Abstract
Spherical high density lipoprotein (sHDL), a key player in reverse cholesterol transport and the most abundant form of HDL, is associated with cardiovascular diseases. Small angle neutron scattering with contrast variation was used to determine the solution structure of protein and lipid components of reconstituted sHDL. Apolipoprotein A1, the major protein of sHDL, forms a hollow structure that cradles a central compact lipid core. Three apoA1 chains are arranged within the low resolution structure of the protein component as one of three possible global architectures: (i) a helical dimer with a hairpin (HdHp), (ii) three hairpins (3Hp), or (iii) an integrated trimer (iT) in which the three apoA1 monomers mutually associate over a portion of the sHDL surface. Cross-linking and mass spectrometry analyses help to discriminate among the three molecular models and are most consistent with the HdHp overall architecture of apoA1 within sHDL.
Keywords: Apolipoproteins, Biophysics, Cholesterol Regulation, Neutron Scattering, Protein Cross-linking, Apolipoprotein A1, Mass Spectrometry Cross-linking, Protein Isotopic Labeling, Small Angle Neutron Scattering, Spherical High Density Lipoprotein
Introduction
Epidemiological studies firmly establish that circulating levels of high density lipoprotein (HDL) cholesterol and apolipoprotein A1 (apoA1),2 the major protein constituent of HDL particles, are inversely associated with atherosclerotic heart disease risk (1–3). Moreover, genetic studies further confirm a strong mechanistic link between HDL and apoA1 and cardiovascular disease (4–8). Defined by its buoyant density characteristics, HDL represent a heterogeneous group of particles with varied lipid composition and protein content that participate in diverse biological functions ranging from lipid transport to innate immune functions. For example, HDL serves as an acceptor of cholesterol from peripheral tissue macrophages and promotes lipid transport through delivery of cholesterol to the liver and steroidogenic tissues (9–11). HDL also mediates systemic anti-inflammatory and anti-oxidant functions (12–14), and HDL-associated proteins can play critical host defense functions (15). ApoA1 represents nearly three-quarters of the protein content of HDL by mass, and it plays a central functional role in facilitating the numerous biological activities of HDL. Typically present at 2–4 molecules/particle depending upon the degree of HDL maturation, apoA1 serves as the fundamental structural element of the particle (16, 17) and is critical for specific interactions with proteins involved in HDL biogenesis (18, 19), maturation and remodeling (20, 21), and recognition by target organ receptors (22, 23).
Because HDL can be generated in a relatively homogenous form, most structural studies of HDL have focused on reconstituted nascent HDL, a particle composed of two molecules of apoA1 associated with phospholipid and free cholesterol (16, 17). Early small angle neutron scattering (SANS) and small angle x-ray scattering (SAXS) studies of nascent HDL particles were reported nearly 3 decades ago and are consistent with an outer protein layer relative to a central lipid core (24, 25). Current structural models of nascent HDL have an anti-parallel apoA1 chain orientation and posit that the protein exists either as a circumferential double belt shape around a central bilayer lipid core comprising phospholipid and free cholesterol (16, 17, 26–31) or as a superhelical shape wrapping around a predominantly micellar lipid core (32, 33). The anti-parallel organization of apoA1 molecules in nascent HDL is supported by numerous biophysical investigations including mass spectrometry/cross-linking (34, 35), fluorescence energy resonance transfer (FRET) (36, 37), and electron spin resonance (ESR) transfer studies (37).
Spherical HDL (sHDL) particles are a more mature form of HDL, whereby large accumulations of neutral lipids, predominantly cholesteryl esters, are carried as cargo by the particles. Despite sHDL being the most abundant circulating form of HDL, relatively few studies have experimentally addressed its structure in detail because of its size and compositional heterogeneity. Indeed, analytical ultracentrifugation (38), nondenaturing gradient gel electrophoresis (39), immunoaffinity chromatography (40), and two-dimensional electrophoresis (41) techniques all reveal heterogeneous sHDL preparations containing typically 3 or 4 apoA1/particle. The ability to form chemically and physically defined reconstituted sHDL by incubating reconstituted nascent HDL with lecithin:cholesterol acyltransferase, the plasma enzyme responsible for the maturation of nascent HDL into spherical particles (42, 43), has permitted some structural studies into the overall architecture of apoA1 on the sHDL surface. Early characterization studies using circular dichroism and fluorescence spectroscopy suggest that the apoA1 in reconstituted nascent HDL and sHDL have similar α-helical content, with sHDL being more resistant to chaotropic denaturants (42). Further, monoclonal antibody studies of different apoA1 epitopes suggest that the overall structure of apoA1 is comparable in reconstituted sHDL and HDL isolated from human plasma (42). However, differences in fluorescence emission (36), particle charge (44), proteolytic sensitivity (45) and 13C NMR studies (46) between reconstituted nascent HDL and sHDL have also been reported, suggesting at a minimum the presence of localized structural changes of the apoA1 in nascent versus sHDL. Despite these observations, recent protein cross-linking/mass spectrometry-based studies of reconstituted nascent HDL of various sizes and monodisperse preparations of sHDL have demonstrated, for the most part, similar apoA1 interchain cross-links within the different particles, suggesting that within the resolution limits of protein cross-linking analysis, apoA1 apparently adopts a relatively consistent general structural framework in HDL particles irrespective of the shape, size, and number of apoA1 molecules present (47). Thus, whether apoA1 undergoes a global reorganization during maturation from a nascent to spherical form remains unknown. Indeed, despite the importance of HDL to both cholesterol metabolism and cardiovascular disease, the overall conformation of apoA1 within sHDL forms has not yet been directly visualized. Because of the lack of direct experimental data on the shape of apoA1 within sHDL, some investigators have produce computational models by means of coarse grained molecular dynamics simulations for sHDL particles containing either two (48, 49) or three (50) truncated apoA1 chains. The relevance of these hypothetical models will remain unclear until they are more fully tested against structural data for apoA1 in sHDL.
Small angle scattering (SAXS for x-ray and SANS for neutron) is a powerful tool for the investigation of macromolecular complexes in solution, providing information on their overall dimensions, morphology, and composition (51–53). SANS with contrast variation yields low resolution structures, yet is particularly useful in revealing the organization of a multicomponent system because it combines structural and compositional information that is difficult to obtain by other approaches (32, 54). During the contrast variation experiment, the scattering length density of solvent is adjusted (by varying the D2O/H2O ratio) to match that of a component within a complex (e.g. protein, lipid, DNA, or RNA), reducing the scattering intensity observed for that component. SANS with contrast variation, especially when coupled with selective deuteration of individual components, can facilitate triangulation of the location of individual components within macromolecular complexes. For example, SANS with contrast variation was the first method to correctly predict the subunit organization of eukaryotic chromosomes (55), the structural orientation of protein and DNA within the nucleosome (56), and the organization of rhodopsin within retinal rod outer segment (57), as well as to triangulate the location of various proteins and RNA within the ribosome (58, 59). Herein we use the combination of contrast variation SANS and selective deuteration of apoA1 to directly visualize the low resolution structure of the protein and lipid core components individually within biologically active reconstituted sHDL. The overall shape observed for apoA1 and its orientation relative to the central lipid phase of sHDL provides structural insights into how the protein component of sHDL accommodates the dynamic changes that occur within the lipid core during particle maturation and remodeling. It also suggests how apoA1 within sHDL functions as a structural scaffolding to facilitate lipid transport functions of the particle during reverse cholesterol transport.
EXPERIMENTAL PROCEDURES
Preparation of Reconstituted Spherical HDL Particles
Human apolipoprotein A1 was purified from plasma from healthy volunteers as described with modifications. Briefly, HDL was isolated from fresh human plasma by density centrifugation from 1.07 to 1.21 g/ml with KBr. Then HDL proteins were precipitated using methanol/chloroform/cold ether sequentially before resuspension in 20 mm Tris, pH 8.5, 6 m urea. Human apoA1 in HDL was purified from HDL protein precipitates using ion-exchange chromatography (Q Sepharose HP HiLoad 26/10 column from GE Healthcare). Human apoA1 was stored in PBS with 3 m guanidine HCl at −80 °C before use. Deuterated recombinant His-tagged human apoA1 (rh-apoA1) was produced in Escherichia coli BL21 pLys strain grown in minimal medium as described previously (27). One liter of the minimal medium consisted of M9 salts, 0.1% NH4Cl, 10 mm MgCl2·6H2O, and 30 mg of thiamine in 99.8% D2O supplemented with trace elements, which included 60 mg of CaCl2·2H2O, 60 mg of FeSO4·7H2O, 11.5 mg of MnCl2·4H2O, 8 mg of CoCl2·6H2O, 7 mg of ZnSO4·7H2O, 30 mg of CuCl2·2H2O, 0.2 mg of H3BO3, 2.5 mg of (NH4)6Mo7O24·4H2O, and 50 mg of EDTA. Bacterial Cells were first grown in LB medium containing 70% D2O overnight at 37 °C and then harvested by centrifugation at 3000 rpm for 30 min. Cells were resuspended in minimal medium after being washed with PBS three times and grown to mid-log phase. Expression of rh-apoA1 was induced by the addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Deuterated rh-apoA1 was purified to homogeneity by nickel chromatography as described (27).
Reconstituted nascent HDL was prepared by the cholate dialysis method (42). For preparation of deuterated reconstituted nascent HDL, deuterated rh-apoA1 was used. An initial ratio of 100:10:1, 1-palmitoyl-2-oleoyl-sn-glycero-3 phosphatidylcholine:cholesterol:apoA1 (mol:mol:mol) was used to generate the reconstituted nascent HDL followed by gel filtration chromatography as described previously (27). Reconstituted spherical HDL was prepared by incubating either deuterated or nondeuterated nascent HDL with isolated human lecithin:cholesterol acyltransferase and isolated human LDL at 37 °C for 24 h (42). Reconstituted spherical HDL was then isolated by sequential ultracentrifugation in the 1.07 < d < 1.21 g/ml density range and further purified by gel filtration chromatography on a Sephacryl S-300 column.
Chemical Cross-linking and Mass Spectrometric Analyses
Chemical cross-linking and mass spectrometric analyses were performed to determine the stoichiometry of apoA1 on spherical HDL. Briefly, bis(sulfosuccinimidyl)suberate (BS3; from Pierce) was dissolved in PBS at a concentration of 8 mg/ml added to reconstituted spherical HDL (0.4 mg/ml apoA1) at a 200:1 molar ratio (BS3 versus apoA1) and incubated for 5 min at 37 °C. The reaction was immediately quenched by the addition of 1 m Tris buffer to a final concentration of 100 mm at pH 7.4. To estimate the number of BS3 mol adducted to apoA1, the content of derivatizable lysine residues remaining on cross-linked delipidated apoA1 was assayed using the sensitive fluorogenic o-phthaldialdehyde reagent and purified apoA1 as the standard (60). The mass of delipidated cross-linked apoA1 from sHDL was measured directly using MALDI-TOF mass spectrometry. Sinapic acid prepared in 50% acetonitrile with 0.1% TFA was mixed with cross-linked apoA1 (1:1 by volume), and ∼ 0.75 μl of the mixture was spotted on a stainless MALDI plate. The spectra were acquired in the positive linear mode on an ABI 4800 proteomics analyzer mass spectrometer. Each spectrum was generated by averaging 3000 laser shots with the laser intensity at 5000. External calibration was performed using bovine serum albumin and monoclonal antibody against IκB kinase as calibrates.
ApoA1 cross-linking within spherical HDL was performed as described (47). Briefly, sHDL (0.5 μg/μl) in PBS, pH 7.4, was incubated with BS3 (10:1 molar ratio) at 4 °C for 24 h. During the first hour the solution was mixed gently every 15 min. The cross-linking reaction was terminated by the addition of Tris HCl at a final concentration of 100 mm and then stored at −80 °C prior to analysis. Cross-linked monomer, dimer, and trimer forms of apoA1 within sHDL were separated on a 12% SDS-PAGE before being transeluted into a 14-channel fraction collector using a mini whole-gel eluter apparatus (Bio-Rad). The fractions containing monomer, dimer, and trimer apoA1 (confirmed by MALDI-TOF analysis as described above) were dialyzed against 20 mm NH4HCO3 and then digested with tryspin (Promega, Madison, WI) at a ratio of 500:1 (w/w). The tryptic digests were separated on a C18 nano column (75 mm × 10 cm, Proxeon) using an EASY-nLC system interfaced with LTQ MS (Thermo Scientific) at a flow rate of 300 nl/min and a gradient of 5–35% acetonitrile in 0.1% formic acid for 45 min. The mass spectra were acquired in a data-dependent mode with a mass scan range of 300 to 2000 daltons. The candidates for cross-linked peptides were searched by inputting masses of peptide list into a Web-based search program, and then the sequences of cross-linked peptides were confirmed by manually analyzing the MS/MS spectra. Only sequence-confirmed cross-links were used.
Compositional Analysis of Reconstituted sHDL
The phospholipid composition of reconstituted spherical HDL was determined by microphosphorus analysis (27, 32). The amount of protein in reconstituted spherical HDL was quantified by stable isotope dilution LC/MS/MS analysis of multiple amino acids (Phe, Tyr, and Leu) assuming the protein sequence of human apoA1. Calculated results of protein mass from all three amino acids agreed within ±7%. The free and total (following KOH saponification) cholesterol concentrations in reconstituted spherical HDL were determined enzymatically using commercial kits from Stanbio Laboratory and Wako Diagnostics. The amount of cholesterol ester in the sHDL preparations was calculated by subtracting free cholesterol from total cholesterol.
Cholesterol Efflux Assay
Cholesterol efflux assays were performed as described (32). Briefly, cholesterol-loaded RAW264.7 cells in 24 well plates were loaded with 0.3 μCi/ml [3H]cholesterol overnight in 1% FBS-DMEM. The next day, ABCA1 expression was induced with 8-bromo-cAMP in DMEM supplemented with 50 mm glucose, 2 mm glutamine, and 0.2% BSA for 16 h at 37 °C. The day after induction, plasma HDL (5 μg/ml protein, final) or reconstituted spherical HDL (5 μg/ml apoA-I, final) in 0.5 ml of serum-free DMEM with or without 8-bromo-cAMP was added to each well. After a 6-h incubation at 37 °C, the medium was removed and centrifuged, and 250 μl was counted as a measure of cholesterol released into the medium. The cells were extracted with hexane/isopropanol (3:2), and radioactivity was counted as a measure of cholesterol retained within the cell. The percent cholesterol efflux was calculated as the radioactivity in the medium/total radioactivity (medium radioactivity plus cell radioactivity). The control was performed using the same procedure without the addition of sHDL.
SR-B1-specific Binding of Spherical HDL
Spherical HDL made with bacterial ApoA1 was iodinated using the iodine monochloride method as described by Brown and colleagues (61). 293-T cells were transfected with vector or SR-B1 using Lipofectamine 2000 according to the manufacturer's instructions. Binding of the radiolabeled reconstituted spherical HDL to SR-B1 was determined by incubating 125I-labeled sHDL with either vector-transfected or SR-B1-transfected cells for 1.5 h at 4 °C (32). The cells were washed twice with 250 mm NaCl, 25 mm Tris, pH 7.4, and once with 250 mm NaCl, 25 mm Tris, pH 7.4, containing 2 mg/ml BSA and then solubilized in 1 ml of 0.1 m NaOH at room temperature for 20 min. Cell-associated radioactivity was subsequently determined with a Gamma 4000 spectrometer (Beckman Coulter). Specific binding was calculated as total binding minus binding in the presence of a 20-fold excess of unlabeled sHDL.
SANS Experiments
Small angle neutron scattering experiments were performed on sHDL samples prepared as described above. SANS data were collected at a controlled temperature (6–8 °C) both on the beamline D22 in Grenoble, France, and on KWS-2, the outstation of the Forschungszentrum Juelich at FRMII in Munich, Germany. All experiments showed consistent results on multiple preparations. Following analysis, the samples were recovered and used for chemical, biochemical, and biological assays. Scattering data were obtained as a function of the scattering angle and corrected by subtraction of incoherent scattering and empty cell before being averaged radially and calibrated in absolute units. All samples and buffers were recorded with neutron beams of λ = 4.5 or 7 Å. The scattering neutron density was determined as the macroscopic cross-section I(q) in units of cm−1 versus the momentum transfer q = (4π/λ)sinθ, where 2θ is the scattering angle. To cover a wide range of q and to ensure good merging statistics, the scattering experiments were performed at 2, 4, and 7 m detector-to-sample distances covering a range of 0.007 < q < 0.45. The buffer was subtracted from the scattering curve, and the resulting scattering curves were processed using the program PRIMUS (62). The experimental radii of gyration of scattering contrast, Rg, of the samples were calculated from the Guinier range (Rgq)2 ∼ 1 of the scattering curve (Equation 1).
The molecular weight calculation of apoA1 was based on Equation 2 using the Icorr(0) of the scattering curve.
The factor 10−3 is required to express protein concentration (c) in the usual way as mg/ml, NA is Avogadro's number, the term in parentheses is the excess scattering length sum of protein atoms calculated from its composition, ρ0 is scattering length density of the solvent, V is protein or lipid volume calculated from its partial specific volume, and m is the molar mass in g/mol (63).
Low Resolution Structure Determination
Low resolution structures of protein and lipid components were generated from their respective corrected intensities, Icorr, by using the program DAMMIN (64). The corrected sample intensities were first fitted in the specified q ranges (protein component, 0.007–0.35 Å−1; lipid component, 0.007–0.24 Å−1) by using the program GNOM (65). GNOM (which uses indirect Fourier transformation (66)) reads one-dimensional experimental scattering intensities (considering errors) and evaluates a distance distribution function p(R). Then the program DAMMIN produces low resolution structures (with maximal extension Dmax) by using the p(R) function. The final structure obtained is consistent with the experimental data. Two qualitative criteria for these fits were checked for each contrast level: 1) a smooth behavior of p(R) and 2) a good agreement between the radii of gyration (Rg) calculated by GNOM and determined by the Guinier approximation (Equation 1). All samples of different contrast level fulfilled these criteria in a satisfactory manner. The output of the GNOM program was used as input data for the program DAMMIN to build the low resolution structures. The DAMMIN default parameters were used for the modeling process. The final models of the protein and lipid components are the average of at least 10 outputs of the program DAMMIN using the program DAMAVER (64). The low resolution structure obtained from the 12% D2O scattering intensities (protein component of sHDL) was used to build the first schematic model of the protein architecture. This model was further optimized by improving the goodness of fit of the calculated scattering intensities versus the experimental data. This was achieved by gradually adjusting the shape of the three apoA1 chains in the model and comparing the calculated scattering intensities with those obtained experimentally.
High Resolution Models
To identify possible arrangements and plausible conformations of the three chains of ApoA1 that can fill the SANS shape of the protein component of sHDL, initially we built straight-line three-dimensional tube-like models made to match the length of a full ApoA1 chain. Next, we bent the tubes to follow the shape of the SANS low resolution structure of the protein and created three different combinations designated (below) as the helical-dimer-hairpin (HdHp), three-hairpin (3Hp), and integrated trimer (iT) models. Then, using these tube-like models we constructed all-atom models of the three possible topologies of the apoA1 trimer (HdHp, 3Hp, and iT). The all-atom models followed accurately the same conformation of the individual chains found in their tube-like counterparts. The apoA1 chains, modeled in an all-α-helix conformation, were oriented with the hydrophobic face of the amphipathic α-helix toward the lipid core and the hydrophilic face toward the outside, similar to the orientation of the amino acid residues found in previous models of nascent HDL (26, 32). These models are very crude, and their geometry was not optimized. That is, energy minimizations or molecular dynamics simulations were not performed either in gas phase or in solution. Thus, these models do not contain accurate information about the solvent accessibility of individual residues. The all-atom models of the ApoA1 trimer were built for the purpose of performing the SANS and cross-link analyses presented below and in supplemental Table 1. We calculated the scattering intensity given off by the all-atom models and the goodness of fit with the experimental scattering data listed in Table 1 using the program CRYSON (67). For the cross-link analysis we performed free rotations of lysine residues within 25 Å of each other for all models. The distances between lysine residues found to be involved in cross-links were measured between the nitrogen atoms of the side chains of the two residues. These distances are listed in supplemental Table 1.3
TABLE 1.
Radius of gyration, dimensions, and goodness of fit for the low resolution SANS structures of spherical reconstituted HDL
| sHDL % D2O |
Rg |
χ2 | Length:Width:Height | |
|---|---|---|---|---|
| Experimental curve | Low resolution structure | |||
| Å | Å | Å | ||
| 0 | 39.1 ± 0.43 | |||
| 12 | 47.3 ± 0.08 | 45.2 | 1.483 | 111:96.6:88.7 |
| 42 | 31.0 ± 0.03 | 30.9 | 2.887 | 88.4:68.2:60.3 |
| 90 | 34.8 ± 0.49 | |||
RESULTS AND DISCUSSION
Chemical, Biochemical, and Biological Activity Characterization of Reconstituted sHDL
Monodisperse and relatively homogeneous preparations of reconstituted sHDL were prepared using the methods initially reported by Rye et al. (43). Preparations of nondeuterated and deuterated sHDL were generated from reconstituted nascent HDL produced with either human apoA1 or deuterated recombinant human apoA1 as outlined under “Experimental Procedures.” Following the final isolation step of gel filtration chromatography, the sHDL preparations were initially characterized by native nondenaturing gradient gel electrophoresis and dynamic light scattering to confirm the relative homogeneity of the preparation. More extensive characterization studies, including those shown in supplemental Fig. 1, were then repeated on samples recovered following SANS analyses to ensure that the properties of the particles did not change during the course of the experiment. sHDL particle preparations studied by SANS and cross-linking/mass spectrometry were initially characterized to confirm homogeneous monodisperse particles with composition and biological activity similar to sHDL isolated from human plasma (supplemental Fig. 1). Reconstituted sHDL particle preparations containing either human plasma-derived apoA1 (h-sHDL) or deuterated recombinant apoA1 (d-sHDL) were each shown to have a hydrodynamic diameter of ∼93 Å as monitored by equilibrium native nondenaturing gradient gel electrophoresis (supplemental Fig. 1, A and C). Chemical compositional analyses of both nondeuterated and deuterated sHDL preparations showed similar lipid composition (phospholipid/free cholesterol/cholesterol ester) and apoA1 content (supplemental Fig. 1B), and cross-linking studies and SDS-PAGE analysis suggested 3 molecules of apoA1/sHDL particle (supplemental Fig. 1D). This result was further confirmed by independent MALDI-TOF mass spectrometry analyses of delipidated cross-linked apoA1 recovered from sHDL, with the bulk of the protein detected possessing mass close (within ∼0.2% predicted) to that anticipated for a trimeric apoA1 particle plus the estimated added mass provided from cross-linking/quenching agents (supplemental Fig. 1E). Analyses of the biological activity of the sHDL preparations used for SANS showed cholesterol efflux activity when incubated with cholesterol loaded macrophages similar to that observed with a common spherical HDL (HDL3) isolated from human plasma (supplemental Fig. 1F), and sHDL preparations showed saturable and specific binding to the HDL receptor, scavenger receptor B1 (supplemental Fig. 1G). Collectively, these results demonstrate that the reconstituted sHDL particles studied by SANS and cross-linking/mass spectrometry in the present studies have a chemical composition, overall stoichiometry, particle size, and biological activity similar to HDL3 isolated from human plasma, and appear to be both monodisperse and homogeneous.
SANS Analyses of sHDL Are Consistent with a Peripheral Trimeric ApoA1 That Cradles an Inner Spheroidal Compact Lipid Core
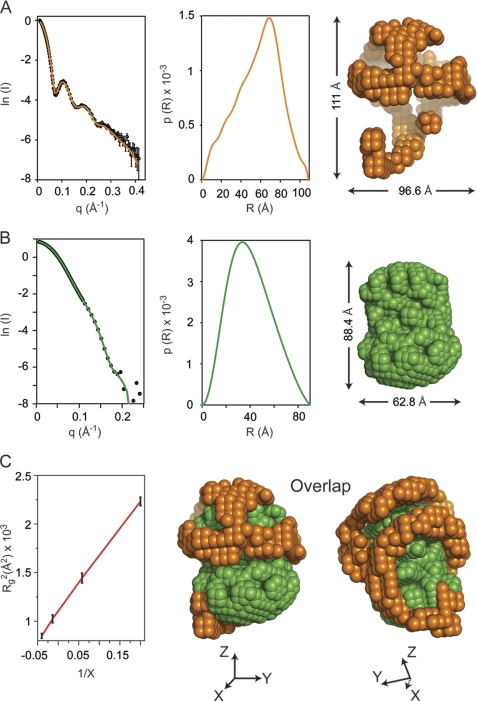
SANS experiments were performed at multiple levels of contrast on sHDL preparations generated using either nondeuterated or deuterated apoA1 (Fig. 1). Samples demonstrated monodisperse characteristics, without detectable traces of aggregation. The molecular weight of the protein within sHDL measured by SANS was consistent with a trimer (8.1–8.6 × 104 atomic mass units). The radius of gyration (Rg) at each contrast level and the overall particle dimensions are listed in Table 1. SANS with contrast variation studies permitted the determination of the individual low resolution structures of the protein and lipid components of sHDL. The scattering signal of the sHDL protein component was enhanced through the incorporation of deuterated apoA1 into the reconstituted sHDL preparations and measurement at the average matching point of the lipid (12% D2O). Fig. 1A, left panel, shows the experimental sHDL scattering intensity data from 12% D2O solution and the superimposed calculated scattering curve for the low resolution structure. The maximum value of the scattering vector (q) is 0.44 Å−1, which corresponds to a resolution in real space of 14.5 Å. The middle panel of Fig. 1A shows the distance distribution function, p(R), which is used to construct the low resolution structure of the protein component of sHDL (Fig. 1A, right panel). To determine the low resolution structure of the lipid component of sHDL, particles reconstituted with nondeuterated apoA1 were measured at 42% D2O contrast level, the matching point of the protein component of sHDL. Fig. 1B shows the experimental data (left panel) and the calculated scattering curve obtained from the low resolution shape envelop of the lipid (right panel). The p(R) function of the lipid component is shown in Fig. 1B, middle panel. Finally, the Stuhrmann plot (68) (where the square of Rg is plotted against the inverse of the difference between the matching point of the whole molecule (18% D2O) and each contrast level) produces a straight-line, with a positive slope (Fig. 1C), indicating two structural features: (i) the centers of gravity of the protein and lipid components within sHDL coincide; and (ii) the component with higher scattering length density (protein) is located at the periphery of the particle. Goodness-of-fit (χ2) analyses for the predicted scattering curves from the low resolution SANS structures of the protein and lipid components of sHDL versus the experimental scattering intensities show excellent agreement for both the protein and lipid components. Overlapping views of the low resolution structures of the protein and lipid components of reconstituted sHDL in two different orientations are shown in Fig. 1C (right). The shape of the combined protein and lipid structures is spheroidal. It is evident from the superposition that the overall architecture of the protein component of sHDL is a hollow structure (orange beads) that cradles the nonspherical compact lipid core (green beads), and the lipid component located inside the particle is only partially surrounded by the outer protein wrapper (Fig. 1C, middle and right).
FIGURE 1.
Small angle neutron scattering results. A, neutron scattering results for sHDL in 12% D2O. Left panel, the scattering intensity as a function of the scattering vector q. The experimental data are plotted with open circles and have error bars attached. The experimental scattering curve displays oscillations with maxima at ∼0.1, 0.2, and 0.25 Å−1. The solid orange line is the scattering intensity calculated from the low resolution structure obtained by deconvoluting the experimental data. Middle panel, the distance distribution function, p(R), plotted as a function of the distance between scattering centers. The p(R) value was obtained by deconvoluting the experimental scattering intensities of sHDL in 12% D2O with the program GNOM. Right panel, low resolution structure of the protein component of sHDL obtained from the p(R) function (middle panel) with the program DAMMIN. B, neutron scattering results for sHDL in 42% D2O. Left panel, the scattering intensity (open circles) for sHDL in 42% D2O. The error bars are very small and only slightly visible at the center of the open circles. The solid green line is the scattering intensity produced by the low resolution structure of the lipid. Middle panel, the p(R) function for the lipid component of sHDL. Right panel, low resolution structure of the lipid component of sHDL obtained from the p(R) function (middle panel). C, left panel, the Stuhrmann plot gives the dependence of Rg2 (where Rg is radius of gyration) on the contrast level (X) in the SANS experiment. A linear graph (red line) indicates that the protein component is located on the outside, and the lipid component is inside as shown by the overlap of the low resolutions structures for the 12% D2O (orange) and 42% D2O (green) in the middle and right panels. X, Y, and Z represent the directions of the Cartesian coordinate system.
Proposed Possible Global Arrangements of ApoA1 Chains in sHDL
In contrast with nascent HDL, the structure of which has been extensively investigated, there are relatively few experimental studies on sHDL structure to date (29, 47). Only one of the recent studies has obtained geometrical constrains among apoA1 molecules within sHDL particles using cross-linking (47), and the other produced only EM images of an entire particle (29). Of note, no studies to date directly visualize the shape of apoA1 in sHDL in sufficient detail to determine the overall architecture of protein within the spherical particle.
Recent mass spectrometry studies by Silva et al. (47) report that cross-links in reconstituted sHDL preparations are for the most part the same as those observed in nascent HDL. However, among the cross-links identified, seven were observed only in reconstituted sHDL (supplemental Table 1) and not in nascent HDL preparations. In that study, several hypothetical arrangements of three apoA1 molecules were proposed to explain the cross-links identified within sHDL. The first arrangement posited was a planar circular belt consisting of three apoA1 molecules, all in an anti-parallel orientation. In the second arrangement, one apoA1 was hypothesized to exist as a hairpin and the other two apoA1 molecules were arranged in a planar circular anti-parallel double belt configuration. In a third arrangement, the three apoA1 molecules were hypothesized to be associated in a clever symmetrical structure retaining the anti-parallel apoA1 chain orientation forming a “trefoil” (47).
The highly symmetric protein models proposed for the three apoA1 molecules based on cross-linking studies (47) clearly do not fit the experimental SANS shape visualized for the protein component of sHDL. However, combining SANS studies with contrast variation and complementary structural information from alternative biophysical approaches such as cross-linking/mass spectrometry data can provide synergistic information that, when considered in combination, imposes considerable constraints on the possible permutations through which the apoA1 may be oriented within sHDL. We note that the SANS resolution for the protein component of sHDL (14 Å) is sufficient to resolve the location of the apoA1 chains within the low resolution structure of sHDL, because the average distance between two apoA1 chains (outer residues) in a belt configuration is about 14 Å. Thus, the low resolution structure of the protein within sHDL visualized by SANS creates a puzzle as to how to arrange three apoA1 molecules within the protein envelop while still accounting for the biophysical attributes reported in prior studies of sHDL preparations. Following other investigators (e.g. Silva et al. (47)) we produced computer-generated three-dimensional schematic models that have all-atom representation, which makes them suitable for distance analysis. We do not claim that these schematics give detailed information about the relative orientation of amino acid residues in apoA1 within sHDL; but we believe that taking into account the spatial constraints imposed by the primary and secondary structure of apoA1 in combination with its low resolution structure resolved by SANS, the schematic model of the protein is a fair description of the overall architecture of the apoA1 trimer in sHDL and thus is appropriate for the MS cross-linking analysis. In this spirit, all protein structures considered included maximal retention of a predominantly anti-parallel orientation for the apoA1 chains in 5/5 helix registry, a necessity given the previously observed crystal structure of the lipid-free truncated mutant (residues 44–243, exon 4) apoA1 (69) and the large numbers of reported FRET, ESR, and cross-link studies of HDL (17, 27, 35, 47, 70–72).
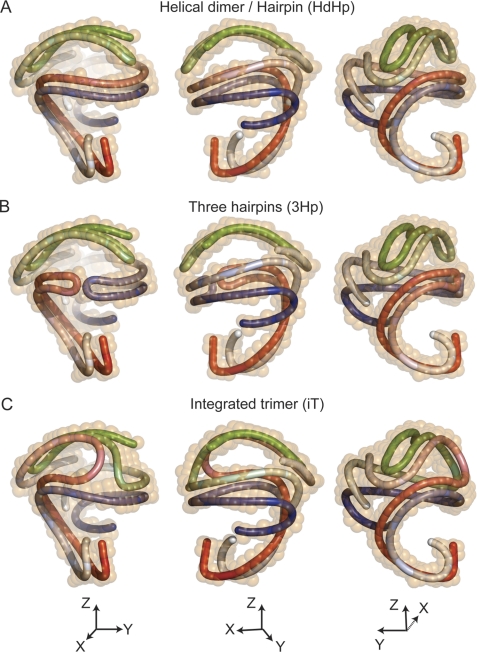
Although it is not immediately recognizable how individual chains of apoA1 might be oriented within the protein low resolution structure, given the length of an apoA1 polypeptide chain, further analyses suggest only a handful of possible global categories of apoA1 configurations that can fit within the structural confines dictated by the protein shape envelop. Fig. 2 shows three general categories of possible arrangements of the apoA1 trimer superimposed on the protein envelop determined by SANS while simultaneously maintaining apoA1 chain anti-parallel orientation. In Fig. 2A the three protein chains (colored red, blue, and green) are arranged as a combination of a helical dimer and a folded-back hairpin. We call this arrangement the “helical-dimer-hairpin” model. The HdHp model for the apoA1 trimer predicts that the apoA1 hairpin folds back, bringing its N terminus close to its helix 5 (h5) domain. Other predicted distinct features are the specific contacts/interactions of the hairpin with the dimer and the fact that the hairpin h5 domain is not aligned with the dimer h5 domains. Another possible global arrangement of the apoA1 trimer within the protein envelope is shown in Fig. 2B as three distinct apoA1 hairpins. Although multiple configurations of this global arrangement are possible, we chose to orient two hairpins (red and blue (Fig. 2B)) such that they interact through their h5 domains (given the cross-linking data for apoA1 in sHDL; see below), whereas the third hairpin (green) has the same orientation as in the HdHp structure in Fig. 2A. This model is called the “three-hairpin” model, because the apoA1 dimer is replaced by two apoA1 monomers that occupy the same volume of the SANS envelope. Further, it can be expected that cross-links that have been reported to discriminate between a double belt model versus the hairpin models for apoA1 of nascent HDL (34, 37, 73) will also discriminate between models HdHp and 3Hp. Finally, we explored the possibility of a third global arrangement whereby the model topologically resembles in part the trefoil model (47), because all three apoA1 monomers associate mutually over a portion of their protein chains. Because the protein envelop visualized by SANS does not share the same symmetry properties with the trefoil model, we call the apoA1 trimeric model that somewhat simulates the trefoil model yet fits within the SANS shape the “integrated trimer” model (Fig. 2C; note that the iT model has only one node instead of two where all three apoA1 chains join together, and the actual conformation of the individual apoA1 chains is very different from that in the trefoil model). In the trefoil model all apoA1 monomers are aligned with their h5 domains (47).
FIGURE 2.
Possible architectures of the apoA1 trimer in spherical HDL. A, superposition of the 12% D2O low resolution structure of sHDL (orange) with a schematic representation of the apoA1 trimer made of a helical dimer (red/blue) and a hairpin (green): the HdHp model. The three chains are gradient-colored (dark N terminus and lighter colored C terminus). The three panels depict different orientations of the HdHp model. B, the architecture of the apoA1 trimer corresponds to three hairpins that fit the 12% D2O low resolution shape: the 3Hp model. The red and blue hairpins interact with their helix 5 domains. The green hairpin retains the same orientation as the one in A. C, in this architecture (iT model) the apoA1 trimer has each chain associated with each other one, producing three dimers. All chains interact together with their helix 5 domains.
Table 2 shows the goodness of fit for each of the hypothetical orientations of apoA1 posited versus the actual experimental SANS data. We used the three-dimensional tube representations shown in Fig. 2 for each model to generate an all-atom model for the protein that was made mostly of a continuous α-helical ribbon. These models do not contain information about the solvent accessibility of individual amino acids, but the orientation of the amino acid residues was consistent with that found in previous models of nascent HDL whereby hydrophobic α-helical surfaces are oriented toward the lipid core and predominantly hydrophilic α-helical apoA1 side chains are oriented toward the aqueous phase (26). The scattering intensities at various scattering angles are shown in Fig. 3A (blue line) for the HdHp model superimposed on the experimental data (open black circles); also shown is the calculated scattering curve corresponding to the low resolution SANS structure (solid orange line). The agreement among all three curves is excellent, as indicated by the goodness of fit (calculated χ2 = 1.192 and 1.483, respectively (Tables 1 and 2)).
TABLE 2.
Radius of gyration, goodness of fit of geometrical constraints, and number of predicted cross-links for the molecular models of apoA1 trimer in spherical reconstituted HDL
| Model | Rg | χ2 | Satisfied cross-linksa |
|---|---|---|---|
| Å | |||
| HdHp | 45.6 | 1.192 | 23 |
| 3Hp | 45.6 | 1.183 | 19 |
| iT | 45.8 | 1.515 | 21 |
a Of 23 reported by Silva et al. (47).
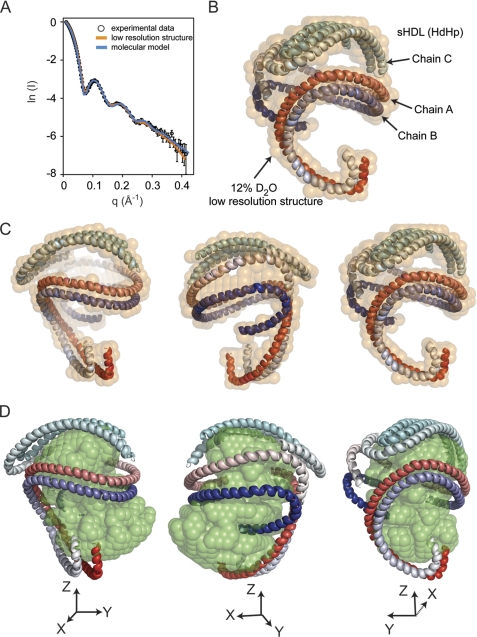
FIGURE 3.
Comparison of experimental neutron scattering intensities with those obtained from the low resolution structure and the molecular model of the apoA1 trimer. A, the fit of scattering intensities produced by the 12% D2O low resolution structure (orange curve) and the molecular model (blue curve) of the apoA1 trimer (HdHp model) to the experimental data. B, the superposition of the molecular model of the apoA1 trimer (HdHp) with the 12% D2O low resolution structure (semitransparent orange beads). The molecular model of the apoA1 trimer is shown as three chains, two of them in a helical dimer conformation, and the third chain forms a hairpin. Because the molecular model does not include any local information about the conformation of the individual amino acid residues, the secondary structure of the chains was assigned to be α-helix for all residues. The three chains are gradient-colored red, blue and cyan. C and D, superposition of the HdHp model with the low resolution structure (orange) of the protein component (12% D2O) of sHDL (C) and the low resolution structure of the lipid component (42% D2O) (D).
A comparison of the goodness of fit of the calculated scattering curves from each of the three global models of apoA1 architecture (HdHp, 3Hp, and iT) versus the actual experimental SANS scattering data reveals that the HdHp and 3Hp models most closely fit the experimental data, whereas the iT model fits less well (Table 2). The all-atom structure for the HdHp model is shown in Fig. 3B superimposed with the low resolution structure of the protein (semitransparent orange beads). Fig. 3C shows the all-atom model HdHp in three different orientations that emphasize the helical conformation of the dimer, and Fig. 3D shows how the all-atom model of the protein wraps around the low resolution structure of the lipid core (semitransparent green beads). The SANS analysis of the three possible overall architectures of the apoA1 trimer in sHDL cannot on its own discriminate between the three global models, but it can select specific conformations of apoA1 that fit the low resolution structure of sHDL. Consequently, the subset of possible conformations can be further analyzed for compliance with geometrical constraints derived from cross-linking studies, and the combined analysis can thus be used to identify the most probable architectures of the apoA1 trimer that satisfy all arrays of biophysical constraints revealed thus far.
Geometrical Constraints Analysis of ApoA1 Chain Cross-linking Data Coupled with SANS Discriminates among the Overall Architectures Posited for ApoA1 in sHDL
The construction of crude all-atom models (HdHp, 3Hp, and iT) for the apoA1 trimer allowed us to perform a geometry constraints analysis of the apoA1 chains by comparing the experimentally reported cross-links for sHDL (35, 47) and those newly determined in our sHDL preparations with those predicted by the models. A summary of this comparison is given in Table 2 and a more detailed description in supplemental Table 1, which lists the individual cross-links identified in our sHDL preparation (93 Å particle, S93) and those predicted by the three proposed architectures of apoA1 trimer in sHDL (i.e. the HdHp, 3Hp, and iT models). Recently, Silva et al. (47) identified 23 cross-links in sHDL particles of 93 Å in diameter (S93) that contain three apoA1 chains. Interchain and intrachain designations for cross-links observed in tryptic digests of cross-linked S93 by Silva et al. (47) were based upon results observed previously with nascent HDL (which contains two apoA1) and a smaller more compact trimeric sHDL form. Thus, cross-linked monomer versus multimeric apoA1 forms were not separated prior to mass spectrometry analysis within S93 sHDL by Silva et al. (47).4 Among the 23 cross-links identified in S93 by Silva et al. (47), 14 were designated as intrachain and 7 as interchain cross-links. Two cross-links were not assigned as either intra- or interchain. We performed similar mass spectrometry/cross-linking analyses of sHDL (S93) preparations but added the step of electroeluting various cross-linked apoA1 bands from sHDL preparations (monomer versus dimer versus trimeric forms confirmed by both SDS-PAGE and MALDI-TOF mass spectrometry (“Experimental Procedures”) as well as performing in-solution tryptic digestion/MS analyses on the isolated cross-linked proteins. This allowed for direct assignment of detected cross-links as either intra- or interchain cross-links within sHDL (supplemental Table 1). It is remarkable that all three all-atom models for the apoA1 trimer in sHDL preparations satisfy the majority of identified cross-links, although the HdHp model appears superior to both the 3Hp and the iT models. For example, the HdHp model satisfies 23 cross-links, the 3Hp model satisfies 19, and the iT model satisfies 21 (Table 2). None of the models satisfies the cross-link Lys208–Lys208 identified by the same group in a 96 Å nascent HDL particle (47) but not predicted by any of the proposed nascent HDL models (e.g. the double belt, the hairpin, the solar flares, and the double super helix models) unless it arises from a particle-particle interaction (27, 32). The previously reported intermolecular cross-link, Lys182–Lys238, was suggested to perhaps represent apoA1 monomers that had undergone a shift from a 5/5 to a 5/2 helix registry (47). All protein models proposed here contain this cross-link in a 5/5 registry, but we predict that it is intrachain. An intrachain designation for this cross-link is in agreement with cross-link MS analyses of electroeluted apoA1 forms examined in the present study (supplemental Table 1). The two cross-links observed by Silva et al. (47) in sHDL that were not assigned as either intra- or interchain (Lys182–Lys239 and N-terminal Lys118) were assigned in this study as inter- and intrachain cross-links, respectively, on the basis of cross-link MS analyses of electroeluted proteins (supplemental Table 1). In the molecular models, Lys182–Lys239 is readily accommodated as an intrachain cross-link, and interchain distances as small as 40 Å are noted, suggesting that an interchain cross-link is also plausible based on the models. The distance corresponding to N-terminal Lys118 in all models was <30 Å for both the intra- and interchain distances in all models (supplemental Table 1).
Although it is remarkable that all three all-atom models for the apoA1 trimer in sHDL preparations satisfy the majority of identified cross-links, there are still significant differences in the cross-links predicted by the models that make the HdHp model appear superior. For example, a more detailed analysis of the data in supplemental Table 1 shows that the 3Hp model can be ruled out as a possible architecture of the apoA1 trimer, as it cannot accommodate most of the interchain cross-links because of its specific topology (all three apoA1 chains self-associate, and only few interchain cross-links would be predicted). As a consequence, four interchain cross-links (Lys40–Lys239, Lys118–Lys140, Lys59–Lys208, and Lys77–Lys195) are not accommodated by this model. On the other hand, the iT model has all three chains associated with each other and thus does not accommodate some of the intrachain cross-links (Lys12–Lys94 and N-terminal Lys106) seen in the HdHp model. Taken together, the combined SANS and MS/cross-link data point to the HdHp model as the most plausible global topology of apoA1 within sHDL.
Comparison of Currently Visualized SANS Shape of ApoA1 within sHDL versus Prior Reported Models Obtained by Molecular Dynamics Simulation and Electron Microscopy Studies
In the absence of direct structural data for the overall shape of protein within sHDL, other investigators have attempted to construct purely theoretical models of the particle by computer simulations. It therefore may be of interest to compare the low resolution (∼14 Å) shape envelop of the protein component within reconstituted sHDL, as visualized by SANS in the present study, with the previous hypothetical models obtained by simulation. Three molecular dynamics simulations of sHDL have been reported to date (48–50). Despite the fact that sHDL typically contains three or more apoA1 chains, these computational models focus predominantly on simulations of sHDL with only two apoA1 chains (48–50). In general, the molecular dynamics simulation studies of sHDL start with the protein in a discoidal belt organization at the beginning of the simulation, suggesting that the overall belt shape of apoA1 becomes more disordered and contorted, with pronounced “kinks” or zigzags in an out-of-plane conformation (48, 49). The protein conformations in the simulations reported are at marked contrast to the SANS shape observed in the present investigation. It should also be noted that prior simulation models of sHDL are also reported to be at marked variance (49) with the three apoA1-containing trefoil structure suggested by Silva et al. (47). It seems likely that one of the major reasons that marked differences exist between the experimentally observed SANS shape (which contains three apoA1 chains) is that only two apoA1 chains are used in the molecular dynamics simulations. It is also worth noting, however, that all simulations and the SANS shapes directly visualized have some similarities. At a very gross level, the topological organization of protein within the particle toward the exterior and a neutral lipid core interior are clear similarities. It is interesting to note that the simulations of an sHDL particle suggest that the N- and C-terminal helices of the apoA1 chains are more flexible and mobile than the central region of the apoA1 chains (49), a finding consistent with the conformational alterations we proposed for apoA1 within a maturing nascent HDL particle to form sHDL (see discussion below and Fig. 4A, top).
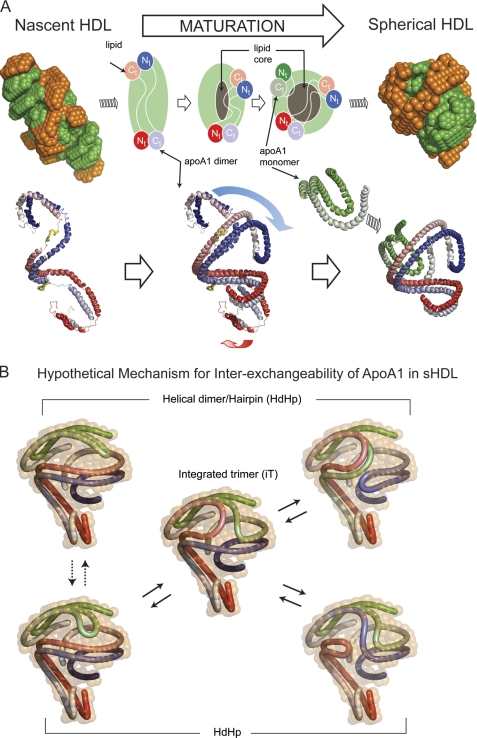
FIGURE 4.
Hypothetical mechanism of conformational change of apoA1 in nascent HDL during maturation into spherical HDL. A, left, superposition of the low resolution structures of protein (orange) and lipid (green) components of nascent HDL (top) and one particular orientation of the double super helix model of the protein component in nascent HDL (bottom left). The two apoA1 chains are gradient-colored red and blue, and the putative lecithin:cholesterol acyltransferase-binding loops (solar flares) are colored yellow. Middle, overlap of the middle domains of nascent HDL and sHDL apoA1 dimers. The overlapping suggests how the N and C termini of the apoA1 dimer in nascent HDL might swing and rearrange during particle maturation into what is found to be the dimer conformation in sHDL. The gray region at the middle of the particle represents the growing core of neutral lipids that accumulate during maturation. Right, superposition of the low resolution structures of protein (orange) and lipid (green) components of spherical HDL (top), and the resultant architecture of apoA1 trimer in spherical HDL (HdHp model, bottom right). B, hypothetical mechanism for apoA1 inter-exchange in sHDL. The hairpin and the apoA1 dimer are shown in different conformations that match the low resolution structure (12% D2O) of the protein. The different configurations suggest that it is reasonable to expect that because of particle dynamics the hairpin can bend such that it aligns its h5 domain with the h5 domain of the dimer (dotted double arrow lines) as a preamble for annealing with the dimer. Thus, the hairpin can exchange with one of the apoA1 monomers of the dimer through a transient integrated trimer-like configuration (center), generating other arrangements of the helical dimer/hairpin combination (solid double arrow lines). This protein reorganization mechanism makes all apoA1 monomers equivalent from an exchange point of view and can, in principle, lead to the integrated trimer (iT model) as illustrated on the right.
During the course of reviewing this manuscript, Zhang et al. (29) published a methodology paper for optimized negative staining electron microscopy (EM) of multiple lipoprotein particles, including some sHDL preparations. Although both cryo-EM and negative staining EM images of sHDL were shown, no structural models were presented, and the low resolution permitted few details of the overall shape of apoA1 within the particles (29). Zhang et al. (29) did note, however, that the images of sHDL show “contiguous high densities near the particle edge and center,” which presumably corresponds to the protein within sHDL. This description is consistent with the low resolution structure of apoA1 within sHDL as visualized by SANS.
The Protein and Lipid Organization in sHDL Provides a General Structural Motif for Lipid Transport
The present study provides the first direct visualization of the individual structural components of sHDL, the major form of HDL in plasma, by means of SANS with contrast variation and a selective isotopic deuterated protein sample. Low resolution images of the protein and lipid components were obtained individually to resolve global time-averaged conformations of each within the whole particle. The overall conformation noted shows how the protein component of sHDL provides structural support, much like a cradle or catcher's mitt, within which the lipid core is carried. This is a highly unusual configuration for known macromolecular complexes and may represent a structural motif replicated in other lipoprotein complexes involved in lipid transport. It is easy to envision how this structural arrangement can enable the lipoprotein to facilitate its lipid cargo-carrying function while also allowing for the dynamic flexibility of a constantly changing particle dimension during cholesterol acceptor and lipid delivery roles. It is intriguing that this sort of global protein and lipid configuration is somewhat similar to observations reported by Weisgraber and colleagues (74) during analysis of apolipoprotein E/diphosphatidylcholine particles in solution using small angle x-ray scattering. In that study, ellipsoidal particles with apolipoprotein E organized in a twisted hairpin conformation were noted, with proposed packing of the phospholipid core being micellar (74).
The low resolution structures observed for sHDL, and previously for nascent HDL with contrast variation SANS (32), suggest a hypothetical path for HDL maturation and reversible remodeling (Fig. 4A). During maturation, cholesterol ester molecules formed by lecithin:cholesterol acyltransferase at the HDL surface partition into the particle core (Fig. 4A, top). Concurrently, the particle changes from a more elliptical nascent HDL shape to a spherical form, consistent with results observed with contrast variation SANS analyses of both nascent HDL and sHDL (Fig. 4A). As can be seen in Fig. 4A (bottom), maturation of the HDL particle is predicted to result in a dramatic change in one of the N and C termini of the helical dimeric apoA1 (folding back upon itself), probably induced by the addition of a third apoA1 chain in the more mature sHDL. This refolding of the apoA1 double chain is suggested based upon the remarkable overlap in SANS shapes noted between nascent HDL and sHDL whereby the helical dimeric apoA1 within nascent HDL closely overlaps with the helical component of the HdHp conformation of sHDL (Fig. 4A, bottom). As noted above, previous simulation studies of sHDL have suggested that the N- and C-terminal regions of apoA1 are more dynamic (49). Similarly, cross-linking studies point toward the N terminus of apoA1 within sHDL folding back and coming in closer proximity to the central region of the apoA1 polypeptide chain (supplemental Table 1).
ApoA1 is an exchangeable apolipoprotein (75, 76) and can convert between lipid-poor and lipid-associated states (77, 78). The exchangeability of ApoA1 and its predilection for forming both highly dynamic amphipathic α-helices and salt bridges seem to be imprinted in its chemical composition (32, 33). A hypothetical mechanism for apoA1 inter-exchangeability on the surface of the sHDL particle is suggested by the present studies. Although this mechanism may be important for sHDL remodeling in plasma at body temperature, the rate of apoA1 exchange should be much lower during the SANS experiment in which samples have been analyzed at 8 °C. For example, assuming that the activation energy barrier for exchange is about 20 kcal/mol, the rate constant at 8 °C is about 30 times smaller than at 37 °C. Fig. 4B is a schematic showing a hypothetical mechanism of how each of the apoA1 of sHDL may be inter-exchangeable through a transient rearrangement of apoA1 conformations posited between the global architectures in the HdHp and iT models. It is conceivable that an apoA1 hairpin chain, following modest rearrangement (Fig. 4B, left), may begin annealing into the apoA1 dimer of HdHp forming a three-way node around the helix 5 domain. This annealing process might be driven by concomitant salt bridge breakage (from intrachain) and formation (in interchain) of the annealing apoA1 monomer while simultaneously the dimer gradually unzips at one end to permit association leading to a transient integrated trimer-like form (Fig. 4B, center). This intermediate configuration may then rearrange into another dimer/hairpin combination (Fig. 4B, right) by a reversal of the annealing process. This mechanism would make it possible for apoA1 chains in the dimer to exchange partners by incorporating the hairpin, thus making all apoA1 polypeptide chains interchangeable on the surface of sHDL. We hypothesize that breakage and formation of a large number of salt bridges within the apoA1 dimer and hairpin may serve as a primary driving force behind the annealing of the dimer-hairpin and dissolution of the annealed trimer, allowing for facile inter-exchangeability of apoA1 within a dynamic sHDL particle. Thus, in the above hypothetical model to explain apoA1 inter-exchangeability, interchain salt bridges are replaced by intrachain salt bridges and vice versa, with little or no energetic penalty.
In summary, we applied SANS with contrast variation and isotopic protein deuteration to the structural interrogation of biologically competent reconstituted sHDL. The low resolution structure of sHDL analyzed by SANS in 12% D2O reveals that the overall global shape occupied by apoA1 has a nonplanar contorted conformation. The low resolution structure of sHDL analyzed by SANS at 42% D2O indicates that the lipid phase of sHDL is a slightly elongated spheroid encased within the protein component that wraps around it, providing a scaffold upon which lipids can associate. The protein and lipid organization revealed may serve as a general structural motif for lipoproteins that mediate lipid transport and help explain the inter-exchangeability of apoA1 within HDL.
Acknowledgments
We are grateful to Dr. Giussepe Zaccai (Institut de Biologie Structurale, CEA-CNRS-UJF, Grenoble, France) for helpful discussions and insightful comments on this manuscript. We also thank Dr. John W. Crabb (Cole Eye Institute, Cleveland Clinic, Cleveland, OH) for generous access to the ABI 4800 MALDI-TOF/TOF mass spectrometer used to analyze the molecular weight of delipidated cross-linked apoA1 of sHDL. Computational resources were provided by the Ohio Supercomputer Center and the National Center for Supercomputer Applications (University of Illinois).
This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL098055, P01 HL076491-055328, and P01 HL087018-02001. This work was also supported in part by a grant from the Foundation Leducq.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
Information on neutron scattering intensities for spherical HDL samples analyzed in 12 and 42% D2O (text files) and low resolution structures of the protein and lipid components of spherical HDL (Protein Data Bank files) is available by contacting the corresponding author (S. L. Hazen).
W. S. Davidson, personal communication.
- apoA1
- apolipoprotein A1
- SANS
- small angle neutron scattering
- SAX
- small angle x-ray scattering
- sHDL
- spherical high density lipoprotein
- h5
- helix 5
- rh-apoA1
- recombinant human apoA1
- BS3
- bis(sulfosuccinimidyl)suberate
- HdHp
- helical-dimer-hairpin
- 3Hp
- three hairpins
- iT
- integrated trimer.
REFERENCES
- 1. Castelli W. P., Doyle J. T., Gordon T., Hames C. G., Hjortland M. C., Hulley S. B., Kagan A., Zukel W. J. (1977) Circulation 55, 767–772 [DOI] [PubMed] [Google Scholar]
- 2. Durrington P. N., Ishola M., Hunt L., Arrol S., Bhatnagar D. (1988) Lancet 1, 1070–1073 [DOI] [PubMed] [Google Scholar]
- 3. Pekkanen J., Linn S., Heiss G., Suchindran C. M., Leon A., Rifkind B. M., Tyroler H. A. (1990) New Engl. J. Med. 322, 1700–1707 [DOI] [PubMed] [Google Scholar]
- 4. Karathanasis S. K., Norum R. A., Zannis V. I., Breslow J. L. (1983) Nature 301, 718–720 [DOI] [PubMed] [Google Scholar]
- 5. Norum R. A., Lakier J. B., Goldstein S., Angel A., Goldberg R. B., Block W. D., Noffze D. K., Dolphin P. J., Edelglass J., Bogorad D. D., Alaupovic P. (1982) New Engl. J. Med. 306, 1513–1519 [DOI] [PubMed] [Google Scholar]
- 6. Plump A. S., Scott C. J., Breslow J. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9607–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rubin E. M., Krauss R. M., Spangler E. A., Verstuyft J. G., Clift S. M. (1991) Nature 353, 265–267 [DOI] [PubMed] [Google Scholar]
- 8. Schaefer E. J., Heaton W. H., Wetzel M. G., Brewer H. B., Jr. (1982) Arteriosclerosis 2, 16–26 [DOI] [PubMed] [Google Scholar]
- 9. Fielding C. J., Fielding P. E. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3911–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oram J. F., Lawn R. M., Garvin M. R., Wade D. P. (2000) J. Biol. Chem. 275, 34508–34511 [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y., Zanotti I., Reilly M. P., Glick J. M., Rothblat G. H., Rader D. J. (2003) Circulation 108, 661–663 [DOI] [PubMed] [Google Scholar]
- 12. Moore R. E., Navab M., Millar J. S., Zimetti F., Hama S., Rothblat G. H., Rader D. J. (2005) Circ. Res. 97, 763–771 [DOI] [PubMed] [Google Scholar]
- 13. Nicholls S. J., Dusting G. J., Cutri B., Bao S., Drummond G. R., Rye K. A., Barter P. J. (2005) Circulation 111, 1543–1550 [DOI] [PubMed] [Google Scholar]
- 14. Pajkrt D., Doran J. E., Koster F., Lerch P. G., Arnet B., van der Poll T., ten Cate J. W., van Deventer S. J. (1996) J. Exp. Med. 184, 1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanhamme L., Paturiaux-Hanocq F., Poelvoorde P., Nolan D. P., Lins L., Van Den Abbeele J., Pays A., Tebabi P., Van Xong H., Jacquet A., Moguilevsky N., Dieu M., Kane J. P., De Baetselier P., Brasseur R., Pays E. (2003) Nature 422, 83–87 [DOI] [PubMed] [Google Scholar]
- 16. Lund-Katz S., Liu L., Thuahnai S. T., Phillips M. C. (2003) Front. Biosci. 8, d1044–1054 [DOI] [PubMed] [Google Scholar]
- 17. Thomas M. J., Bhat S., Sorci-Thomas M. G. (2008) J. Lipid Res. 49, 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oram J. F., Vaughan A. M. (2000) Curr. Opin. Lipidol. 11, 253–260 [DOI] [PubMed] [Google Scholar]
- 19. Wang N., Lan D., Chen W., Matsuura F., Tall A. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9774–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barter P. J., Rye K. A. (2001) Curr. Opin. Lipidol. 12, 377–382 [DOI] [PubMed] [Google Scholar]
- 21. Zannis V. I., Chroni A., Krieger M. (2006) J. Mol. Med. 84, 276–294 [DOI] [PubMed] [Google Scholar]
- 22. Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. (1996) Science 271, 518–520 [DOI] [PubMed] [Google Scholar]
- 23. Wang N., Weng W., Breslow J. L., Tall A. R. (1996) J. Biol. Chem. 271, 21001–21004 [DOI] [PubMed] [Google Scholar]
- 24. Atkinson D., Small D. M., Shipley G. G. (1980) Ann. N.Y. Acad. Sci. 348, 284–298 [DOI] [PubMed] [Google Scholar]
- 25. Atkinson D., Smith H. M., Dickson J., Austin J. P. (1976) Eur. J. Biochem. 64, 541–547 [DOI] [PubMed] [Google Scholar]
- 26. Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. (1999) J. Biol. Chem. 274, 31755–31758 [DOI] [PubMed] [Google Scholar]
- 27. Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. (2007) Nat. Struct. Mol. Biol. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 28. Jones M. K., Zhang L., Catte A., Li L., Oda M. N., Ren G., Segrest J. P. (2010) J. Biol. Chem. 285, 41161–41171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang L., Song J., Cavigiolio G., Ishida B. Y., Zhang S., Kane J. P., Weisgraber K. H., Oda M. N., Rye K. A., Pownall H. J., Ren G. (2011) J. Lipid Res. 52, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. (2010) J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lagerstedt J. O., Cavigiolio G., Budamagunta M. S., Pagani I., Voss C. J., Oda M. N. (2011) J. Biol. Chem. 286, 2966–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., DiDonato J. A., Hazen S. L. (2009) J. Biol. Chem. 284, 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gogonea V., Wu Z., Lee X., Pipich V., Li X. M., Ioffe A. I., DiDonato J. A., Hazen S. L. (2010) Biochemistry 49, 7323–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 35. Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. (2005) Biochemistry 44, 8600–8607 [DOI] [PubMed] [Google Scholar]
- 36. Li H. H., Lyles D. S., Pan W., Alexander E., Thomas M. J., Sorci-Thomas M. G. (2002) J. Biol. Chem. 277, 39093–39101 [DOI] [PubMed] [Google Scholar]
- 37. Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. (2006) J. Biol. Chem. 281, 20418–20426 [DOI] [PubMed] [Google Scholar]
- 38. Anderson D. W., Nichols A. V., Pan S. S., Lindgren F. T. (1978) Atherosclerosis 29, 161–179 [DOI] [PubMed] [Google Scholar]
- 39. Blanche P. J., Gong E. L., Forte T. M., Nichols A. V. (1981) Biochim. Biophys. Acta 665, 408–419 [DOI] [PubMed] [Google Scholar]
- 40. Cheung M. C., Segrest J. P., Albers J. J., Cone J. T., Brouillette C. G., Chung B. H., Kashyap M., Glasscock M. A., Anantharamaiah G. M. (1987) J. Lipid Res. 28, 913–929 [PubMed] [Google Scholar]
- 41. Francone O. L., Gurakar A., Fielding C. (1989) J. Biol. Chem. 264, 7066–7072 [PubMed] [Google Scholar]
- 42. Jonas A., Wald J. H., Toohill K. L., Krul E. S., Kézdy K. E. (1990) J. Biol. Chem. 265, 22123–22129 [PubMed] [Google Scholar]
- 43. Rye K. A., Hime N. J., Barter P. J. (1995) J. Biol. Chem. 270, 189–196 [DOI] [PubMed] [Google Scholar]
- 44. Davidson W. S., Sparks D. L., Lund-Katz S., Phillips M. C. (1994) J. Biol. Chem. 269, 8959–8965 [PubMed] [Google Scholar]
- 45. Dalton M. B., Swaney J. B. (1993) J. Biol. Chem. 268, 19274–19283 [PubMed] [Google Scholar]
- 46. Sparks D. L., Phillips M. C., Lund-Katz S. (1992) J. Biol. Chem. 267, 25830–25838 [PubMed] [Google Scholar]
- 47. Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Catte A., Patterson J. C., Bashtovyy D., Jones M. K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemelä P., Karttunen M., Marrink S. J., Vattulainen I., Segrest J. P. (2008) Biophys. J. 94, 2306–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shih A. Y., Sligar S. G., Schulten K. (2009) J R Soc. Interface 6, 863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vuorela T., Catte A., Niemelä P. S., Hall A., Hyvönen M. T., Marrink S. J., Karttunen M., Vattulainen I. (2010) PLoS Comput. Biol. 6, e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 52. Stuhrmann H. B. (2008) Acta Crystallogr. A 64, 181–191 [DOI] [PubMed] [Google Scholar]
- 53. Guilbaud J. B., Saiani A. (2010) Chem. Soc. Rev. [Epub ahead of print] [Google Scholar]
- 54. Serdyuk I. N., Zaccai N. R., Zaccai J. (2007) Methods in Molecular Biophysics: Structure, Dynamics, Function, pp. 794–837, Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 55. Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. (1975) Nature 253, 245–249 [DOI] [PubMed] [Google Scholar]
- 56. Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. (1977) Nucleic Acids Res. 4, 3199–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saibil H., Chabre M., Worcester D. (1976) Nature 262, 266–270 [DOI] [PubMed] [Google Scholar]
- 58. May R. P., Nowotny V., Nowotny P., Voss H., Nierhaus K. H. (1992) EMBO J. 11, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ramakrishnan V. (1986) Science 231, 1562–1564 [DOI] [PubMed] [Google Scholar]
- 60. Peng D. Q., Wu Z., Brubaker G., Zheng L., Settle M., Gross E., Kinter M., Hazen S. L., Smith J. D. (2005) J. Biol. Chem. 280, 33775–33784 [DOI] [PubMed] [Google Scholar]
- 61. Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 62. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 63. Jacrot B., Zaccai G. (1981) Biopolymers 20, 2413–2426 [Google Scholar]
- 64. Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 66. Glatter O. (1977) J. Appl. Crystallogr. 12, 415–421 [Google Scholar]
- 67. Svergun D. I., Richard S., Koch M. H., Sayers Z., Kuprin S., Zaccai G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stuhrmann H. B. (1970) Acta Crystallogr. A 26, 297–306 [Google Scholar]
- 69. Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jonas A. (2002) in Biochemistry of Lipids (Vance D. E., Vance J. E. eds) pp. 483–504, Elsevier Science Publishers B. V., Amsterdam [Google Scholar]
- 71. Davidson W. S., Hilliard G. M. (2003) J. Biol. Chem. 278, 27199–27207 [DOI] [PubMed] [Google Scholar]
- 72. Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., Brouillette C. G. (1998) Biochemistry 37, 11714–11725 [DOI] [PubMed] [Google Scholar]
- 73. Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. (2004) Biochemistry 43, 11717–11726 [DOI] [PubMed] [Google Scholar]
- 74. Peters-Libeu C. A., Newhouse Y., Hall S. C., Witkowska H. E., Weisgraber K. H. (2007) J. Lipid Res. 48, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 75. Breslow J. L. (1991) Annu. Rev. Med. 42, 357–371 [DOI] [PubMed] [Google Scholar]
- 76. Wang J., Sykes B. D., Ryan R. O. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Oda M. N., Forte T. M., Ryan R. O., Voss J. C. (2003) Nat. Struct. Biol. 10, 455–460 [DOI] [PubMed] [Google Scholar]
- 78. Wang L., Atkinson D., Small D. M. (2005) J. Biol. Chem. 280, 4154–4165 [DOI] [PubMed] [Google Scholar]