily defined by the residues Trp207, Phe325, and Trp329, whereas the corresponding substrate-binding site in gQC is significantly extended because of an ∼15 Å movement in the indole ring of Trp231 relative to that of Trp207 in sQC. This implies that these two QCs have distinct substrate binding modes, although Trp231 Nϵ1 makes a hydrogen bond to the Pro256-O of a neighboring symmetry-related gQC molecule, and this may stabilize the observed conformation here. In solution, flexibility of the loop also suggests its functional role. On the other hand, Glu225 of gQC could be superimposed very well with the catalytic glutamate residue (Glu201) of sQC (Fig. 3C) (4). The catalytically essential hydrogen-bond network (Glu201···Asp305···Asp248) proposed for sQCs (5) is also conserved in gQC (Glu225···Asp326···Asp269). These findings suggest that gQC shares a conserved catalytic mechanism with sQC.
Structures of the Inhibitor-bound gQC and sQC
Because the bound cacodylate molecule interfered with the inhibitor binding during the soaking experiments, the inhibitor-bound structures of gQC were obtained only from the crystals grown at pH 6.0. The clear omit density maps for the bound inhibitors allowed us to precisely locate their positions in the gQC structures (Fig. 5). In general, the bound inhibitor does not induce large conformational changes in gQC structure (average r.m.s. deviation = 0.3 Å for all Cα atoms), except for a loop movement in the active site of the 1-benzylimidazole-bound and PBD150-bound gQC structures (compare Fig. 6, C and D with Fig. 6A). The inhibitors use one of their imidazole nitrogen atoms to bind the catalytic zinc ion by replacing the zinc-coordinated water molecule. In the structure of N-ω-acetylhistamine-bound gQC (Fig. 6B), the inhibitor has an orientation almost parallel to the backbone of Val324–Asp326, stabilized by two hydrogen bonds to Asp269 Oδ2 (distance = 2.76 Å) and Glu325 O (2.70 Å) in addition to the zinc coordination bond. When compared with the structures of N-ω-acetylhistamine-bound sQC (Fig. 6, F and J), the inhibitor adopts almost identical orientations in these two enzymes but loses a hydrogen bond in gQC (Fig. 6B) because of the slightly outward positioning of Val324 and Glu325 in gQC when compared with the corresponding Ile303 and Gln304 in sQC. Our inhibition assay demonstrated that the inhibitor binds gQC ∼3.4-fold weaker when compared with sQC (Table 2), presumably reflecting the different numbers of hydrogen bonding.
FIGURE 5.
The QC inhibitors used in the present study. A, chemical structure of the QC inhibitors. B, the Fo − Fc stimulated annealing omit densities for the inhibitors bound in various QC structures, as indicated, are overlaid with the final refined models. The contour levels of the densities are also indicated.
FIGURE 6.
Comparison of the binding modes of QC inhibitors in gQC and sQC. A, the free-form gQC. B, the N-ω-acetylhistamine-bound gQC (AH-gQC). To enhance the clarity of the hydrogen bondings, the side chain of Glu325 is sliced out of view. C, the 1-benzylimidazole-bound gQC (BI-gQC). Note that the loop Lys229–Lys234 shows two distinct conformations, with the one identical to that of free-form enzyme being colored in orange. D, the PBD150-bound gQC. Note that the loop Lys229–Lys234 shows a large conformational change when compared with that of free-form enzyme. E, the free-form sQC (open form). F, the N-ω-acetylhistamine-bound sQC (open form). Note that the inhibitor forms three hydrogen bonds with sQC, in contrast to the two with gQC as observed in panel B. For clarity, the side chain of Gln304 is sliced out of view. G, the 1-benzylimidazole-bound sQC (open form). The inhibitor shows fewer hydrophobic contacts with sQC when compared with that with gQC as observed in panel C. H, the PBD150-bound sQC (open form). Note that the indole ring of Trp207 rotates to an opposite orientation when compared with that of free-form enzyme. I, the free-form sQC (closed form). J, the N-ω-acetylhistamine-bound sQC (closed form). The inhibitor exhibits a binding mode as observed in panel F. For clarity, the side chain of Gln304 is also sliced out of view. K, the 1-benzylimidazole-bound sQC (closed form). The inhibitor shows a binding mode as observed in panel G, except for the additional contact with Trp207. L, the PBD150-bound sQC (closed form). Except in the free-form QCs, only the residues that bind to the inhibitors or show a large positional change are labeled. The structures for free-form, N-ω-acetylhistamine-bound, and 1-benzylimidazole-bound sQCs are from the previous report (4).
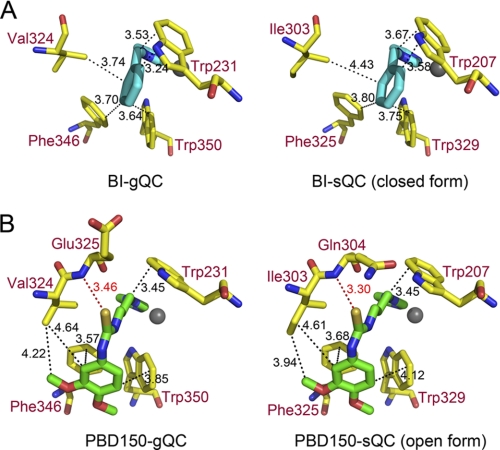
The structure of 1-benzylimidazole-bound gQC revealed two distinct conformations in the loop Lys229–Lys234 of gQC (Fig. 6C), with Trp231 in this loop moving ∼17 Å. The bulky phenyl ring of the inhibitor is closely surrounded by the hydrophobic Val324, Phe346, and Trp350, with distances of 3.64–3.74 Å (Fig. 7A). This inhibitor also attracts the hydrophobic Trp231 to attach (distance ∼3.4 Å) through a large loop movement in the enzyme. In addition, the imidazole ring of the inhibitor makes van der Waals contacts with Leu270 with a distance of 3.8 Å (Fig. 6C). By contrast, the structures of 1-benzylimidazole-bound sQC revealed fewer hydrophobic contacts between the inhibitor and sQC (4), primarily because of Ile303 being farther from the bound inhibitor when compared with the corresponding Val324 in gQC (Fig. 7A). Moreover, the Trp207-inhibitor interaction in sQC, which corresponds to the Trp231-inhibitor interaction in gQC, can only be found in the closed form, but not the open form, structure of sQC (compare Fig. 6, G and K). This is due to the opposite orientations in the indole ring of Trp207. The ∼2.3-fold stronger effect of the inhibitor toward gQC than sQC may reflect the distinct extents in hydrophobic interaction between the inhibitor and these two QCs (Table 2).
FIGURE 7.
A close-up view of the interactions of 1-benzylimidazole and PBD150 with gQC and sQC. A, comparison of 1-benzylimidazole (BI) bound to gQC and sQC. The possible hydrophobic contacts are drawn with black dashed lines, with distances being indicated in angstroms. B, comparison of PBD150 bound to gQC and sQC. The possible hydrophobic contacts and hydrogen bonds are drawn with dashed lines colored in black and red, respectively, with distances being indicated in angstroms.
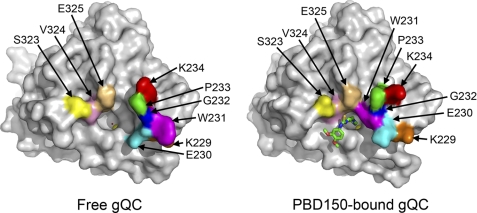
In the structure of PBD150-bound gQC, the Lys229–Lys234 loop in gQC underwent a large conformational change (compare Fig. 6D with Fig. 6A and also see Fig. 8). When compared with the structure of 1-benzylimidazole-bound gQC (Fig. 6C), similar conformational change in this loop was observed, except for the different Lys229, Glu230, and Lys234 side-chain positionings. The sulfur atom in PBD150 forms a hydrogen bond to Glu325-N (Figs. 6D and 7B), with a reasonable distance (3.46 Å) (43). The phenyl ring of the inhibitor partially stacks with the phenyl group of Phe346 and also makes van der Waals contacts with the indole ring of Trp350 and the side chain of Val324 (Fig. 7B). Moreover, the propyl chain of the inhibitor is bound with Trp231 through a large loop movement in the enzyme. Similar to 1-benzylimidazole, the imidazole ring of PBD150 also makes van der Waals contacts with Leu270 (distance = 3.85 Å) (supplemental Fig. S5). In the structures of PBD150-bound sQC (Fig. 6, H and L), the inhibitor adopts an almost identical binding mode as observed in the gQC structure, except for the slightly stronger hydrophobic interaction to Ile303 when compared with the corresponding interaction to Val324 in gQC (Fig. 7B). Notably, Trp207 in both the open form and the closed form structures of sQC shows an identical orientation that binds to the propyl chain of PBD150. Surprisingly, our inhibition assay showed that PBD150 is ∼19-fold more effective toward sQC than gQC (Table 2), which seems unlikely to reflect the limited difference in the inhibitor modes. Probably, the relatively neutral surface around the active site of sQC favors the exposed and hydrophobic phenyl group of PBD150, in contrast to the negatively charged surface around the active site of gQC (Fig. 3B).
FIGURE 8.
Surface representation showing the large loop movement in the active site of gQC upon binding to PBD150. The residues that undergo a large positional change upon inhibitor binding are labeled and painted with various colors.
DISCUSSION
As mentioned in previous reports (13, 15), QC inhibitors may have potential application in the treatment of the diseases associated with aberrantly higher QC activity, such as Alzheimer disease. Previous studies and developments of QC inhibitors were focused on the sQC target. However, gQC may cause side effects by those QC inhibitors because limited information is available about the QC-involved pathways, and numerous hormones and chemokines require QC for their maturation. Therefore, understanding the structure-function relationship of gQC and sQC would be useful for the discovery of more specific QC inhibitors and for the understanding of physiological functions of these two QCs.
Here, we describe the atomic resolution structures of gQC at physiological pH conditions, which reveal a relatively wide open and negatively charged active site when compared with that of sQC. We also report the structures of PBD150-bound gQC and sQC. Moreover, in addition to our published structures of sQC bound to the inhibitors N-ω-acetylhistamine and 1-benzylimidazole (4), the structures of gQC in complex with these two inhibitors are described as well. On the basis of these inhibitor-bound structures, we have observed distinct inhibitor binding modes between gQC and sQC, which are consistent with the results from our inhibitor assays reported here. Interestingly, the cacodylate molecule found in the active site of gQC may allow us to suggest a new starting model, other than the imidazole ring, for the discovery of potent QC inhibitors.
Unexpectedly, our inhibition assays showed that the inhibitor PBD150 is ∼19-fold more effective toward sQC than gQC. This is likely due to different hydrophobic and charge interactions between this inhibitor and these two enzymes. Likewise, two distinct Drosophila QCs, named DromeQC and isoDromeQC, were previously identified and characterized in which DromeQC had an ∼32-fold weaker sensitivity toward the PBD150 inhibition when compared with that of isoDromeQC (44). By aligning the sequences of Drosophila QCs with the gQC and sQC sequences (supplemental Fig. S6A), we found that the residues contributing to the hydrophobic interactions with PBD150 are completely conserved in these QCs, except for Val324 (referring to the gQC sequence). Surprisingly, DromeQC has also a valine residue at this position, but this valine is substituted by an isoleucine in sQC and isoDromeQC. This finding supports our structural observation that the Val/Ile exchange between gQC and sQC leads, in part, to their different sensitivities toward the PBD150 inhibition. Moreover, the structure model of DromeQC appears to have a more negatively charged surface around its active site when compared with the structure of isoDromeQC (supplemental Fig. S6B), primarily because of the Asp/Ser substitution, consistent with our hypothesis.
On the basis of the strong inhibition activity of 1-benzylimidazole when compared with N-ω-acetylhistamine, the active site of QCs appears to prefer the binding of hydrophobic inhibitors, which is consistent with the preference for QC substrates (3). In fact, more than half of the contacts between PBD150 and QCs are due to hydrophobic interactions. These findings presumably reflect that the hydrophobic pocket in the active site of QCs is shared by the inhibitors and substrates. The hydrophobic propyl chain of PBD150 allows Trp207 in sQC to bind it. This propyl group also attracts Trp231 in gQC to touch through a large loop movement in the enzyme, implicating that the “cap-like” tryptophan residue in QCs plays a critical role in the inhibitor binding. Previously, Buchholz et al. (22) showed that replacing the propyl chain of PBD150 by an ethyl group reduced the inhibition activity by ∼300-fold, likely due to a loss of hydrophobic interactions with the tryptophan residue. Moreover, as judged from the length of the propyl chain plus the imidazole ring in PBD150, Trp231 in gQC, and also Trp207 in sQC, may be able to bind the second residue of QC substrates during catalysis. Namely, a similar loop movement may occur in gQC upon substrate binding.
Recently, Buchholz et al. (23) further improved the potency of PBD150 by an order of magnitude by adding a methyl group at the C5 atom of the imidazole ring (supplemental Fig. S5). By examining the imidazole environment of the bound PBD150, we found that the C4 and C5 atoms of the imidazole ring are surrounded by the side chain of a leucine residue, i.e. Leu270 in gQC and Leu249 in sQC, and a small space adjacent to the C5 atom still remains (supplemental Fig. S5). In this regard, our structures clearly indicate that the additional methyl group in PBD150 creates a more extensive hydrophobic contact with the leucine residue. This finding also indicates the obvious importance of hydrophobic interaction in the inhibitor binding to QCs.
Tissue distribution analysis of gQC in mouse model revealed that the enzyme is ubiquitously expressed (25), suggesting an important physiological role of the enzyme. The co-retention of gQC with glycosyltransferases in the Golgi apparatus implies a “housekeeping” protein maturation system, responsible for glycosylation and N-terminal pGlu formation of proteins. In addition, the existence of gQC may complement the physiological functions of sQC, whose expression was mainly identified in the brain tissues (8). These two QCs may be responsible for catalysis in different tissues, i.e. sQC in neuronal tissues and gQC in peripheral tissues.
Acknowledgments
We thank Dr. Tzu-Ping Ko of the Institute of Biological Chemistry at Academia Sinica (Taipei, Taiwan) for helpful discussion of the manuscript. We are grateful to Hui-Ling Shr and Ren-Han Hsu of the Core Facility for Protein Production and X-Ray Structural Analysis (Taipei, Taiwan) for crystallization screening. We are also grateful for the access to the synchrotron radiation beamlines 13B1 and 13C1 (National Synchrotron Radiation Research Center, Hsinchu, Taiwan).
This work was supported by the Academia Sinica and Core Facility for Protein Production and X-Ray Structural Analysis of the National Research Program for Genomic Medicine (Grant NSC97-3112-B-001-035-B4 to A. H.-J. W.).
The atomic coordinates and structure factors (codes 3PB4, 3PB6, 3PB7, 3PB8, 3PB9, 3PBB, and 3PBE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- pGlu
- pyroglutamate
- QC
- glutaminyl cyclase
- gQC
- Golgi-resident QC
- sQC
- secretory QC
- βNA
- 2-naphthylamide
- r.m.s.
- root mean square.
REFERENCES
- 1. Goren H. J., Bauce L. G., Vale W. (1977) Mol. Pharmacol. 13, 606–614 [PubMed] [Google Scholar]
- 2. Van Coillie E., Proost P., Van Aelst I., Struyf S., Polfliet M., De Meester I., Harvey D. J., Van Damme J., Opdenakker G. (1998) Biochemistry 37, 12672–12680 [DOI] [PubMed] [Google Scholar]
- 3. Schilling S., Manhart S., Hoffmann T., Ludwig H. H., Wasternack C., Demuth H. U. (2003) Biol. Chem. 384, 1583–1592 [DOI] [PubMed] [Google Scholar]
- 4. Huang K. F., Liu Y. L., Cheng W. J., Ko T. P., Wang A. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13117–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang K. F., Wang Y. R., Chang E. C., Chou T. L., Wang A. H. (2008) Biochem. J. 411, 181–190 [DOI] [PubMed] [Google Scholar]
- 6. Seifert F., Schulz K., Koch B., Manhart S., Demuth H. U., Schilling S. (2009) Biochemistry 48, 11831–11833 [DOI] [PubMed] [Google Scholar]
- 7. Busby W. H., Jr., Quackenbush G. E., Humm J., Youngblood W. W., Kizer J. S. (1987) J. Biol. Chem. 262, 8532–8536 [PubMed] [Google Scholar]
- 8. Sykes P. A., Watson S. J., Temple J. S., Bateman R. C., Jr. (1999) FEBS Lett. 455, 159–161 [DOI] [PubMed] [Google Scholar]
- 9. Wintjens R., Belrhali H., Clantin B., Azarkan M., Bompard C., Baeyens-Volant D., Looze Y., Villeret V. (2006) J. Mol. Biol. 357, 457–470 [DOI] [PubMed] [Google Scholar]
- 10. Schilling S., Stenzel I., von Bohlen A., Wermann M., Schulz K., Demuth H. U., Wasternack C. (2007) Biol. Chem. 388, 145–153 [DOI] [PubMed] [Google Scholar]
- 11. Huang W. L., Wang Y. R., Ko T. P., Chia C. Y., Huang K. F., Wang A. H. (2010) J. Mol. Biol. 401, 374–388 [DOI] [PubMed] [Google Scholar]
- 12. Guevara T., Mallorquí-Fernández N., García-Castellanos R., García-Piqué S., Ebert Petersen G., Lauritzen C., Pedersen J., Arnau J., Gomis-Rüth F. X., Solà M. (2006) Biol. Chem. 387, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 13. Schilling S., Zeitschel U., Hoffmann T., Heiser U., Francke M., Kehlen A., Holzer M., Hutter-Paier B., Prokesch M., Windisch M., Jagla W., Schlenzig D., Lindner C., Rudolph T., Reuter G., Cynis H., Montag D., Demuth H. U., Rossner S. (2008) Nat. Med. 14, 1106–1111 [DOI] [PubMed] [Google Scholar]
- 14. Ezura Y., Kajita M., Ishida R., Yoshida S., Yoshida H., Suzuki T., Hosoi T., Inoue S., Shiraki M., Orimo H., Emi M. (2004) J. Bone. Miner. Res. 19, 1296–1301 [DOI] [PubMed] [Google Scholar]
- 15. Batliwalla F. M., Baechler E. C., Xiao X., Li W., Balasubramanian S., Khalili H., Damle A., Ortmann W. A., Perrone A., Kantor A. B., Gulko P. S., Kern M., Furie R., Behrens T. W., Gregersen P. K. (2005) Genes Immun. 6, 388–397 [DOI] [PubMed] [Google Scholar]
- 16. Thouënnon E., Elkahloun A. G., Guillemot J., Gimenez-Roqueplo A. P., Bertherat J., Pierre A., Ghzili H., Grumolato L., Muresan M., Klein M., Lefebvre H., Ouafik L., Vaudry H., Plouin P. F., Yon L., Anouar Y. (2007) J. Clin. Endocrinol Metab. 92, 4865–4872 [DOI] [PubMed] [Google Scholar]
- 17. Muthusamy V., Duraisamy S., Bradbury C. M., Hobbs C., Curley D. P., Nelson B., Bosenberg M. (2006) Cancer. Res. 66, 11187–11193 [DOI] [PubMed] [Google Scholar]
- 18. Cynis H., Scheel E., Saido T. C., Schilling S., Demuth H. U. (2008) Biochemistry 47, 7405–7413 [DOI] [PubMed] [Google Scholar]
- 19. Schilling S., Appl T., Hoffmann T., Cynis H., Schulz K., Jagla W., Friedrich D., Wermann M., Buchholz M., Heiser U., von Hörsten S., Demuth H. U. (2008) J. Neurochem. 106, 1225–1236 [DOI] [PubMed] [Google Scholar]
- 20. Saido T. C., Iwatsubo T., Mann D. M., Shimada H., Ihara Y., Kawashima S. (1995) Neuron 14, 457–466 [DOI] [PubMed] [Google Scholar]
- 21. Hook V., Schechter I., Demuth H. U., Hook G. (2008) Biol. Chem. 389, 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buchholz M., Heiser U., Schilling S., Niestroj A. J., Zunkel K., Demuth H. U. (2006) J. Med. Chem. 49, 664–677 [DOI] [PubMed] [Google Scholar]
- 23. Buchholz M., Hamann A., Aust S., Brandt W., Böhme L., Hoffmann T., Schilling S., Demuth H. U., Heiser U. (2009) J. Med. Chem. 52, 7069–7080 [DOI] [PubMed] [Google Scholar]
- 24. Gillis J. S. (2006) J. Transl. Med. 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cynis H., Rahfeld J. U., Stephan A., Kehlen A., Koch B., Wermann M., Demuth H. U., Schilling S. (2008) J. Mol. Biol. 379, 966–980 [DOI] [PubMed] [Google Scholar]
- 26. Stephan A., Wermann M., von Bohlen A., Koch B., Cynis H., Demuth H. U., Schilling S. (2009) FEBS J. 276, 6522–6536 [DOI] [PubMed] [Google Scholar]
- 27. Huang K. F., Liu Y. L., Wang A. H. (2005) Protein. Expr. Purif. 43, 65–72 [DOI] [PubMed] [Google Scholar]
- 28. Segel I. H. (1993) in Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme System, pp. 100–118, John Wiley & Sons, Inc., New York [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cowtan K. D., Zhang K. Y. (1999) Prog. Biophys. Mol. Biol. 72, 245–270 [DOI] [PubMed] [Google Scholar]
- 33. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 34. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37. Engh R. A., Huber R. (1991) Acta Crystallogr. A 47, 392–400 [Google Scholar]
- 38. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 39. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 40. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 41. Chevrier B., Schalk C., D'Orchymont H., Rondeau J. M., Moras D., Tarnus C. (1994) Structure 2, 283–291 [DOI] [PubMed] [Google Scholar]
- 42. Greenblatt H. M., Almog O., Maras B., Spungin-Bialik A., Barra D., Blumberg S., Shoham G. (1997) J. Mol. Biol. 265, 620–636 [DOI] [PubMed] [Google Scholar]
- 43. Valdés-Martínez J., Hernández-Ortega S., Rubio M., Li D. T., Swearingen J. K., Kaminsky W., Kelman D. R., West D. X. (2004) J. Chem. Crystallogr. 34, 533–540 [Google Scholar]
- 44. Schilling S., Lindner C., Koch B., Wermann M., Rahfeld J. U., von Bohlen A., Rudolph T., Reuter G., Demuth H. U. (2007) Biochemistry 46, 10921–10930 [DOI] [PubMed] [Google Scholar]
- 45. Tusnády G. E., Simon I. (2001) Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 46. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]