Abstract
Cysteine (Cys) plays a major role in growth and survival of the human parasite Entamoeba histolytica. We report here the crystal structure of serine acetyltransferase (SAT) isoform 1, a cysteine biosynthetic pathway enzyme from E. histolytica (EhSAT1) at 1.77 Å, in complex with its substrate serine (Ser) at 1.59 Å and inhibitor Cys at 1.78 Å resolution. EhSAT1 exists as a trimer both in solution as well as in crystal structure, unlike hexamers formed by other known SATs. The difference in oligomeric state is due to the N-terminal region of the EhSAT1, which has very low sequence similarity to known structures, also differs in orientation and charge distribution. The Ser and Cys bind to the same site, confirming that Cys is a competitive inhibitor of Ser. The disordered C-terminal region and the loop near the active site are responsible for solvent-accessible acetyl-CoA binding site and, thus, lose inhibition to acetyl-CoA by the feedback inhibitor Cys. Docking and fluorescence studies show that EhSAT1 C-terminal-mimicking peptides can bind to O-acetyl serine sulfhydrylase (EhOASS), whereas native C-terminal peptide does not show any binding. To test further, C-terminal end of EhSAT1 was mutated and found that it inhibits EhOASS, confirming modified EhSAT1 can bind to EhOASS. The apparent inability of EhSAT1 to form a hexamer and differences in the C-terminal region are likely to be the major reasons for the lack of formation of the large cysteine synthase complex and loss of a complex regulatory mechanism in E. histolytica.
Keywords: Crystal Structure, Enzyme Structure, Oxidative Stress, Parasite Metabolism, Protein Assembly, Protein-Protein Interactions, Enzyme Regulation
Introduction
In bacteria and plants, l-cysteine is synthesized from l-Serine by two key enzymes serine acetyltransferase (SAT)3 and O-acetyl serine sulfhydrylase (OASS). SAT converts l-Serine to O-acetyl serine by transferring an acetyl group from acetyl-CoA. Later, OASS converts the O-acetyl serine (OAS) to l-cysteine by adding a sulfide. This cysteine biosynthetic pathway contributes significantly to the incorporation of inorganic sulfur into organic compounds. SAT is in low abundance compared with OASS and is the rate-limiting component in this pathway (1).
The intracellular level of Cys is maintained by complex kinetics and strictly regulated by two known mechanisms. In the first, SAT is inhibited by Cys through a feedback mechanism. The second mechanism involves association of SAT and OASS to form a cysteine synthase complex. In the complex form, SAT activity increases, whereas OASS activity is reduced (2). This results in excess production of OAS, which causes the dissociation of the cysteine synthase complex and subsequent down-regulation of SAT activity (3).
The structures of SAT from Escherichia coli (EcSAT) and Haemophilus influenzae (HiSAT) have been reported (4, 5) and also in complex with cysteine and acetyl-CoA (6). These structures exist as hexamers in the form of dimer of trimers. The solution studies and structures of OASS reveal that it stays as a dimer (7–9). Biochemical and modeling studies indicated that one SAT hexamer and two OASS dimers interact to form the cysteine synthase complex (10). The molecular details of the cysteine synthase complex are still not very clear. Various biochemical and structural studies have revealed that C-terminal end of SAT interacts with the active site of OASS (11, 27).
Entamoeba histolytica is the enteric protozoan parasite that causes amoebic colitis and extra-intestinal abscesses in approximately 50 million people and kills about 70,000 each year (12). Cysteine was reported as the major thiol in E. histolytica (13) and is assumed to play a major role in oxidative stress defense mechanisms in this glutathione-deficient organism. Cysteine has also been shown to be necessary for survival, growth, elongation, and attachment to matrix (14, 15). The cysteine biosynthetic pathway could be crucial target to develop new chemotherapeutics (16). Similar to other bacteria and plants, l-cysteine is synthesized by serine acetyltransferase (EhSAT) and O-acetyl serine sulfhydrylase (EhOASS) in E. histolytica. But unlike in other bacteria and plants, EhSAT and EhOASS do not interact with each other and do not form the cysteine synthase complex (14).
The fact that the E. histolytica genome encodes three forms of SAT suggests the importance of this enzyme for its survival. The EhSAT1 and EhSAT2 isoforms are very similar (78% identity), whereas the EhSAT3 isoform has very low sequence homology with EhSAT1 and EhSAT2 (48% identity). These three isoforms show differences in sensitivity to the feedback inhibitor l-cysteine (17). Till now there is no structural information available as to how sensitivity for feedback inhibition of cysteine is lost for some mutants/isoforms and why cysteine synthase complex does not form in E. histolytica.
Here for the first time we report the structure of EhSAT at 1.77 Å resolution as well as its structure bound with its substrate serine at 1.59 Å resolution and its inhibitor cysteine at 1.78 Å resolution. EhSAT1 in these structures is trimeric rather than hexameric. The structures with Ser and Cys indicate how Cys can be a competitive inhibitor with Ser and does not inhibit acetyl-CoA. The docking and fluorescence binding studies with EhOASS clearly show that if the EhSAT1 had different C-terminal sequence, EhSAT1 and EhOASS could interact with each other. The C-terminal mutant of EhSAT1 does inhibit EhOASS, confirming our docking studies. These new structural features and biochemical studies suggest abolition of cysteine synthase complex formation to ensure ample supply of cysteine for the survival of the organism.
EXPERIMENTAL PROCEDURES
Cloning of EhSAT1
The serine acetyltransferase 1 (GenBankTM accession no. Ab023954) coding sequence was amplified by PCR from genomic DNA of E. histolytica strain HM1:IMSS using the forward and reverse primers 5′-CCGGCTAGCATGGACAATTACATTTATTC-3′ and 5′-CCGCTCGAGAATCGATGGTGAATTTGC-3′, respectively, The EhSAT1 gene was cloned into pET21c vector (Novagen) between Nhe1 and Xho1 sites with the C-terminal His6 tag.
Overexpression and Purification
The recombinant plasmid pET21c-containing EhSAT1 insert was transformed into E. coli BL21 (DE3) cells (Novagen). Freshly transformed BL21 cells were grown in LB media supplemented with 100 μg ml−1 ampicillin at 37 °C to an A600 of 0.5. Then the culture was induced by 1 mm isopropyl β-D-1-thiogalactopyranoside (Sigma) for overexpression of EhSAT1, and the culture was allowed to grow for another 4 h at 30 °C. The cells were harvested by centrifugation at 6000 × g for 5 min at 4 °C. The harvested cells were suspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5% (v/v) glycerol, 5 mm β-mercaptoethanol, 100 μm phenylmethanesulfonyl fluoride) containing 0.1% (v/v) Triton X-100 (U. S. Biochemical Corp.) and lysed with 3 cycles of flash-freezing in liquid nitrogen and thawing. The cell lysate was sonicated on ice and centrifuged at 15,000 rpm for 20 min at 4 °C. The clear supernatant containing EhSAT1 was passed through nickel-Sepharose column (GE Healthcare), which was pre-equilibrated with lysis buffer containing 10 mm imidazole. The column was washed with 5 volumes of wash buffer (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 40 mm imidazole) at room temperature. The bound protein was eluted with elution buffer (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5% (v/v) glycerol, 5 mm β-mercaptoethanol, 100 μm phenylmethanesulfonyl fluoride, and 150 mm imidazole) and collected in 1.5-ml fractions. The fractions with optimum absorbance at 280 nm were checked for homogeneity on SDS-PAGE, concentrated, and subjected to gel filtration on a HiLoad Superdex 200G 16/60 column (GE Healthcare) at a flow rate of 0.5 ml min−1. The column was pre-equilibrated and eluted with buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% (v/v) glycerol, 5 mm β-mercaptoethanol. The elution profile shows that the EhSAT1 exists as trimer (supplemental Fig. 1). The purity of the protein was assessed on SDS-PAGE and concentrated using Centricon tubes (Amicon) to a final concentration of 12 mg ml−1 as estimated by the Bradford method.
Crystallization
The purified and concentrated EhSAT1 was subjected to crystallization trials using various screens. Hanging drops were prepared in 24-well plates by mixing 3 μl of protein solutions with 3 μl of reservoir solution and equilibrated against 500 μl of reservoir solution. The crystallization trials were carried out at 289 and 277 K. Several crystallization conditions were tested with polyethylene glycol (PEG), ammonium sulfate, and 2-methyl-2,4-pentanediol (Sigma). After optimization of the physicochemical parameters, the best crystals of EhSAT1 were obtained using 1.0 m ammonium sulfate as precipitant in 100 mm Tris pH 7.4–8.0 buffer containing 150 mm NaCl and 5% (v/v) glycerol. One of the substrate l-Ser was used for co-crystallization with EhSAT1. The EhSAT1-Ser complex was subjected to several crystallization conditions. The best crystals were obtained in 1.2 m ammonium sulfate, 100 mm Tris, pH 8.3, 150 mm NaCl, 5% glycerol (v/v), and 10 mm l-Ser, which is very similar to native condition. The feedback inhibitor, l-cysteine, was also used for co-crystallization with EhSAT1. The EhSAT1-Cys complex was subjected to several crystallization conditions. The best crystals were obtained in 6% (w/v) PEG 2000, 100 mm Tris-HCl, pH 7.0, 2 mm cysteine, and 80 mm trimethylamine N-oxide, which is quite different from native conditions.
Data Collection and Processing
Crystals of native EhSAT1 as well as EhSAT1-Ser complex were equilibrated in a reservoir solution with sequential increases of 5, 10, 20, and 25% (v/v) glycerol. These crystals were mounted in cryoloops and flash-frozen in a liquid nitrogen stream at 100 K. EhSAT-Cys complex crystals were transferred into a reservoir solution with a sequential increase of PEG 2000 to reach 30% (w/v) final concentration. Initial data were collected at a home source (Advanced Instrumentation Research Facility) on a Bruker Microstar generator and MAR imaging plate. The high resolution diffraction data were collected at the European Synchrotron Radiation Facility beamline BM14. The data sets were indexed, integrated, and scaled using HKL-2000 data processing software (18).
Structure Determination and Refinement of EhSAT1
The EhSAT1 structure was determined by a molecular replacement method using E. coli SAT as the search model (PDB ID 1T3D) (4). The EhSAT1 was crystallized in the R3 space group with one molecule in an asymmetric unit. Initially, molecular replacement trials were performed with whole EcSAT as a search model. The solution obtained did not improve much after several cycles of model building or by any autobuilding methods. Later, about half of the molecule from the N-terminal side, which had very less sequence homology, was removed, and only half of the molecule from the C-terminal side was used for molecular replacement with MOLREP (19). The solution was then fed to ARP/wARP (20) for model building, which yielded about a 65% structure with good side-chain fitting. The rest of the molecule was then traced into the electron density in COOT (21) and refined by iterative model building using the COOT graphics package combined with REFMAC (22). The final model was well refined with good quality electron density and crystallographic R and Rfree values of 20.8 and 24.9%, respectively. Electron density for about 40 amino acids at the C terminus was not found. The sulfate and water molecules were added to the molecule, guided by consideration of Fo − Fc density at a >3 σ contour level. Initially, the water molecules were traced by ARP/wARP solvent (20) and then were checked manually with electron density and hydrogen bonding interaction with the protein.
Structure Determination and Refinement of EhSAT1-Cys Complex
The native EhSAT1 structure was used as a model for solving the structure of EhSAT1-Cys complex by molecular replacement using MOLREP (19), which yielded a very good solution with a correlation coefficient of 91% and R factor of 38.8%. The model was refined with REFMAC (22) and further improved with iterative model building with COOT (21). Electron density improved after keeping cysteine at the active site. Further refinement and model building cycles yielded a well traced model with good refinement statistics (Table 1).
TABLE 1.
Table showing data solution and refinement statistics
Values in the parentheses are for last resolution shell. Rfree factor was calculated with a subset of 5% randomly selected reflections. ESU, estimated standard uncertainties.
Structure Determination and Refinement of EhSAT1-Ser Complex
The native EhSAT1 structure was used as model for solving the structure of EhSAT1-Ser complex by molecular replacement using MOLREP (19), which yielded a very good solution with a correlation coefficient of 97.4% and an R factor of 37.0%. The model was refined with REFMAC (22) and further improved with iterative model building with COOT (21). Electron density improved after keeping Ser at the active site, but the refinement statistics never improved better than an R factor of 27% and Rfree factor of 30%. However, after the inclusion of intensity-based twin refinement in Refmac5, the R and Rfree dropped dramatically to 16.0 and 18.17%, respectively, just after 10 cycles of refinement. It showed two twin operators, namely, -H-K, H, L and -H-K, K, -L with twin fraction of 0.562 and 0.438, respectively. Structure factors and refined coordinates have been deposited in the Protein Data Bank with accession numbers 3P1B for native, 3P47 for cysteine complex, and 3Q1X for serine complex structures.
Deletion of the C-terminal Region of EhSAT1
Two deletion constructs were prepared of the EhSAT1 molecule based on the crystallographic structure. One construct was 795 bp, and other was 807 bp in length corresponding to 36- and 40-amino acid deletions from the C-terminal end. These were also cloned in pET21c and expressed in BL21. These deletion constructs were not expressed very well; they were not stable and precipitated even at low concentrations.
Cloning of C-terminal Mutant of EhSAT1
The EhSAT1 was modified at the C-terminal end with last four amino acids mutated to DWSI instead of native sequence of SPSI. The mutated gene was cloned in pET28a (Novagen) with the N-terminal His tag using reverse primer 5′-CCGCTCGAGTCAAATCGACCAGTCATTTGC-3′ and same forward primer that was used for native EhSAT1. The plasmid containing the EhSAT1-DWSI construct was transformed into E. coli BL21 (DE3) cells, and the expression was checked. The protein was purified using the same method used for native EhSAT1 as described above. Purified protein was concentrated and used for inhibition studies with EhOASS.
SAT Assay
The activities of different constructs were assayed in a reaction mixture containing 0.1 mm acetyl-CoA and 1 mm DTNB in 50 mm Tris buffer, pH 8.0. Different concentrations of l-Ser from 1 to 10 mm were used. 2.5 μg of enzyme was used for each assay. The increase in absorbance due to formation of the 5-thio(2-nitrobenzoate) (TNB) (was measured at 412 nm using a visible light spectrophotometer.
Designing SAT C-terminal Mimicking Peptides to Study the Possibility of Cysteine Synthase Complex Formation
The Mycobacterium tuberculosis OASS structure with the C-terminal peptide of SAT (23) was taken as model to design better binding peptides to E. histolytica OASS. The native EhOASS structure was used to design good affinity binding peptides. Taking the lead from SAT C-terminal sequences, Ile was kept at the C-terminal end, and a four-amino acid peptide sequence library of 64 peptides were generated with different permutations and combinations of different amino acids at three positions. Docking studies were done with this peptide library against EhOASS structure. Five peptides were short-listed considering various energy parameters for inhibition studies and binding affinity studies with EhOASS (supplemental Table 1).
Inhibition Studies of EhOASS with SAT C-terminal Mimicking Peptides
The enzyme EhOASS was expressed and purified as described earlier (9). O-Acetyl l-serine, DTNB, DTT, and HEPES were purchased from Sigma. Five shortlisted peptides from docking studies and the tetrapeptide of C-terminal EhSAT1, SPSI, were purchased from GenPro Biotech. The sulfhydrylase activity was monitored using TNB as an alternative substrate. The disappearance of TNB was monitored continuously at 412 nm using a UV-visible spectrophotometer Ultrospec 21000pro (GE Healthcare). A typical assay contained the following in final concentrations: 100 mm HEPES, pH 7.0, 0.5 mm OAS, 0.05 mm TNB, and 25 μg of EhOASS. A decrease in enzyme activity was monitored over a fixed interval of time in the presence of 0.5 mm peptide. The pattern of the standard reaction was compared with the ones with the peptides and percentage decrease in activity calculated using the equation 100 − ((decrease in absorbance for reactions with peptide/decrease in absorbance for standard reaction) × 100).
Inhibition Studies of EhOASS with C-terminal Mutant EhSAT1 (DWSI-EhSAT1)
DWSI-EhSAT1 mutant protein in a buffer containing 150 mm NaCl, 5% (v/v) glycerol, and 50 mm HEPES, pH 7, was taken for inhibition studies with EhOASS. Because protein concentration was limiting compared with OAS concentration, inhibition was studied at lower concentrations of OAS (10 μm) and DWSI-EhSAT1 (4 μm) in a reaction mixture of 400 μl; EhOASS concentration was kept at 12.5 μg. The reaction was run in duplicate. A decrease in enzyme activity was monitored, and percentage inhibition was calculated as in case of the peptides.
Binding Affinity Estimation to EhOASS with C-terminal Mimicking Peptides
The binding affinity of various designed peptides to EhOASS was estimated using fluorescence emission of PLP following the methodology described earlier (24, 25) with minor modifications. The fluorophor PLP in the active site of OASS absorbs at 412 nm and emits at 510 nm. Titration of EhOASS with the peptides, which bind to the active site, leads to an increase in emission at 510 nm. Fluorescence measurements were carried out using a Cary400 Scan fluoro-spectrophotometer (Varian Inc.). Emission spectra upon excitation at 412 nm (slit excitation = 5 nm, slit emission = 10 nm) of a solution containing 250 μg/ml EhOASS, 100 mm HEPES, pH 7.0) was recorded. Fluorescence peak was measured in the absence of the peptide (F). Difference in fluorescence arising due to the addition of peptide, ΔF, was also measured and corrected for dilution. ΔF/F was plotted versus peptide concentration. The dependence on peptide concentration of fluorescence emission at 510 nm upon excitation at 412 nm was fitted to a binding isotherm for one binding site using the Sigmaplot software. The equation was I = Imax[L]/(Kdiss + [L]), where I = (change in fluorescence intensity at a given peptide concentration/original intensity without any peptide), Imax = maximum change in fluorescence intensity, [L] is the peptide concentration, and Kdiss is the dissociation constant of the complex.
RESULTS
Overall Structure of EhSAT1
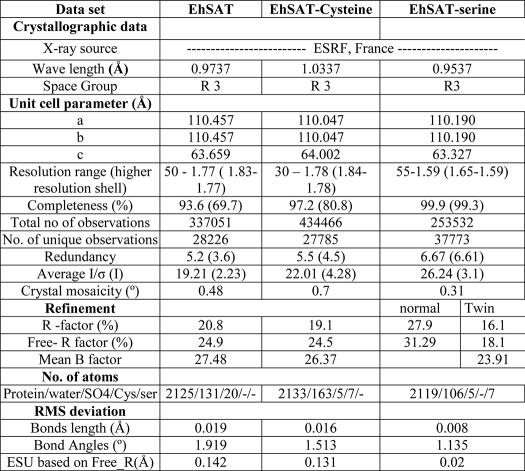
The final EhSAT1 structure consisting of residues 1–268 was refined to a 20.8% R factor and 24.9% Rfree factor. In the native EhSAT1 structure, 37 residues of the C-terminal end and 3 residues in the loop near the active site were not traced due to high disorder in this region. The final model consists of 269 residues, 169 waters, and 4 sulfate ions. Each asymmetric unit contains a single molecule of EhSAT1 in the R3 space group. Three molecules from three asymmetric units in the R3 space group form a SAT trimer (Fig. 1), which is consistent with the gel filtration profile. The crystallographic axis in the R3 space group and symmetry axis in the biological assembly of molecule are the same.
FIGURE 1.
The trimeric arrangement of EhSAT1. Three molecules of EhSAT1 (shown in different colors) interact with each other to form trimers displayed as a ribbon diagram with a side view, generated by PyMOL (32). A, shown is the cleft formed near the third β-helical coil between the interfaces of two molecules is the active site, where Ser is shown. B, shown is a top view of the trimers, with a three-fold axis perpendicular to the page. C, a hexameric structure E. coli SAT (4) is shown.
Structure of EhSAT1-Ser Complex
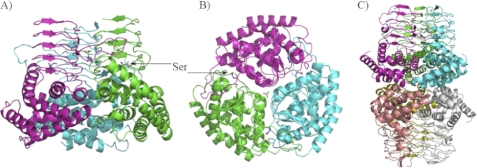
l-Ser, which is one of the substrates of EhSAT1, was co-crystallized with the enzyme to understand how the substrate binds to the enzyme active site. In the crystal structure Ser was well ordered in the active site (Fig. 2). The loop, near the active site third coil, which is disordered in the native structure, is ordered in the Ser complex structure. The Ser bound to the active site stabilizes the loop.
FIGURE 2.
A, EhSAT1 monomer shows bound ligand. The N-terminal region of the protein is arranged to form an α-helical-rich domain, and the C-terminal region is arranged to form a left-handed β-helical domain, characteristic of O-acetyltransferases. B, shown is a magnified view of the active site with bound Ser superposed with 2Fo− Fc electron density (green) at 1.2 sigma level in EhSAT1-Ser complex structure. (C) The magnified view of the active site with Cys superposed with 2Fo− Fc electron density (green color) at 1.4δ level in EhSAT1-Cys complex structure. In Fig. B and C, the bound amino acids were also superposed with 2Fo− Fc electron density map at 3δ level, shown in pink. D, interactions of l-Serine bound at the active site of the EhSAT1 molecule are shown. The complete active site is formed between two molecules of trimer; each molecule is shown in a different color. The carboxyl group of Ser makes a salt bridge with the side chain of Arg-222A and a hydrogen bond with side chain of His-208A and the water molecule. The hydroxyl group makes hydrogen bonds with the side chains of His-223A and His-180B. The amino group of serine makes a salt bridge with the carboxyl group of the Asp-114B and Asp-179B. A couple of water molecules also occupies the active site area, making hydrogen bonds with Asp-179B. E, interactions with the inhibitor l-cysteine bound at active site are shown. The position of the cysteine is identical to the serine in active site with minor changes. The carboxyl oxygen interacts with the amide group of the Arg-222A and also to His-223A and His-208A. The -SH group interacts with the His-180B, His-223A, and a water molecule. The -NH2 of cysteine bonds with the carboxyl groups of the Asp-114B and Asp-179B.
Each Ser makes several interactions with two adjacent chains in the active site. The carboxyl group of Ser makes very strong salt bridges with Arg-A222 and hydrogen-bonds with His-A208 and a water molecule. The hydroxyl group makes two hydrogen bonds with His-B180 and His-A223. And the amino group makes salt bridges with Asp-B114 and Asp-B179 (Fig. 2D). There are two more water molecules in the active site, interacting with Asp-B179.
Structure of the EhSAT-Cys Complex
The cysteine is seen to be bound to the active site in the EhSAT-Cys complex structure at exactly the same place as that of serine but with small reorientation of side chains and water molecules. The carbonyl oxygens of the bound cysteine form strong salt bridges with the amide groups of Arg-222A, His-223A, and His-208A (Fig. 2E). The sulfhydryl forms hydrogen bonds with His-223A, His-180B, and a water molecule. The amino group of cysteine forms hydrogen bonds with the carbonyl oxygen of Asp-114B and Asp-179B. The Asp-B179 is bound to only one water molecule compared with two waters in the Ser-bound structure. The differences compared with the Ser-bound structure may be due to the larger size of sulfur in Cys than oxygen of Ser and also due to the longer C-S bond. This large sulfur also displaces water atoms from the active site, which were present in the Ser-bound structure. The very similar binding of Cys and Ser in the active site clearly proves that Cys inhibits SAT by competing with Ser and not with acetyl-CoA.
Deletion Mutants of EhSAT1 and Activity
In several studies, deletion mutants from the C-terminal region were generated on EcSAT and checked for activity and cysteine feedback inhibition sensitivity. It was found that a deletion of 10 amino acids from the C-terminal region partially altered the inhibition sensitivity to cysteine (26), whereas a 20-residue deletion shows lowered susceptibility for cysteine inhibition (27). Also, the complex formation ability of SAT with OASS was lost when the C-terminal 10–25 amino acids were deleted (26).
The C-terminal deletion mutants in EhSAT1 were made because 1) about 40 residues from the C-terminal region in each of the three structures of EhSAT1 were disordered, 2) the C-terminal region does not participate in acetyl-CoA binding and its inhibition kinetics, and 3) it is known from other organisms that the C-terminal region of SAT interacts with OASS active site (10, 11, 25, 26), but in E. histolytica it is reported that EhSAT1 and EhOASS do not interact (14). Hence, it was expected that the C-terminal end may not play a role in the structure and function of EhSAT1, and this region may be dispensable. To test this, two deletion mutants were generated by deleting either 40 or 36 amino acids from the C-terminal end of the EhSAT1. The deletion mutants were not very stable, aggregated readily to make insoluble precipitate, and showed very low activity. This indicates that the C-terminal region, although not forming any kind of structure, nevertheless plays a role in enhancing the solubility and stability of the protein.
C-terminal Mimicking Peptides of SAT Show Inhibition and Binding to EhOASS
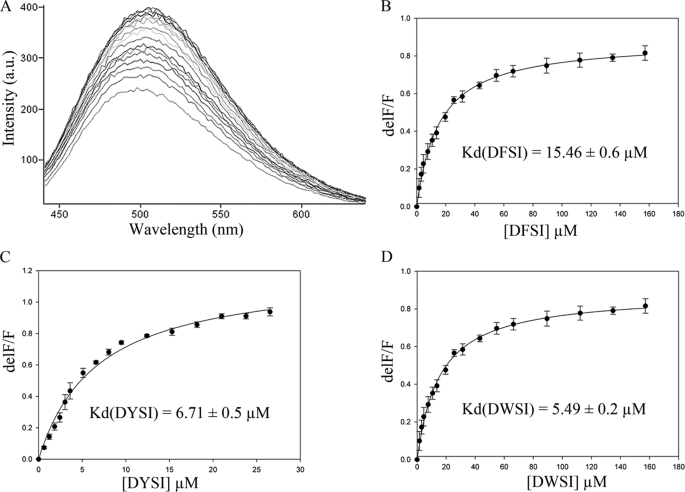
The EhSAT1 C-terminal was expected to bind OASS active site, as it has Ile at the C-terminal end, similar to SAT from E. coli, SAT from Arabidopsis thaliana, SAT from H. influenza, and SAT from M. tuberculosis. But was reported earlier that EhSAT1 and EhOASS do not interact with each other (13). To understand why EhSAT1 does not interact with EhOASS, the C terminus of the SAT mimicking peptide library was generated to study the binding affinity with EhOASS and inhibition activity. Different combinations of the tetra-amino acid peptide library were generated keeping Ile at the C-terminal end for docking studies. The four amino acid peptides were docked into the active site of EhOASS, keeping the M. tuberculosis OASS (MtbOASS) structure in complex with the tetra-peptide as a model (23). On the basis of energy parameters, five peptides were shortlisted (supplemental Table 1) and acquired for binding affinity estimation. These five peptides and the peptide SPSI derived from C-terminal EhSAT1 sequence were taken for inhibition studies and binding affinity estimation. Of six peptides, only three peptides (DFSI, DWSI, and DYSI) showed better inhibition of EhOASS activity, and other peptides including SPSI inhibited the EhOASS activity to a much lesser extent (Table 2). The binding affinity experiments with these peptides to EhOASS showed that the peptides which showed better inhibition, DFSI, DWSI, and DYSI, resulted in noticeable change in the fluorescence emission of PLP, indicating that these peptides bound strongly to the active site. The peptide SPSI, derived from EhSAT1 C-terminal sequence, inhibited about 30% activity of EhOASS, but it did not show any noticeable change in the fluorescence of PLP. The titration of DFSI, DWSI, and DYSI peptides and change in the fluorescence emission followed a typical Michaelis-Menten curve (Fig. 3). These peptides, DFSI, DWSI, and DYSI, showed micromolar binding affinity to the EhOASS.
TABLE 2.
Inhibition of EhOASS with peptides mimicking the SAT C-terminal sequences
| Peptide | % Inhibition |
|---|---|
| DPTI | 42.17 ± 5.7 |
| DTTI | 29.96 ± 4.0 |
| DFSI | 50.41 ± 0.7 |
| DYSI | 52.17 ± 0.9 |
| DWSI | 54.64 ± 2.8 |
| SPSI (derived from EhSAT1) | 30.19 ± 1.6 |
| DWSI-EhSAT1a | 62.4 ± 3 |
| EhSAT1a | 17.5 ± 3 |
a These reactions were performed at lower molar ratio of inhibitor compared to substrate, as mentioned in methods.
FIGURE 3.
Changes in fluorescence spectrum of EhOASS to study the binding affinity of peptides. A, shown is a typical fluorescence spectrum of EhOASS with titration of peptide. The fluorescence emission of PLP in the active site was monitored by changes in fluorescence at 508 nm versus DFSI concentration (B), DYSI concentration (C), and DWSI concentration (D). a.u., arbitrary units.
DWSI-EhSAT1 Shows Better Inhibition to EhOASS
Based on the results of docking, inhibition, and binding studies of SAT C-terminal mimicking peptides, the peptide DWSI was found to bind and inhibit EhOASS better. To test whether EhSAT1 with DWSI at its C-terminal end instead of natural sequence SPSI would inhibit EhOASS, we mutated the C-terminal end of EhSAT1 to DWSI (EhSAT1-DWSI). The activity of EhOASS was monitored in the presence of DWSI-EhSAT1, and it was found to decrease by about 62% (Table 2), indicating better inhibition of EhOASS compared with DWSI peptide, whereas the native EhSAT1 inhibited EhOASS activity only by 17%.
DISCUSSION
Sequence comparison of EhSAT1 with other SATs shows very high diversity in the N-terminal domain region (supplemental Fig. 2). The structure of N-terminal domain is helix-rich, with eight helices, which is similar to EcSAT and HiSAT. However, the overall orientation of the N-terminal domain is completely different in comparison with EcSAT/HiSAT (see details below). The C-terminal domain sequence and structure is well conserved with a typical left-handed five-coil β-helix domain. This domain also includes the active site, which is located between dimeric interfaces near the third coil (Fig. 1). The loop near the third coil is partly disordered, and three residues could not be built even after reducing the electron density to 0.5δ level. The active site in the native structure is occupied with a sulfate ion making interactions with Arg-222, His-223, His-208, and a water molecule.
Comparison with Other SAT Structures
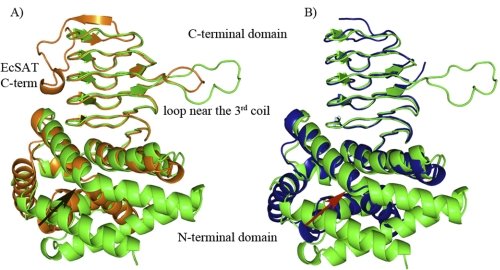
This is the first time any serine acetyltransferase was observed as a trimer in both solution as well as in the crystal structure. Both EcSAT and HiSAT were observed as hexamers in crystal structures (4, 5) as well as in electron microscopic images (28), where two trimers interact with each other in 2-fold axis symmetry. Otherwise, the overall architecture of each protomer of EhSAT1 is similar to each protomer of EcSAT (r.m.s.d. of 1.04 Å for 166 residues) and HiSAT (r.m.s.d. of 1.03 Å for 165 residues). All the protomers consist of helix-rich N-terminal domain and left-handed parallel β-helix containing C-terminal domain. The structure of the C-terminal domain is highly similar in the different isoforms; this region is also conserved and contains the active site. Active sites are located between two adjacent C-terminal domains of trimer, amounting to three active sites for each trimer. In contrast, about 100 residues of the N-terminal domain of EhSAT1 are quite different compared with EcSAT (Fig. 4A) and HiSAT (Fig. 4B). The major difference begins at about residue 97, between helix 4 and helix 5 (Fig. 4 and supplemental Fig. 2). In EcSAT and HiSAT, a small loop connects these helices, whose axes are oriented about 55° relative to one another. In EhSAT1, helix 4 and 5 are almost continuous, with just a kink near Pro-97 between them. The N-terminal region also differs due to the insertion of 7 residues (80–86) in EhSAT1 compared with EcSAT/HiSAT (supplemental Fig. 2). In EhSAT1, Cys-88 and Cys-145 are in close proximity (3.81 Å) to each other and can form a disulfide bridge. This kind of disulfide bridge is not possible in other SATs, as these residues are not conserved.
FIGURE 4.
Superposition of the EhSAT1 monomer on EcSAT (A) and HiSAT monomer (B). These images clearly reveal the difference in the N-terminal region (with r.m.s.d. of 11.22Å (A) and 6.420Å (B), respectively), which could be the major cause of loss of hexamerization in EhSAT1. The maximum structural deviation occurs between helix 4 and 5, which is indicated with an arrow.
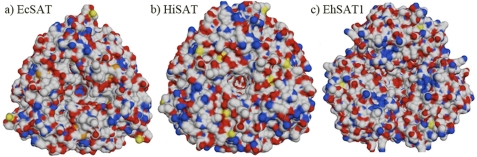
The N-terminal domain appears to determine the oligomeric state of SAT. The r.m.s.d. between N-terminal domain of EhSAT with EcSAT and HiSAT is 8.2 and 6.42 Å, respectively (Fig. 4). As noted above, the N-terminal region of EhSAT1 has very low sequence similarity with other SATs. The first 100 residues of the N-terminal domain have no sequence similarity with EcSAT and HiSATs. Moreover, there is an insertion of about eight residues in the middle of N-terminal domain of EhSAT1. Although the number of helices is the same in the N-terminal domain of all SAT structures, the orientation of first four helices is quite different. In contrast to HiSAT and especially EcSAT, where the N-terminal domains from the trimeric portion of the oligomer form a quite symmetrical triangle, the arrangement is relatively asymmetric in EhSAT1 (Fig. 5). Moreover, in contrast to the relatively hydrophobic surface of the trimer in EcSAT/HiSAT, which appears to drive a dimerization of trimers, the same surface in EhSAT1 is relatively charged. Taken together, these structural differences may be the reason that EhSAT1 does not form a hexamer.
FIGURE 5.
The charge distribution on the surfaces of EcSAT (a), HiSAT (b), and EhSAT1 (c) molecules from the N-terminal side view of trimers on space filled models. EcSAT and HiSAT have more hydrophobic surface compared with EhSAT1 and may be responsible for two trimers of EcSAT and HiSAT to interact and form hexamers.
Differences in Acetyl-CoA Binding Site
The loop near the third coil, which is part of the active site, has an eight-amino acid-long insertion unique to EhSAT1 as compared with EcSAT and HiSAT. This insertion could influence the activity of the enzyme and the interactions with the C-terminal region. In the HiSAT-Cys complex structure, Thr-181 of loop3, near the active site, forms a hydrogen bond with Gln-254 of the C-terminal region of the next molecule (supplemental Fig. 3). In this structure, the C-terminal region is well traced up to residue 257, and only 10 residues are disordered at the C-terminal end. In HiSAT-Co-A complex structure, Thr-181 interacts with Co-A and, thus, cannot interact with the C-terminal Gln-254; due to this, the C-terminal region was unstable and could not be traced in the structure (6). But in the case of EhSAT1, neither Thr nor Gln is conserved in the sequence, and other residues do not compensate for these interactions. This lack of interactions of C-terminal region may be responsible for it being highly disordered in all EhSAT1 structures. Thus, the acetyl-CoA binding site is open and accessible in all three structures of EhSAT1 including the EhSAT1-Cys complex structure.
Mutation of Met-256 in E. coli SAT to Ile makes the enzyme less susceptible to cysteine inhibition (27). Several amino acids around Met-256 are thought be involved in the conformational changes necessary for cysteine inhibition sensitivity (29, 30). Met at this position is not conserved in EhSAT1, and this region in the structures of EhSAT1 is not stable and could not be traced, but this enzyme was shown to be sensitive to cysteine by competitive inhibition (17). In EhSAT1, competitive inhibition by Cys might be simply due to just competing to the binding site and the binding energy differences between Cys and Ser.
In the case of HiSAT, it is clearly observed that in the Cys-bound structure, the C-terminal end of the protein interacts with the third β-coil and buries the acetyl-CoA binding site (6) (supplemental Fig. 3), thus, causing complex kinetics in which Cys appears to compete with acetyl-CoA (31) (Table 3). It may be noted that the inhibition constant, Ki(Cys) for EcSAT is quite low compared with EhSAT1, as the C-terminal region must also be involved in EcSAT inhibition covering the acetyl-CoA binding site, but it is not involved in EhSAT1. In the case of EcSAT/HiSAT, the inhibition is a combined effect of Cys competition at the active site and allosteric binding of the C-terminal region near the active site by burying the acetyl-CoA binding site. Note that the loop of the third coil and the C-terminal end of EhSAT1 are quite different compared with EcSAT and HiSAT. There are no interactions between these regions in EhSAT1, which were observed in the HiSAT structure. In the case of EhSAT1, the kinetics should be mere competitive inhibition of Cys to Ser and should not have any effect on acetyl-CoA kinetics, and this is clearly observed in kinetic studies reported by Hussain et al. (17).
TABLE 3.
Comparison of Cys inhibition kinetics
HiSAT kinetic constants could not be compared, as the experiments were performed at different pH values.
EhSAT1 C-terminal Mimicking Peptides Interact with EhOASS
It is known from earlier studies on E. coli, H. influenza, and M. tuberculosis enzymes that the C terminus of SAT interacts with the active site of OASS (10, 11), where Ile is present at the C-terminal end. Although the EhSAT1 has Ile at the C-terminal end, it has been reported that it will not interact with EhOASS (13, 14). To investigate further, we got peptide “SPSI” derived from the EhSAT1 C-terminal sequence and tested for binding and inhibition studies of EhOASS. The peptide SPSI showed about 30% competitive inhibition but did not show any noticeable change in PLP fluorescence, indicating that it does not bind strongly to EhOASS active site. To further understand the reason for the loss of interactions between these proteins, we generated a library of tetra-peptides mimicking the C-terminal sequence of SAT by keeping Ile at the end, and docking studies were performed to the active site of EhOASS.
Docking studies of SAT C-terminal mimicking peptides with EhOASS structure suggested that several peptides could have better binding energy than the EhSAT1-derived C-terminal peptide, SPSI. Five peptides that have best binding energy to EhOASS were obtained and studied for the inhibition activity and binding affinity against the EhOASS. Three peptides, DFSI, DWSI, and DYSI, show better inhibition and binding affinity against the EhOASS compared with SPSI. It may be noted that DFSI is the C-terminal sequence of M. tuberculosis SAT. The docking studies suggest that the Asp and aromatic amino acid makes several interactions with the EhOASS active site cavity. It appears that if the C-terminal end of EhSAT1 had any of these peptide sequences (DFSI/DWSI/DYSI), then the EhSAT1 would have interacted with EhOASS to form a multienzyme complex. But this multienzyme complex would not be a decamer as observed in other cysteine synthase complexes because the SAT in other organisms is a hexamer and EhSAT1 is a trimer.
Earlier, we proposed on the basis of EhOASS structure that the C-terminal-end helix of EhOASS may be responsible for loss of EhOASS and EhSAT1 interactions (9), as this helix was in a completely different orientation compared with other OASS structures, and also, it covered a groove near the active site. But binding studies with the C-terminal mimicking peptides of SAT to EhOASS clearly shows that the loss of interactions is mainly due to differences in the C-terminal residues of EhSAT.
C-terminal-modified EhSAT1 (DWSI-EhSAT1) Inhibits EhOASS Efficiently
It is known from earlier studies that EhSAT1 does not interact with EhOASS (13, 14). The inhibition and binding studies of EhOASS with EhSAT1-derived peptide SPSI also showed that SPSI does bind to the EhOASS active site, confirming earlier studies. But different SAT C-terminal mimicking peptides, especially DWSI, showed better inhibition of EhOASS and also high binding affinity. We wanted to check if the C terminus of EhSAT1 was mutated to DWSI, would this mutant protein bind to EhOASS? As expected, the EhOASS activity was inhibited by about 62% in the presence of DWSI-EhSAT1, suggesting that if a different C-terminal sequence was present in EhSAT1, it could have interacted with EhOASS, and E. histolytica could have a complex regulatory cysteine synthesis mechanism.
Implications on E. histolytica
E. histolytica is deficient of catalase and glutathione systems, which are generally involved in antioxidative mechanisms in many organisms. Cysteine was shown to be the major thiol involved in antioxidative defense of E. histolytica, and it also plays a vital role in the survival of this organism including adherence to matrix, elongation, motility, and growth (13). The EhSAT1 structure reveals that E. histolytica has evolved in a way that the complex regulatory mechanisms are turned off with a simple modification of the N-terminal and C-terminal regions and an insertion in the loop near the active site that still maintained its enzyme activity. The variations in the C-terminal region and insertion in the third coil loop near the active site leads to an exposed acetyl-CoA binding site, as the C-terminal region in EhSAT1 does not cover this region. Thus, Cys binding does not inhibit the acetyl-CoA binding and competes only with Ser for the active site. Differences in the N-terminal region of EhSAT1 compared with that of other SATs lead to trimer formation and loss of hexamer forming ability. This trimer in turn cannot form a large decameric cysteine synthase complex (hexamer of SAT and two dimers of OASS), thus, leading to the loss of OAS and sulfide regulation on this pathway. The pathway is simply regulated by feedback inhibition of Cys, thus, making sure that the organism has enough Cys to protect from oxidative stress.
Conclusion
EhSAT1 exists as a trimer both in solution and in crystal structure. The differences in the N-terminal domain and its charge distribution may be responsible for not forming hexamers as seen in EcSAT and HiSAT.
The C-terminal end and third β-coil loop, which are quite different from EcSAT and HiSAT, do not interact with each other in the EhSAT1-Cys complex structure, making the acetyl-CoA binding site solvent-accessible. These differences are responsible for loss of inhibition kinetics to acetyl-CoA by Cys. Ser and Cys bind to almost the same location in the active site, with minor differences, clearly indicating that Cys inhibits SAT by competitive inhibition with Ser. In EcSAT and HiSAT, the Cys inhibits both acetyl-CoA binding as well as serine binding, whereas Cys just competitively inhibits Ser binding to EhSAT1, which is clearly explained by the EhSAT-Cys complex structure.
Even though Ile is there at the C-terminal end, EhSAT1 cannot bind strongly with the EhOASS active site. If the C-terminal of EhSAT1 had ended with DXSI (X is any aromatic amino acid), instead of SPSI, EhSAT1 could have interacted with EhOASS strongly and could have formed a multienzyme complex. But this multienzyme complex would have been a pentamer rather than a decamer, as observed in other organisms. This loss of a complex regulatory mechanism (explained here by structural features that abolish hexamer formation, by Cys not inhibiting acetyl-CoA binding, and by cysteine synthase complex formation) ensure that there is enough Cys available in the E. histolytica for its survival.
Acknowledgments
We thank Dr. Garib Murshudov, York University, for helpful suggestions on twinning refinement at CCP4 workshop, Delhi, the European Synchrotron Radiation Facility, beam line 14 staff, the Dept. of Biotechnology, Government of India for access to the beam line, and Dr. Jerry Brown, Brandeis University, for critical reading and editing of the manuscript. S. K. thanks the University Grants Commission. We also thank Prof. Alok Bhattacharya, Jawaharlal Nehru University for support and genomic DNA.
This work was supported by the Council for Scientific and Industrial Research, Government of India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
The atomic coordinates and structure factors (codes 3P1B, 3P47, and 3Q1X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SAT
- serine acetyltransferase
- OASS
- O-acetyl serine (OAS) sulfhydrylase
- DTNB
- 5,5′-dithio-bis(2-nitrobenzoic acid)
- PLP
- pyridoxal-5′-phosphate
- TNB
- 5-thio(2-nitrobenzoate)
- Ec-
- E. coli
- Hi-
- H. influenzae
- Eh
- E. histolytica
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Smith I. K., Thompson J. F. (1971) Biochim. Biophys. Acta 227, 288–295 [DOI] [PubMed] [Google Scholar]
- 2. Saito K., Yokoyama H., Noji M., Murakoshi I. (1995) J. Biol. Chem. 270, 16321–16326 [DOI] [PubMed] [Google Scholar]
- 3. Kredich N. M., Becker M. A., Tomkins G. M. (1969) J. Biol. Chem. 244, 2428–2439 [PubMed] [Google Scholar]
- 4. Pye V. E., Tingey A. P., Robson R. L., Moody P. C. (2004) J. Biol. Chem. 279, 40729–40736 [DOI] [PubMed] [Google Scholar]
- 5. Gorman J., Shapiro L. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1600–1605 [DOI] [PubMed] [Google Scholar]
- 6. Olsen L. R., Huang B., Vetting M. W., Roderick S. L. (2004) Biochemistry 43, 6013–6019 [DOI] [PubMed] [Google Scholar]
- 7. Krishna C., Jain R., Kashav T., Wadhwa D., Alam N., Gourinath S. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Becker M. A., Kredich N. M., Tomkins G. M. (1969) J. Biol. Chem. 244, 2418–2427 [PubMed] [Google Scholar]
- 9. Chinthalapudi K., Kumar M., Kumar S., Jain S., Alam N., Gourinath S. (2008) Proteins 72, 1222–1232 [DOI] [PubMed] [Google Scholar]
- 10. Feldman-Salit A., Wirtz M., Hell R., Wade R. C. (2009) J. Mol. Biol. 386, 37–59 [DOI] [PubMed] [Google Scholar]
- 11. Zhao C., Moriga Y., Feng B., Kumada Y., Imanaka H., Imamura K., Nakanishi K. (2006) Biochem. Biophys. Res. Commun. 341, 911–916 [DOI] [PubMed] [Google Scholar]
- 12. WHO/PAHO/UNESCO (1997) Epidemiol. Bull. 18, 13–14 [Google Scholar]
- 13. Nozaki T., Asai T., Kobayashi S., Ikegami F., Noji M., Saito K., Takeuchi T. (1998) Mol. Biochem. Parasitol. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 14. Nozaki T., Asai T., Sanchez L. B., Kobayashi S., Nakazawa M., Takeuchi T. (1999) J. Biol. Chem. 274, 32445–32452 [DOI] [PubMed] [Google Scholar]
- 15. Nozaki T., Ali V., Tokoro M. (2005) Adv. Parasitol. 60, 1–99 [DOI] [PubMed] [Google Scholar]
- 16. Agarwal S. M., Jain R., Bhattacharya A., Azam A. (2008) Int. J. Parasitol. 38, 137–141 [DOI] [PubMed] [Google Scholar]
- 17. Hussain S., Ali V., Jeelani G., Nozaki T. (2009) Mol. Biochem. Parasitol. 163, 39–47 [DOI] [PubMed] [Google Scholar]
- 18. Otwinoski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 20. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Schnell R., Oehlmann W., Singh M., Schneider G. (2007) J. Biol. Chem. 282, 23473–23481 [DOI] [PubMed] [Google Scholar]
- 24. Cook P. F., Wedding R. T. (1976) J. Biol. Chem. 251, 2023–2029 [PubMed] [Google Scholar]
- 25. Campanini B., Speroni F., Salsi E., Cook P. F., Roderick S. L., Huang B., Bettati S., Mozzarelli A. (2005) Protein Sci. 14, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mino K., Hiraoka K., Imamura K., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (2000) Biosci. Biotechnol. Biochem. 64, 1874–1880 [DOI] [PubMed] [Google Scholar]
- 27. Denk D., Böck A. (1987) J. Gen. Microbiol. 133, 515–525 [DOI] [PubMed] [Google Scholar]
- 28. Hindson V. J., Moody P. C., Rowe A. J., Shaw W. V. (2000) J. Biol. Chem. 275, 461–466 [DOI] [PubMed] [Google Scholar]
- 29. Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (1999) Biosci. Biotechnol. Biochem. 63, 168–179 [DOI] [PubMed] [Google Scholar]
- 30. Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (2000) Biosci. Biotechnol. Biochem. 64, 1628–1640 [DOI] [PubMed] [Google Scholar]
- 31. Kredich N. M., Tomkins G. M. (1966) J. Biol. Chem. 241, 4955–4965 [PubMed] [Google Scholar]
- 32. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 33. Hindson V. J. (2003) Biochem. J. 375, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson C. M., Huang B., Roderick S. L., Cook P. F. (2004) Arch. Biochem. Biophys. 429, 115–122 [DOI] [PubMed] [Google Scholar]