Abstract
Considerable attention has recently been paid to the N-Myc downstream-regulated gene (NDRG) family because of its potential as a tumor suppressor in many human cancers. Primary amino acid sequence information suggests that the NDRG family proteins may belong to the α/β-hydrolase (ABH) superfamily; however, their functional role has not yet been determined. Here, we present the crystal structures of the human and mouse NDRG2 proteins determined at 2.0 and 1.7 Å resolution, respectively. Both NDRG2 proteins show remarkable structural similarity to the ABH superfamily, despite limited sequence similarity. Structural analysis suggests that NDRG2 is a nonenzymatic member of the ABH superfamily, because it lacks the catalytic signature residues and has an occluded substrate-binding site. Several conserved structural features suggest NDRG may be involved in molecular interactions. Mutagenesis data based on the structural analysis support a crucial role for helix α6 in the suppression of TCF/β-catenin signaling in the tumorigenesis of human colorectal cancer, via a molecular interaction.
Keywords: Cell Differentiation, Cellular Regulation, Myc, Tumor Suppressor, X-ray Crystallography, Apoptosis, NDRG Family, NDRG2
Introduction
Myc directs numerous biological functions, such as cell proliferation, cell growth, apoptosis, and differentiation, via transcriptional regulation of its target genes (1). Among the target genes, Myc-repressed genes have been of particular interest, because several of these genes have been found to possess tumor-suppressive and anti-metastatic properties (2). The human N-Myc downstream-regulated gene family (NDRG) is a novel class of Myc-repressed genes that has gained a lot of attention in recent years. This family consists of NDRG1, NDRG2, NDRG3, and NDRG4 and includes alternative splicing isoforms of NDRG2 and NDRG4. The NDRG family is also found in other multicellular organisms and is highly conserved between species. Studies of the function and regulation of the NDRG family indicate that it plays multiple roles. In particular, recent data suggest that NDRG proteins may function as tumor suppressors and may also play important roles in the development of other diseases. For example, NDRG1 has been recognized as a metastasis suppressor in prostate (3), breast (4), and colon (5) cancers. In addition, NDRG1 is known to be responsible for the demyelinating neuropathy associated with hereditary motor and sensory neuropathy-Lom (6). NDRG2 has also been identified as a potential tumor suppressor. Its expression is negatively correlated with cancer progression, and it was significantly diminished in the cancerous tissues of patients suffering from breast (7), lung (8), colon (9, 10), skin (11), thyroid (12), liver (13), oral (14), and gastric (15) cancers. In addition, NDRG2 is recognized to play a role in the development of Alzheimer disease. Up-regulation of this gene is associated with disease pathogenesis of the human brain (16). Involvement of NDRG3 and NDRG4 in several cancers has been studied as well, although conflicting results have been reported (17–19). Thus, members of the NDRG family are considered to be important to tumorigenesis and may be used as biomarkers in many cancers, as well as in other diseases.
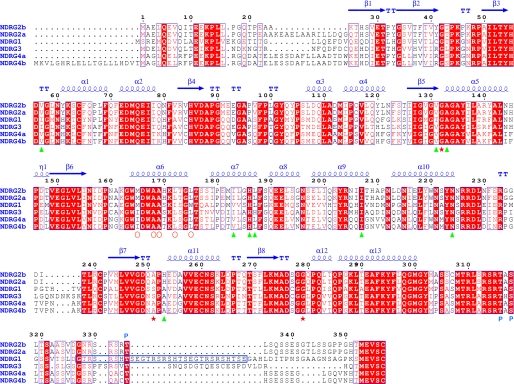
The human NDRG family members exhibit 53–65% amino acid sequence identity to each other, with most variation in the N- and C-terminal regions. The most notable difference is that NDRG1 contains three decapeptide sequence (GTRSRSHTSE) repeats at its C terminus, whereas the remaining members have only a single repeat, which also lacks the last two residues Ser and Glu (Fig. 1). Although the C-terminal regions of NDRG proteins contain several modified residues that enable phosphorylation by different protein kinases (20), and are predicted to have an unfolded structure, a Bayesian computational algorithm established that the NDRG proteins may belong to the α/β-hydrolase (ABH)5 superfamily (Fig. 2a) (21). The amino acid sequences of NDRG proteins are best conserved in the region spanning this ABH domain. Therefore, the ABH domain of the NDRG family may play an important role in its biological activity, although functional motifs within the ABH domain have not yet been identified.
FIGURE 1.
Structure-related functional sequence conservation between hNDRG family members. Elements of the secondary structure of hNDRG2 are shown above the alignments. The numbering is based on hNDRG2, which is labeled NDRG2b in this figure. The other isoform of hNDRG2 is labeled NDRG2a. hNDRG4 also consists of two isoforms, a and b. Red stars represent residues located at the corresponding positions of the conserved catalytic residues Ser, Asp, and His, respectively, in the α/β-hydrolase family proteins. Green triangles represent residues involved in the pseudo-active site. Red open circles indicate the hydrophobic residues on the helix α6. P indicates phosphorylation sites. The long blue, open box indicates the three decapeptide sequence repeats of hNDRG1. Strictly conserved residues are highlighted with solid red boxes. Biological sources and accession codes for the sequences are as follows: NDRG2b, N-Myc downstream-regulated gene 2 isoform b (gi:42544224); NDRG2a, N-Myc downstream-regulated gene 2 isoform a (gi:42544222); NDRG1, N-Myc downstream-regulated gene 1 (gi:48145801); NDRG3, N-Myc downstream-regulated gene 3 (gi:12083721); NDRG4a, N-Myc downstream-regulated gene 4 isoform a (gi:13430864); and NDRG4b, N-Myc downstream-regulated gene 4 isoform b (gi:194440722). Sequence alignments were assembled using T-COFFEE software and visualized using ESPript software, both located on the ExPASy Proteomics Server.
FIGURE 2.
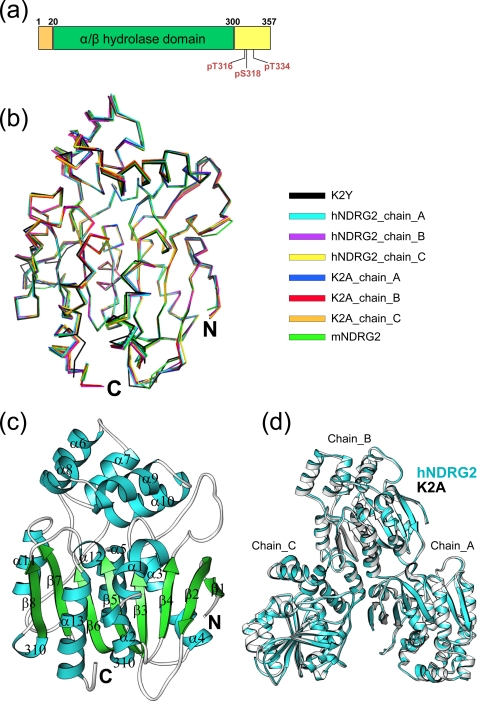
Crystal structure of NDRG2. a, schematic representation of the domain structure of human NDRG2. Phosphorylation sites are indicated. b, superimposition of each chain of the hNDRG2 and K2A structures in the asymmetric unit, as well as K2Y and mNDRG2. c, ribbon representation of K2Y. α and 310 helices are shown in cyan, β-strands in green, and loops in gray. d, hNDRG2 (cyan) and K2A (gray) molecules in the asymmetric unit.
Although the importance of the NDRG family in cellular function has garnered much attention over the last decade, efforts to decipher its function at the molecular level have been largely unsuccessful. In addition, although amino acid sequence analysis suggests that the NDRG members belong to the ABH superfamily, this still needed to be confirmed by an experimentally determined structure. Therefore, to understand the intrinsic structural properties and the molecular basis for the biological function of the NDRG family, we have performed a crystallographic analysis of the human and mouse NDRG2 proteins. Here, we describe the structural details of NDRG2 and suggest its involvement in molecular interactions. Because sequences and structures of mouse and human NDRG2 are virtually identical, our analysis is focused on the human homologue.
EXPERIMENTAL PROCEDURES
Gene Cloning and Surface Entropy Reduction
The gene encoding human NDRG2 isoform (NDRG2b, gi: 42544224, hereafter referred to as hNDRG2) was supplied by Dr. Jae Wha Kim at Korea Research Institute of Bioscience and Biotechnology. The DNA fragment encoding the human NDRG2 protein (residues 23–304) was amplified by PCR and subcloned into the pPosKJ expression vector (22). This vector produced NDRG2(23–304) fused to a hexahistidine tag and bacterial hemoglobin (His6-VHb) at its N terminus. To improve crystal quality of the human NDRG2, surface entropy reduction (SER) (23) was introduced into the NDRG2(23–304) protein to induce epitopes that favor the formation of crystal contacts. The program SER predicted candidate residues Lys45 and Lys47 for engineering. Double mutations at these positions were introduced by replacing these residues with alanine (K45A/K47A, referred to as K2A) or tyrosine (K45Y/K47Y, referred to as K2Y), using QuikChange (Stratagene), according to the manufacturer's protocol. The gene encoding mouse NDRG2 (mNDRG2, gi: 15277976) shares 95% amino acid sequence identity with human NDRG2, except for the extra 14 N-terminal residues from 26 to 39. The DNA fragment encoding the mNDRG2 protein (residues 40–313 corresponding to residues 26–299 of hNDRG2) was amplified by PCR from a clone obtained from the IMAGE consortium. The PCR product was subcloned into plasmid pMH4, which encodes an expression and purification tag (MGSDKIHHHHHH) at the N terminus. The cloning junctions were confirmed by DNA sequencing.
Protein Expression and Purification
Escherichia coli Rosetta-gami (DE3) cells harboring the overexpression plasmids of hNDRG2(23–304) and its mutants were grown in LB/ampicillin medium at 37 °C until the cultures reached an A600 between 0.6 and 0.8. The temperature was lowered to 25 °C, and protein expression was induced by addition of 0.25 mm isopropyl β-d-thiogalactopyranoside for 15 h. The cells were then harvested by centrifugation at 5,000 × g for 10 min at 4 °C. The cell pellets were resuspended in ice-cold buffer A (50 mm Tris-HCl, pH 8.0, 300 mm NaCl), and the cell suspension was ultrasonicated. The crude cell extracts were centrifuged at 11,000 × g for 1 h at 4 °C. Cell lysate containing His6-VHb fused proteins was loaded onto a nickel-nitrilotriacetic acid-agarose (Qiagen) column that was pre-equilibrated with buffer A. The column was intensively washed with buffer A, and the resin-bound proteins were eluted with buffer A containing 250 mm imidazole. The His6-VHb was then released from the protein by incubation with recombinant tobacco etch virus protease (Invitrogen) at 10 °C for 15 h, followed by size-exclusion chromatography and nickel-nitrilotriacetic acid affinity column chromatography. After purification, the recombinant native and mutant proteins contained a three-residue cloning artifact (Gly-His-Met) in their N termini. The purified proteins were dialyzed against 50 mm Tris-HCl, pH 8.0, concentrated to 10–20 mg/ml, and stored at −80 °C until used. Size-exclusion chromatography on a Superdex-75 10/30 column (Amersham Biosciences) equilibrated with 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl showed the molecular mass of the proteins to be ∼26 kDa (data not shown), indicating that hNDRG2(23–304) and its mutants exist as monomers in solution. The mNDRG2 expression was performed in a modified Terrific Broth using the E. coli strain GeneHogs® (Invitrogen). Lysozyme was added to the culture at the end of fermentation to a final concentration of 250 μg/ml. Bacteria were lysed by sonication after a freeze-thaw procedure in lysis buffer (50 mm Tris-HCl, pH 7.9, 50 mm NaCl, 10 mm imidazole, 0.25 mm tris(2-carboxyethyl)phosphine hydrochloride (TCEP)), and the cell debris was pelleted by centrifugation at 3,400 × g for 60 min. The soluble fraction was applied to a nickel resin (Amersham Biosciences) pre-equilibrated with lysis buffer. The nickel resin was washed with wash buffer (50 mm potassium phosphate, pH 7.8, 40 mm imidazole, 300 mm NaCl, 10% (v/v) glycerol, 0.25 mm TCEP), and the protein was eluted with buffer (20 mm Tris-HCl, pH 7.9, 300 mm imidazole, 10% (v/v) glycerol, 0.25 mm TCEP). Buffer exchange was performed to remove imidazole from the eluate, and the protein in buffer Q (20 mm Tris-HCl, pH 7.9, 5% (v/v) glycerol, 0.25 mm TCEP) containing 50 mm NaCl was applied to a Resource Q column (Amersham Biosciences) pre-equilibrated with the same buffer. The protein was eluted using a linear gradient of 50–500 mm NaCl in buffer Q. The appropriate fractions were pooled, further purified using a Superdex 200 size-exclusion column (Amersham Biosciences) with elution in crystallization buffer (20 mm Tris-HCl, pH 7.9, 150 mm NaCl, 0.25 mm TCEP), and concentrated to 17.6 mg/ml for crystallization assays.
Crystallization
The initial crystallization of hNDRG2 was performed using commercially available sparse-matrix screening kits, using the sitting drop vapor-diffusion method. Initial crystals of hNDRG2(23–304) were produced using 18% PEG 8000, 0.2 m calcium acetate, and 0.1 m sodium cacodylate, pH 6.5, and x-ray diffraction-quality crystals appeared using 9% PEG 8000, 0.2 m calcium acetate, and 0.1 m sodium cacodylate, pH 6.4–7.5. Initial crystals of the K2A mutant appeared using 20% PEG 8000, 0.2 m calcium acetate, and 0.1 m MES, pH 6.0, and were further improved with 16% PEG 8000, 0.2 m calcium acetate, and 0.1 m MES, pH 6.8–7.1. Remarkable results were obtained from the K2Y mutant protein, which produced hits under 13 different coarse screening conditions (data not shown). The best crystals of this mutant were obtained using 1.5 m NaCl and 0.1 m imidazole, pH 7.0–8.0. The mNDRG2 protein was crystallized using the nanodroplet vapor-diffusion method (24) with standard Joint Center for Structural Genomics crystallization protocols (25). The crystallization reagent consisted of 20% PEG 400, 0.2 m magnesium chloride, and 0.1 m HEPES, pH 7.5.
Data Collection, Structural Determination, and Refinement
Diffraction data from a native mNDRG2 crystal were collected at the Advanced Light Source (Berkeley, CA) on beamline 8.3.1. Data were integrated and reduced using MOSFLM (26) and then scaled with the program SCALA (27). The diffraction data were indexed in hexagonal space group P3121, with one molecule in the asymmetric unit. The structure was determined by molecular replacement (MR) using the program MOLREP (28). A homology model based on an FFAS (29) alignment between mNDRG2 and the C–C bond hydrolase MhpC from E. coli (PDB code 1U2E) that share 13% sequence identity was constructed with the modeling program WHATIF (30) and was used in the MR searches. Structure refinement was performed using REFMAC (31) and COOT (32). The refinement resulted in Rfree and Rcryst factors of 18.3 and 14.5%, respectively. The model contains 278 amino acid residues, 314 water molecules, two magnesium ions, a benzoic acid, and a PEG molecule. The refined structure of mNDRG2 is substantially different from the structure of MhpC used to build the MR search model, with a root mean square deviation (r.m.s.d) of 3.0 Å of the Cα atoms over 244 structurally aligned residues. Diffraction data for the hNDRG2(23–304) crystals were collected at Pohang Accelerator Laboratory beamline 4A at 2.8 Å resolution. All of the x-ray diffraction data were processed and scaled using the HKL2000 software package (33). The crystal belongs to the space group P212121, and three molecules are located in the asymmetric unit. The engineered K2A and K2Y crystals resulted in significantly improved diffraction. Diffraction data for K2A were collected at Pohang Accelerator Laboratory beamline 6C at 2.0 Å resolution. The crystal also belongs to the space group P212121, with three molecules in the asymmetric unit. Diffraction data for K2Y were collected at the same beamline (i.e. 6C) at 2.15 Å resolution. The K2Y crystal was indexed to space group P3121 with one molecule per asymmetric unit. The structure of K2Y was solved by the MR method using the mNDRG2 model, with the program MOLREP. The model was manually built into electron density maps with the program COOT (32). Translation-libration-screw refinement of the isotropic displacement parameters was carried out with REFMAC (31) in the final cycles of refinement. The cycles of manual rebuilding and refinement resulted in Rfree and Rcryst factors of 20.8 and 17.3%, respectively. The model contains 281 amino acid residues, 137 water molecules, a chloride ion, and two imidazole molecules. The hNDRG2(23–304) and K2A structures were solved using the K2Y model, with the program MOLREP. The same procedures were performed for refinement of the hNDRG2(23–304) and K2A structures. The refinement of native protein resulted in Rfree and Rcryst factors of 25.9 and 20.1%, respectively. The model contains 843 amino acid residues, 93 water molecules, and 3 acetate molecules. The refinement of K2A resulted in Rfree and Rcryst factors of 23.3 and 18.0%, respectively. The model contains 843 amino acid residues, 399 water molecules, a calcium ion, 4 acetate molecules, and 3 glycerols. All models satisfied the quality criteria limits of the program PROCHECK (34). The crystallographic data statistics are summarized in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
The numbers in parentheses describe the relevant value for the highest resolution shell.
| Dataset | hNDRG2 | hNDRG2_K2A | hNDRG2_K2Y | mNDRG2 |
|---|---|---|---|---|
| Experimental data | ||||
| Beamline | 4A(MXW) | 6C(MXII) | 6C(MXII) | ALS 8.3.1 |
| Wavelength | 0.9795 Å | 1.23985 Å | 1.23985 Å | 1.02 Å |
| Space group | P212121 | P212121 | P3121 | P3121 |
| Cell dimensions | ||||
| a | 86.41 Å | 86.23 Å | 93.10 Å | 46.37 Å |
| b | 88.90 Å | 88.05 Å | 93.10 Å | 46.37 Å |
| c | 126.80 Å | 126.91 Å | 90.21 Å | 214.75 Å |
| Resolution | 2.80 Å (2.90–2.80 Å) | 2.00 Å (2.07–2.00 Å) | 2.15 Å (2.23–2.15 Å) | 1.70 Å (1.74–1.70 Å) |
| No. of total reflections | 160,865 | 291,415 | 227,387 | 176,642 |
| No. of unique reflections | 24,361 | 63,329 | 24,942 | 28,650 |
| Redundancy | 6.6(6.3) | 4.6(3.1) | 9.1(6.6) | 6.2(2.6) |
| Completeness | 99.6% (99.0%) | 96.5% (92.2%) | 99.9% (99.8%) | 92.9% (61.8%) |
| Rsyma | 10.5% (41.5%) | 13.6% (44.3%) | 8.0% (49.6%) | 5.0% (40.5%) |
| I/σ(I) | 19.1(4.5) | 11.17(1.83) | 28.16(2.78) | 21.2(2.9) |
| Refinement | ||||
| Resolution | 40.00–2.81 Å | 30.00–2.00 Å | 30.0–2.15 Å | 30.0–1.70 Å |
| Reflections in work/test sets | 23,070/1,242 | 60,071/3,217 | 23,642/1,268 | 27,201/1,449 |
| Rcryst/Rfreeb,c | 20.1%/25.9% | 18.0%/23.3% | 17.3%/20.8% | 14.5%/18.3% |
| r.m.s. deviations | ||||
| Bond lengths | 0.029 Å | 0.011 Å | 0.015 Å | 0.016 Å |
| Bond angles | 2.33° | 1.34° | 1.46° | 1.56° |
| Model composition | ||||
| Protein residues | 843 | 843 | 281 | 278 |
| Ligands | 3 acetates | 4 acetates, 3 glycerols, | 2 imidazoles, | 1 benzoic acid, 1 PEG, |
| 1 calcium ion | 1 chloride ion | 2 magnesium ions | ||
| Waters | 93 | 399 | 137 | 314 |
| Geometry | ||||
| Most favored regions | 90.3% | 92.7% | 92.0% | 95.3% |
| Additional allowed regions | 9.7% | 7.3% | 8.0% | 4.7% |
| PDB accession code | 2XMQ | 2XMR | 2XMS | 2QMQ |
a Rsym = Σ|Ii − 〈I〉|/ΣI, where Ii is the intensity of the ith observation, and 〈I〉 is the mean intensity of the reflections.
b Rcryst = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitude, respectively.
c Rfree = Σ‖Fobs| − |Fcalc‖/Σ |Fobs|, where all reflections belong to a test set of randomly selected data.
Preparation of NDRG2 Plasmids and Transfection
The human colon adenocarcinoma SW620 cells and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 2 mm glutamine, 1% penicillin/streptomycin, and 10% fetal bovine serum (FBS; HyClone, Logan, UT) and kept at 37 °C in a humidified incubator that was maintained with 5% CO2. LiCl (Sigma) at 20 mm concentration was added to the cell culture for 6–12 h after transfection. The full-length cDNA for a 357-residue-long hNDRG2 was cloned from a Jurkat cDNA library via PCR amplification. The deletion hNDRG2Δ164–175 or site-directed hNDRG2L172D mutants were also generated via PCR. The PCR products were subsequently cloned into pcDNA3 expression vector (Invitrogen) and verified by sequencing. Cells at 80% confluence were plated 1 day before transfection and then transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The amount of DNA for transfection was used at a 1.5 μg per well in a 6-well plate.
Luciferase Reporter Assay
To assess the modulation of TCF/β-catenin signaling by introduction of hNDRG2, we used the TOPflash luciferase reporter assay system, which contains a luciferase reporter plasmid with three copies of the optimal T-cell factor/lymphoid enhancer-binding factor (TCF/LEF)-binding sites upstream of the minimal thymidine kinase promoter. To conduct the luciferase reporter assay, the cells were transfected with TOPflash (or FopFlash, which harbors mutant TCF binding sites) luciferase reporter plasmid (Upstate Biotechnology, Inc.), β-galactosidase plasmid for the quantification of transfection efficiency, and pcDNA3-construct plasmids at a 1.5 μg/well. After 2 days, the cells were lysed with buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100) on ice for 30 min. The cleared lysates were then transferred to each of the wells in the 96-well plates, and luciferase assay reagent was added. The light intensity was determined using a plate-reading luminometer (Turner Designs, Sunnyvale, CA), and luciferase activity was calculated relative to β-galactosidase activity.
RT-PCR Analysis
Four groups of tissue pairs, including normal and cancerous region from patients with colon cancer, were obtained from tumor tissue bank of a local hospital. Prepared colon cancer tissues were lysed, and total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The RNAs were quantified, and 5 μg of RNA was used for RT-PCR to generate each cDNAs using a ProStar first-Strand RT-PCR kit (Stratagene, La Jolla, CA). β-Actin product was used as a reaction standard. PCR products of hNDRG2, β-actin, cyclin D1, and fibronectin were obtained from a reaction standard using its specific primer as follows: hNDRG2, sense 5′-ATGGCAGCGCAGAAGGACCAG-3′ and antisense 5′-TCACACGGGTTCCATCTGCAG-3′; β-actin, sense 5′-AGCCGTGGCCATCTCTTGCTCGAAG-3′ and antisense 5′-GCCATGTACGTTGCTATCCAGGCTG-3′; cyclin D1, sense 5′-AACTACCTGGACCGCTTCCT-3′ and antisense 5′-CCACTTGAGCTTGTTCACCA-3′; and fibronectin, sense 5′-CGGGAATCTTCTCTGTCAGC-3′ and antisense 5′-GCCATGACAATGGTGTGAAC-3′.
Immunoprecipitation
To detect the association of β-catenin with hNDRG2, immunoprecipitation experiments were carried out as described previously (10).
Protein Data Bank Accession Codes
The atomic coordinates and structure factor amplitudes of the hNDRG2(23–304), K2A, K2Y, and mNDRG2(40–313) proteins have been deposited in the Protein Data Bank (35) under the accession codes 2XMQ, 2XMR, 2XMS, and 2QMQ, respectively.
RESULTS
Overall Structure of the NDRG2 Protein
Many trials aimed at the production of soluble protein of the recombinant full-length hNDRG2 or mNDRG2 using various expression systems were unsuccessful. Therefore, we used the truncated constructs hNDRG2 (residues 23–304) and mNDRG2 (residues 40–313) to characterize their structures. mNDRG2 was determined at a resolution of 1.7 Å by the Joint Center for Structural Genomics and provided the first structure for the NDRG family (Pfam database (36) ID: PF03096), thereby facilitating structure determination of additional family members, including hNDRG2. Subsequently, the structures of the hNDRG2 K2Y and K2A mutants were determined by the MR method at 2.15 and 2.0 Å resolution, respectively. All NDRG2 structures are essentially identical to each other (Fig. 2b) and consist of two domains: a large canonical α/β-hydrolase fold domain and a small cap-like domain (Fig. 2c). The large domain consists of an archetypal, eight-stranded β-sheet, consisting of a β-hairpin structure (β1 and β2) and a six-stranded, parallel β-sheet (β3–β8). Eight α-helices (α1–α5 and α11–α13) and two 310 helices form the outer solvent-exposed layer surrounding the β-sheets. The small cap-like domain (Ala161 to Arg227) consists of five α-helices (α6–α10). hNDRG2 and its K2A mutant each contain three molecules in the asymmetric unit (Fig. 2d). Each structure of hNDRG2, K2A, and K2Y in the asymmetric unit is essentially the same, and the r.m.s.d values of their Cα atoms are all less than 0.36 Å (Fig. 2b).
Evolutionary Comparison between NDRG2 and Its Structural Homologues
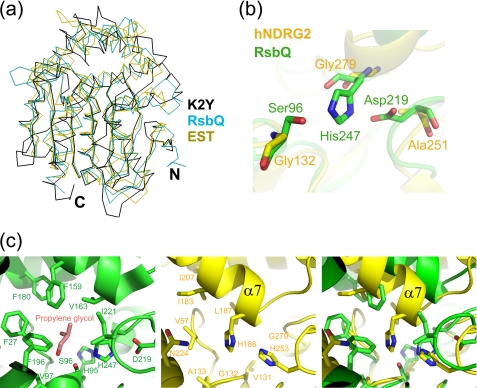
A structural similarity search conducted using DALI (37) revealed that NDRG2 is similar to a number of α/β-hydrolases, with Z scores ranging from 2.8 to 25.5, confirming that it belongs to the ABH superfamily. The r.m.s.d values for these structures vary from 2.7 to 4.3 Å for 92 to 245 aligned Cα atoms. The pairwise sequence identities between NDRG2 and its structural homologues range from 5 to 17%. NDRG2 is most similar to the Bacillus subtilis stress-response regulator, RsbQ (PDB ID code 1WOM; Z = 25.5) (38), and Pseudomonas putida IFO12996 esterase, EST (PDB ID code 1ZOI; Z = 25.0) (Fig. 3a) (39). The most striking difference between NDRG2 and other members of ABH superfamily is the absence of the classical catalytic triad, Ser-His-Asp. Superimposition of the protein structures reveals that the corresponding residues in hNDRG2 are Gly132–Gly279–Ala251 (Fig. 3b). The same corresponding residues are found in mNDRG2. Gly132 is located at the tip of a β-strand-turn-helix structural motif (β5 and α5), in an equivalent position to catalytic residue Ser96 in RsbQ. Gly279, corresponding to residue His247 in RsbQ, is located in a loop between β8 and α12, and Ala251, corresponding to the general base Asp219, is located on a loop between β7 and α11. Another remarkable difference between the proteins is that, although RsbQ and EST form a pocket for substrate binding in the active site, NDRG2 does not possess a pocket in this region (Fig. 3c). Indeed, RsbQ forms two cavities, a large cavity and a small cavity, near the catalytic residues. In the RsbQ structure, the large cavity trapped propylene glycol or a phenylmethanesulfonyl group, which suggests that its potential substrate is small and hydrophobic (38). When the RsbQ and NDRG2 structures were superimposed, no cavity was detected in the NDRG2 protein. Helix α7 of NDRG2 extends into the pseudo-active site, and residues on the helix (e.g. His186 and Leu187) occupy the pseudo-active site, preventing the generation of a cavity (Fig. 3c).
FIGURE 3.
Comparison between NDRG2 and its structural homologues. a, structural comparison of proteins in the α/β-hydrolase family. K2Y, the B. subtilis stress-response regulator (RsbQ, PDB code 1WOM), and P. putida IFO12996 esterase (EST, PDB code 1ZOI) proteins are displayed in black, cyan, and olive, respectively. b, superposition of the pseudo-active site residues of hNDRG2 with the catalytic residues of RsbQ. The active triad of RsbQ is shown as a green carbon skeleton. The corresponding residues of hNDRG2 are displayed as a yellow carbon skeleton. c, comparison of the active site in RsbQ with the corresponding site in hNDRG2. The active site in RsbQ is displayed in green, and the pseudo-active site in hNDRG2 is shown in yellow.
Thus, the structure of NDRG2 suggests that this protein may be a nonenzymatic homologue of the ABH superfamily. The same evolutionary characteristics are also present in other NDRG family members (Fig. 1). The Gly-Gly-Ala triad is conserved in all members, with the only exception being the substitution of Ala to Ser in NDRG1 and -3. In addition, His186, the crucial residue for disruption of the substrate-binding site, is absolutely conserved.
To establish whether or not hNDRG2(23–304) possesses hydrolase activity, an assay was performed to measure the ability of this protein to hydrolyze the chromogenic ρ-nitrophenyl butyrate substrate, as described previously (40). Under these experimental conditions, the protein did not have any detectable hydrolase activity (data not shown). This result further supports the notion that the presence of an ABH domain in NDRG2 does not per se signify enzymatic function.
Structural Analysis
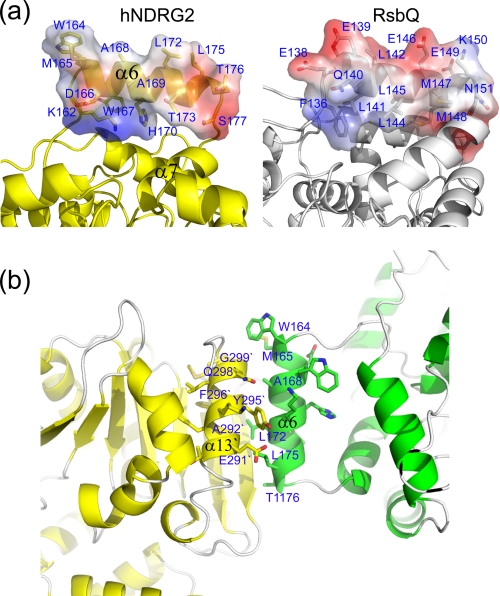
Instead of having enzymatic activity, nonenzymes have been found to have a binding role, interacting with metal ions, small ligands, proteins, or other biological macromolecules (41). Structural inspection of hNDRG2 suggests a potential molecular interaction site. Intriguingly, helix α6 in the structure exhibits a unique feature. As in typical hydrolases, for example lipases and esterases, helix α6, together with other helices in the cap-like domain of NDRG2, is supposed to be involved in hydrophobic pocket formation near the active site. In the case of RsbQ, the corresponding helix is involved in formation of the hydrophobic pocket. The residue distribution of the RsbQ helix shows that hydrophobic residues face toward the inside hydrophobic pocket, whereas charged residues are exposed to the solvent (Fig. 4a); however, the residue distribution of helix α6 of NDRG2 differs. Charged residues face toward the inside, whereas hydrophobic residues are exposed to the solvent (Fig. 4a). Thus, helix α6 of NDRG2 does not contribute to the formation of a hydrophobic pocket. In addition, helix α6 somewhat stands out from the main body of NDRG2 and is easily accessible for interaction with its binding target. These structural features of the helix α6 are conserved in all NDRG family members (Fig. 1). Crystallographic analysis of K2Y indicates that helix α6 forms a contact with helix α13 in the symmetry-related molecule and is mainly involved in molecular packing (Fig. 4b). Hydrophobic interactions between nonpolar residues are prominent at the interface between the helices. The same interface conformation between the helices is seen both in the asymmetric unit of K2A and in the native structures (Fig. 2d). These observations suggest that helix α6 may be an important motif for molecular interactions with NDRG2.
FIGURE 4.
Structural analysis of the cap-like domain of NDRG2. a, comparison of the residue distribution of helix α6 of NDRG2 with the corresponding helix in RsbQ. The surface is represented in a range of colors that indicate electrostatic potential, with red being negative and blue being positive. b, analysis of interaction interface of the symmetry-related K2Y molecules.
Impact of the Helix α6 in TCF/β-Catenin Signaling
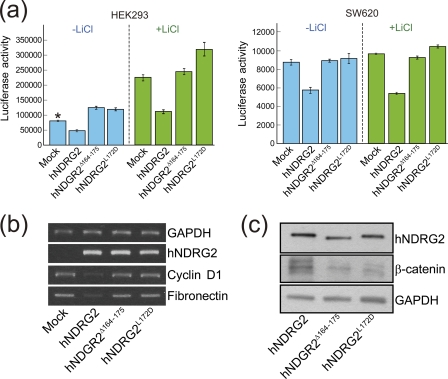
TCF/LEF is one of the key signaling pathways in tumorigenesis, cell growth, motility, and differentiation (42–46). The stability and intracellular localization of β-catenin, which is known to be regulated by the kinase activity of glycogen synthase kinase-3β (GSK-3β), is critical in the TCF/LEF signal pathway (42–46). In our previous study, an immunoprecipitation assay demonstrated that hNDRG2 associates with β-catenin and may play a pivotal role as a tumor suppressor by the attenuation of TCF/β-catenin signaling in SW620 cell lines (10). Based on the study, we evaluated the effect of the helix α6 on the function of hNDRG2 using TCF/β-catenin signaling system in SW620 colon cancer cells, as well as in HEK293 cells, via TOPflash luciferase reporter assay. As shown in Fig. 5a, TCF/LEF activity was prominently reduced in the hNDRG2-transfected HEK293 and SW620 cells. However, the helix α6 deletion mutant hNDRG2Δ164–175 was not able to inhibit the TCF/LEF transcriptional activity. We further assessed the residues on the helix α6. As stated previously, a surprising number of hydrophobic residues are distributed on the outside surface of the helix. Those residues are Trp164, Met165, Ala168, Ala169, Leu172, and Leu175 (Fig. 4a). Among these residues, Leu172 is located in the middle of the helix and fully exposed to solvent, suggesting that it may be critical for the function of hNDRG2. The single point mutation (L172D) was introduced into hNDRG2, and we evaluated its effect on the modulation of TCF/β-catenin signaling in HEK293 and SW620. Remarkably, this single mutation abolished the function of hNDRG2 on TCF/LEF activity (Fig. 5a). Furthermore, the effect of helix α6 on the function of hNDRG2 for TCF/β-catenin signaling in SW620 colon cancer cells was verified by RT-PCR of TCF/LEF target genes. Cyclin D1 and fibronectin are well known genes transcriptionally activated by TCF/LEF signaling. As shown in Fig. 5b, the genes were down-regulated by hNDRG2. In contrast, no expression change of the genes was found with the mutants hNDRG2Δ164–175 and hNDRG2L172D. We carried out a further immunoprecipitation assay to assess the involvement of helix α6 in association with β-catenin. Both hNDRG2Δ164–175 and hNDRG2L172D mutants revealed significantly diminished association ability with β-catenin (Fig. 5c). Therefore, these results clearly indicate that helix α6 plays a vital role for the function of hNDRG2 in regulating TCF/LEF transcriptional activity through association with β-catenin.
FIGURE 5.
Impact of the helix α6 of NDRG2 on TCF/β-catenin signaling. a, luciferase reporter assay was carried out to evaluate TCF/LEF transcriptional activity. The mutant plasmids hNDRG2Δ164–175 and hNDRG2L172D transfected into the HEK293 and SW620 cells with or without LiCl did not affect the TCF/LEF transcriptional activity, whereas the native hNDRG2 attenuated the TCF/LEF activity. Mean ± S.D. values from three independent experiments performed in duplicate are shown (*, p < 0.05). b, target genes of TCF/LEF, cyclin D1 and fibronectin were assessed by RT-PCR. Introduction of NDRG2 into the SW620 cells resulted in down-regulation of the target genes, although no regulation of TCF/LEF target genes was observed in the mutant introduced cells. c, hNDRG2 was immunoprecipitated with an anti-NDRG2 antibody, and the precipitant was analyzed by SDS-PAGE and Western blot analysis using anti-β-catenin antibody.
DISCUSSION
Although the NDRG family has been studied in a number of cell and animal systems and has been shown to influence diverse cellular processes, such as the modulation of cell differentiation and proliferation, little study has been made, so far, of its function and mechanism at the molecular level. In this study, we present crystal structures of the human and mouse NDRG2 proteins. The C-terminal region of NDRG family members, ranging from residues ∼300 to 360, is predicted to be unfolded, is known to contain modified residues (20), and is involved in the regulation of numerous cellular activities. The function of the N-terminal fragment, comprising residues ∼1 to 20, is currently unknown. Accordingly, we assessed several constructs of NDRG2 and identified a truncated NDRG2, spanning residues 23–304 for hNDRG2 and residues 40–313 for mNDRG2 that expressed and yielded soluble protein. The mNDRG2 crystals diffracted to 1.7 Å resolution, whereas the human protein diffracted to a resolution of 2.8 Å. To improve the quality of the hNDRG2 crystals, the SER method (23) was employed. SER-engineered K2A and K2Y crystals gave much improved diffraction to 2.0 and 2.15 Å resolution, respectively. Assessment of the crystal contacts of the mutants provides a possible explanation for the effects of the SER mutations (supplemental Fig. 1a). In both structures of the mutants, the mutated residues are directly involved in the formation of a new packing interface on the surface of the protein (supplemental Fig. 1b).
The high resolution NDRG2 structures further allowed us to speculate on the molecular function. Although NDRG2 shows high structural similarity to the ABH superfamily, the catalytic triad is not conserved and the active site pocket is blocked by helix α7. Thus, NDRG2 may be nonenzymatic. The existence of enzymatic and noncatalytic proteins within the same superfamily is a common occurrence (41), and these homologues probably evolved from a common ancestor. The ABH fold (that contains the ABH superfamily) is one of the most adaptable and prevalent protein folds known (47). It forms a stable scaffold for a wide variety of enzymes. Like most examples identified (41), the nonenzymatic NDRG2 may be derived from an ancestral ABH catalytic precursor. We imagine that an ancestral ABH gene duplicated, mutated, and combined through evolution to generate NDRG2, which became necessary for life. Often, nonenzymatic homologues are found to regulate cellular events by interacting with other biological molecules (41).
Recently, Wang et al. (8) reported that the expression of NDRG2 is up-regulated by hypoxia-inducible factor 1 in tumor cell lines under hypoxic conditions, with concomitant translocation of NDRG2 from the cytoplasm into the nucleus, and eventually induced apoptosis. The authors mapped NDRG2 and showed that the noncatalytic ABH domain (residues 1–257) is responsible for the translocation. They further concluded that residues 101–178 of NDRG2 may be critical for the translocation. The study suggested that NDRG2 may have a distinct regulatory motif for translocation because it does not have a common type of nuclear import element, such as a nuclear location signal (8). Knowledge of the three-dimensional structure of the NDRG2 allowed us to visualize the impact of this region on translocation. This region is composed of a part of a long loop (Gly101 to Ser106), helices α3 to α6 and strands β5 and β6. In other words, all elements, except for α6, form part of the ABH scaffold and probably do not participate directly in its biological function. Accordingly, helix α6 may be attributed to the translocation. As discussed previously, α6 possesses an unusual structural feature, in which its hydrophobic surface is exposed to solvent. Interestingly, a sequence “172LXXL175”, which is similar to the “LXXLL” motif involved in many examples of nuclear translocations and interactions of nuclear receptors (48, 49), is located on the helix surface. Based on these structural observations, we thus suggest that helix α6 may be an important motif for nuclear translocation of NDRG2 under hypoxic conditions, being either directly or indirectly responsible for this event. A detailed investigation of helix α6 should now be undertaken to gain a better understanding of the translocation mechanism of NDRG2 under cell stress.
The TCF/β-catenin signaling pathway has been implicated in the regulation of colonic epithelial cell proliferation and cell differentiation (50–52). It is known that the intracellular concentration of β-catenin is regulated by its ubiquitin-dependent degradation, which occurs via interaction with adenomatous polyposis coli tumor suppressor protein and phosphorylation at its N terminus through the interaction with glycogen synthase kinase-3β (GSK-3β) (53–56). The accumulation of β-catenin in the cytoplasm results in the formation of the β-catenin-TCF complex, its nuclear translocation, and consequently, stimulation of tumor formation through up-regulation of c-Myc and cyclin D1 (57, 58).
In our previous study, we showed that NDRG2 plays a role as a tumor suppressor by inducing the down-regulation of TCF/β-catenin signaling in human SW620 colon cancer cells (10). These results showed that NDRG2 diminishes the phosphorylation of GSK-3β, which enhances its kinase activity, and subsequently down-regulates β-catenin in SW620 cells. The decreased intracellular level of β-catenin induces the reduction of nuclear β-catenin, which then results in the attenuation of TCF/LEF activity and subsequent down-regulation of cyclin D1 and fibronectin. Phosphorylation of hNDRG2 at position Thr334 is known to be performed by Akt kinase and was critical for the regulation of TCF/β-catenin signaling. We also observed that the exogenously introduced hNDRG2 is localized mainly in the plasma membrane and cytosol in the SW620 colon cancer cell line, with a small portion of hNDRG2 in the nucleus. The cellular localization of hNDRG2 in the TCF/β-catenin signaling pathway has been to date unclear.
In this study, we showed that helix α6 may play a pivotal role for hNDRG2 function in the TCF/β-catenin signaling (Fig. 5, a and b). We further demonstrated that the helix is critical for the association with β-catenin (Fig. 5c). It is not clear, however, whether the association is mediated by direct interaction between hNDRG2 and β-catenin or by involvement of other interaction molecules. Indeed, in our ongoing study concerning the regulation TCF/β-catenin signaling by hNDRG2, yeast two-hybrid screening showed that hNDRG2 directly interacts with α-catenin, and immunoprecipitation revealed that it also associates with E-cadherin and β-catenin in human colon adenocarcinoma cells.6
From structural and cell biological points of view, hNDRG2 may be implicated in molecular interactions and in the cellular localization as well. In particular, helix α6 may play a vital role in those functions. Therefore, further studies should be conducted to elucidate molecular mechanisms underlying hNDRG2-mediated suppression of TCF/β-catenin signaling in the tumorigenesis of human colorectal cancer.
In conclusion, we have made a substantial effort to improve our understanding of the role of NDRG family proteins at the molecular level. Since the initial discovery of the family members, numerous studies have demonstrated that they function as tumor suppressors and are involved in many diseases (59, 60). Such observations have emphasized the importance of the biological role of the family, and yet the molecular mechanisms underlying their function remain to be clarified. Therefore, our structural study of NDRG2 may provide a platform for the functional studies of NDRG family proteins and lead to the development of novel strategies for anti-cancer therapies.
Acknowledgments
We thank Drs. Kyung-Jin Kim and Ghyung-Hwa Kim at Pohang Accelerator Laboratory for help with human NDRG2 data collection. We also thank all the members of the Joint Center for Structural Genomics for their general contributions to the mouse NDRG2 protein production and structure determination. The Joint Center for Structural Genomics is supported by the National Institutes of Health, NIGMS Protein Structure Initiative Grant U54 GM074898. Beamline 8.3.1 was funded by the National Science Foundation, University of California and Henry Wheeler. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and the National Institutes of Health (NCRR Biomedical Technology Program and NIGMS). Portions of this work were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. Portions of this work were carried out at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract DE-AC02-05CH11231.
This work was supported by the 21C Frontier Microbial Genomics and Applications Center Program (to M. H. K.) funded by the Ministry of Education, Science, and Technology and in part by Korea Healthcare Technology R&D Project Grant A090509 (to J. W. K.) funded by the Ministry for Health, Welfare and Family Affairs, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (codes 2XMQ, 2XMR, 2XMS, and 2QMQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
H. B. Kang and J. W. Kim, unpublished data.
- ABH
- α/β-hydrolase
- SER
- surface entropy reduction
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- MR
- molecular replacement
- TCF/LEF
- T-cell factor/lymphoid enhancer-binding factor.
REFERENCES
- 1. Vervoorts J., Lüscher-Firzlaff J., Lüscher B. (2006) J. Biol. Chem. 281, 34725–34729 [DOI] [PubMed] [Google Scholar]
- 2. O'Connell B. C., Cheung A. F., Simkevich C. P., Tam W., Ren X., Mateyak M. K., Sedivy J. M. (2003) J. Biol. Chem. 278, 12563–12573 [DOI] [PubMed] [Google Scholar]
- 3. Bandyopadhyay S., Pai S. K., Gross S. C., Hirota S., Hosobe S., Miura K., Saito K., Commes T., Hayashi S., Watabe M., Watabe K. (2003) Cancer Res. 63, 1731–1736 [PubMed] [Google Scholar]
- 4. Bandyopadhyay S., Pai S. K., Hirota S., Hosobe S., Takano Y., Saito K., Piquemal D., Commes T., Watabe M., Gross S. C., Wang Y., Ran S., Watabe K. (2004) Oncogene 23, 5675–5681 [DOI] [PubMed] [Google Scholar]
- 5. Guan R. J., Ford H. L., Fu Y., Li Y., Shaw L. M., Pardee A. B. (2000) Cancer Res. 60, 749–755 [PubMed] [Google Scholar]
- 6. Okuda T., Higashi Y., Kokame K., Tanaka C., Kondoh H., Miyata T. (2004) Mol. Cell. Biol. 24, 3949–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park Y., Shon S. K., Kim A., Kim K. I., Yang Y., Cho D. H., Lee M. S., Lim J. S. (2007) Biochem. Biophys. Res. Commun. 363, 361–367 [DOI] [PubMed] [Google Scholar]
- 8. Wang L., Liu N., Yao L., Li F., Zhang J., Deng Y., Liu J., Ji S., Yang A., Han H., Zhang Y., Zhang J., Han W., Liu X. (2008) Cell. Physiol. Biochem. 21, 239–250 [DOI] [PubMed] [Google Scholar]
- 9. Kim Y. J., Yoon S. Y., Kim J. T., Choi S. C., Lim J. S., Kim J. H., Song E. Y., Lee H. G., Choi I., Kim J. W. (2009) Int. J. Cancer 124, 7–15 [DOI] [PubMed] [Google Scholar]
- 10. Kim Y. J., Yoon S. Y., Kim J. T., Song E. Y., Lee H. G., Son H. J., Kim S. Y., Cho D., Choi I., Kim J. H., Kim J. W. (2009) Carcinogenesis 30, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hummerich L., Müller R., Hess J., Kokocinski F., Hahn M., Fürstenberger G., Mauch C., Lichter P., Angel P. (2006) Oncogene 25, 111–121 [DOI] [PubMed] [Google Scholar]
- 12. Zhao H., Zhang J., Lu J., He X., Chen C., Li X., Gong L., Bao G., Fu Q., Chen S., Lin W., Shi H., Ma J., Liu X., Ma Q., Yao L. (2008) BMC Cancer 8, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee D. C., Kang Y. K., Kim W. H., Jang Y. J., Kim D. J., Park I. Y., Sohn B. H., Sohn H. A., Lee H. G., Lim J. S., Kim J. W., Song E. Y., Kim D. M., Lee M. N., Oh G. T., Kim S. J., Park K. C., Yoo H. S., Choi J. Y., Yeom Y. I. (2008) Cancer Res. 68, 4210–4220 [DOI] [PubMed] [Google Scholar]
- 14. Furuta H., Kondo Y., Nakahata S., Hamasaki M., Sakoda S., Morishita K. (2010) Biochem. Biophys. Res. Commun. 391, 1785–1791 [DOI] [PubMed] [Google Scholar]
- 15. Assämäki R., Sarlomo-Rikala M., Lopez-Guerrero J. A., Lasota J., Andersson L. C., Llombart-Bosch A., Miettinen M., Knuutila S. (2007) Genes Chromosomes Cancer 46, 564–576 [DOI] [PubMed] [Google Scholar]
- 16. Mitchelmore C., Büchmann-Møller S., Rask L., West M. J., Troncoso J. C., Jensen N. A. (2004) Neurobiol. Dis. 16, 48–58 [DOI] [PubMed] [Google Scholar]
- 17. Wang W., Li Y., Li Y., Hong A., Wang J., Lin B., Li R. (2009) Int. J. Cancer 124, 521–530 [DOI] [PubMed] [Google Scholar]
- 18. Schilling S. H., Hjelmeland A. B., Radiloff D. R., Liu I. M., Wakeman T. P., Fielhauer J. R., Foster E. H., Lathia J. D., Rich J. N., Wang X. F., Datto M. B. (2009) J. Biol. Chem. 284, 25160–25169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melotte V., Lentjes M. H., van den Bosch S. M., Hellebrekers D. M., de Hoon J. P., Wouters K. A., Daenen K. L., Partouns-Hendriks I. E., Stessels F., Louwagie J., Smits K. M., Weijenberg M. P., Sanduleanu S., Khalid-de Bakker C. A., Oort F. A., Meijer G. A., Jonkers D. M., Herman J. G., de Bruïne A. P., van Engeland M. (2009) J. Natl. Cancer Inst. 101, 916–927 [DOI] [PubMed] [Google Scholar]
- 20. Burchfield J. G., Lennard A. J., Narasimhan S., Hughes W. E., Wasinger V. C., Corthals G. L., Okuda T., Kondoh H., Biden T. J., Schmitz-Peiffer C. (2004) J. Biol. Chem. 279, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 21. Shaw E., McCue L. A., Lawrence C. E., Dordick J. S. (2002) Proteins 47, 163–168 [DOI] [PubMed] [Google Scholar]
- 22. Kwon S. Y., Choi Y. J., Kang T. H., Lee K. H., Cha S. S., Kim G. H., Lee H. S., Kim K. T., Kim K. J. (2005) Plasmid 53, 274–282 [DOI] [PubMed] [Google Scholar]
- 23. Goldschmidt L., Cooper D. R., Derewenda Z. S., Eisenberg D. (2007) Protein Sci. 16, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santarsiero B. D., Yegian D. T., Lee C. C., Spraggon G., Gu J., Scheibe D., Uber D. C., Cornell E. W., Nordmeyer R. A., Kolbe W. F., Jin J., Jones A. L., Jaklevic J. M., Schultz P. G., Stevens R. C. (2002) J. Appl. Crystallogr. 35, 278–281 [Google Scholar]
- 25. Lesley S. A., Kuhn P., Godzik A., Deacon A. M., Mathews I., Kreusch A., Spraggon G., Klock H. E., McMullan D., Shin T., Vincent J., Robb A., Brinen L. S., Miller M. D., McPhillips T. M., Miller M. A., Scheibe D., Canaves J. M., Guda C., Jaroszewski L., Selby T. L., Elsliger M. A., Wooley J., Taylor S. S., Hodgson K. O., Wilson I. A., Schultz P. G., Stevens R. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leslie A. G. W. (1992) Joint CCP4 + ESF-EACBM Newsletter Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 27. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 28. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 29. Rychlewski L., Jaroszewski L., Li W., Godzik A. (2000) Protein Sci. 9, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vriend G. (1990) J. Mol. Graph. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaneko T., Tanaka N., Kumasaka T. (2005) Protein Sci. 14, 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elmi F., Lee H. T., Huang J. Y., Hsieh Y. C., Wang Y. L., Chen Y. J., Shaw S. Y., Chen C. J. (2005) J. Bacteriol. 187, 8470–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim M. H., Kang B. S., Kim S., Kim K. J., Lee C. H., Oh B. C., Park S. C., Oh T. K. (2008) Proteins 70, 578–583 [DOI] [PubMed] [Google Scholar]
- 41. Todd A. E., Orengo C. A., Thornton J. M. (2002) Structure 10, 1435–1451 [DOI] [PubMed] [Google Scholar]
- 42. Mariadason J. M., Bordonaro M., Aslam F., Shi L., Kuraguchi M., Velcich A., Augenlicht L. H. (2001) Cancer Res. 61, 3465–3471 [PubMed] [Google Scholar]
- 43. Rockman S. P., Currie S. A., Ciavarella M., Vincan E., Dow C., Thomas R. J., Phillips W. A. (2001) J. Biol. Chem. 276, 45113–45119 [DOI] [PubMed] [Google Scholar]
- 44. Chakrabarty S., Radjendirane V., Appelman H., Varani J. (2003) Cancer Res. 63, 67–71 [PubMed] [Google Scholar]
- 45. Liu J. J., Huang B. H., Zhang J., Carson D. D., Hooi S. C. (2006) J. Cell. Physiol. 207, 287–292 [DOI] [PubMed] [Google Scholar]
- 46. Shelton D. N., Sandoval I. T., Eisinger A., Chidester S., Ratnayake A., Ireland C. M., Jones D. A. (2006) Cancer Res. 66, 7571–7577 [DOI] [PubMed] [Google Scholar]
- 47. Nardini M., Dijkstra B. W. (1999) Curr. Opin. Struct. Biol. 9, 732–737 [DOI] [PubMed] [Google Scholar]
- 48. Zelko I., Sueyoshi T., Kawamoto T., Moore R., Negishi M. (2001) Mol. Cell. Biol. 21, 2838–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aranda A., Pascual A. (2001) Physiol. Rev. 81, 1269–1304 [DOI] [PubMed] [Google Scholar]
- 50. Bienz M., Clevers H. (2000) Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 51. Moon R. T., Bowerman B., Boutros M., Perrimon N. (2002) Science 296, 1644–1646 [DOI] [PubMed] [Google Scholar]
- 52. Henderson B. R., Fagotto F. (2002) EMBO Rep. 3, 834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salic A., Lee E., Mayer L., Kirschner M. W. (2000) Mol. Cell 5, 523–532 [DOI] [PubMed] [Google Scholar]
- 54. Cohen P., Frame S. (2001) Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 55. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 56. Behrens J., Jerchow B. A., Würtele M., Grimm J., Asbrand C., Wirtz R., Kühl M., Wedlich D., Birchmeier W. (1998) Science 280, 596–599 [DOI] [PubMed] [Google Scholar]
- 57. Tetsu O., McCormick F. (1999) Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 58. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 59. Kovacevic Z., Richardson D. R. (2006) Carcinogenesis 27, 2355–2366 [DOI] [PubMed] [Google Scholar]
- 60. Yao L., Zhang J., Liu X. (2008) Acta Biochim. Biophys. Sin. 40, 625–635 [DOI] [PubMed] [Google Scholar]