Abstract
Almost all viral pathogens utilize a cytoskeleton for their entry and intracellular transport. In HIV-1 infection, binding of the virus to blood resting CD4 T cells initiates a temporal course of cortical actin polymerization and depolymerization, a process mimicking the chemotactic response initiated from chemokine receptors. The actin depolymerization has been suggested to promote viral intracellular migration through cofilin-mediated actin treadmilling. However, the role of the virus-mediated actin polymerization in HIV infection is unknown, and the signaling molecules involved remain unidentified. Here we describe a pathogenic mechanism for triggering early actin polymerization through HIV-1 envelope-mediated transient activation of the LIM domain kinase (LIMK), a protein that phosphorylates cofilin. We demonstrate that HIV-mediated LIMK activation is through gp120-triggered transient activation of the Rack-PAK-LIMK pathway, and that knockdown of LIMK through siRNA decreases filamentous actin, increases CXCR4 trafficking, and diminishes viral DNA synthesis. These results suggest that HIV-mediated early actin polymerization may directly regulate the CXCR4 receptor during viral entry and is involved in viral DNA synthesis. Furthermore, we also demonstrate that in resting CD4 T cells, actin polymerization can be triggered through transient treatment with a pharmacological agent, okadaic acid, that activates LIMK and promotes HIV latent infection of resting CD4 T cells. Taken together, our results suggest that HIV hijacks LIMK to control the cortical actin dynamics for the initiation of viral infection of CD4 T cells.
Keywords: Actin, Cell Surface Receptor, Chemotaxis, Cytoskeleton, HIV, CD4, CXCR4, Cofilin, LIMK1, gp120
Introduction
Infection by the human immunodeficiency virus (HIV) causes severe depletion of blood CD4 T cells (1, 2). The early interaction between HIV and T cells, particularly virus binding to its receptors, plays an important role in viral infection and pathogenesis. This interaction mediates viral fusion and entry (3, 4). It also initiates intracellular signaling cascades that are important for the early steps of the HIV life cycle (5–9). For example, it has recently been shown that at the earliest time of HIV infection, viral binding to the chemokine coreceptor, CXCR4, activates an actin depolymerization factor cofilin to increase the cortical actin dynamics in resting T cells, facilitating viral intracellular migration (9).
The cortical actin is a common structure that is targeted by most viruses for entry and intracellular transport (10, 11). In HIV-1 infection, the direct involvement of the cortical actin in early stages of viral infection has been suggested in HIV-mediated CD4-CXCR4 receptor clustering (5–7, 12–14), subsequent viral DNA synthesis (9, 15), and intracellular migration (9). It has been shown that the initial binding of gp120 to surface CD4 promotes localized aggregation of the CD4 and CXCR4 receptor, which appears to be dependent on the actin-cross-linking protein filamin (7) and the ezrin-radixin-moesin protein moesin (5, 6). Filamin-A interacts directly with the cortical actin and with both CD4 and CXCR4 and is activated by early signaling through these receptors (6). Similarly, moesin is activated early through CD4 signaling and may serve to anchor the cortical actin to the plasma membrane (5, 6). Following viral fusion and entry, the cortical actin also presents itself as an immediate barrier for viral intracellular migration, which requires gp120-CXCR4 signaling to activate an actin depolymerizing factor cofilin to overcome this restriction (9). Current models suggest that the concerted action of filamin, moesin, and cofilin in regulating the cortical actin dynamics plays a critical role in viral entry and early postentry processes (11, 16–18), although the molecular details of the spatiotemporal regulation are poorly delineated.
At the earliest time of HIV-1 infection, binding of the virus to blood resting CD4 T cells initiates a rapid, transient cortical actin polymerization followed by actin depolymerization (9, 19), a process mimicking the chemotactic response initiated by SDF-1 binding to its natural receptor CXCR4 (9, 19, 20). Nevertheless, the direct role of this early actin polymerization in HIV infection is unknown, and the signaling molecules involved remain unidentified. In this article, we demonstrate that the initial actin polymerization is triggered by HIV-1 gp120-mediated transient activation of the LIM domain kinase 1 (LIMK1),3 a kinase that phosphorylates and inactivates cofilin. We also report that LIMK1-mediated actin polymerization directly regulates CXCR4 receptor internalization that may facilitate early CD4-CXCR4 receptor clustering and viral entry. In addition, LIMK1-mediated actin polymerization is also critical for postentry viral DNA synthesis. These results demonstrated an important role of LIMK1 and early actin polymerization in the initiation of HIV-1 infection of CD4 T cells.
EXPERIMENTAL PROCEDURES
Preparation of Resting CD4 T Cells and Macrophages from Peripheral Blood
Resting CD4 T cells were purified from peripheral blood by two rounds of negative selection as described previously (21). Briefly, for the first round depletion, we used monoclonal antibodies against human CD14, CD56, HLA-DR, HLA-DP, and HLA-DQ (BD Biosciences). For the second-round depletion, we used monoclonal antibodies against human CD8, CD11b, and CD19 (BD Biosciences). Antibody-bound cells were depleted by using Dynabeads Pan Mouse IgG (Invitrogen). Cells were rested overnight before infection or treatment. In some occasions, CD4 T cells were prestimulated with anti-CD3/CD28 beads (2 beads per cell) for 1 day or cultured in IL-7 (Pepro Tech) (10 ng/ml) for 4 days before HIV infection. Blood monocyte-derived macrophages were prepared as described previously (22).
Virus Preparation and Infection of T Cells
Virus stocks of HIV-1NL4–3, NL(AD8), and YU2 were prepared by transfection of HeLa cells with cloned proviral DNA as described (21). Single-cycle replicating viruses, HIV-1(VSV-G) and HIV-1(Env), were prepared as described previously (23). Levels of p24 in viral supernatant were measured using the PerkinElmer Alliance p24 antigen ELISA kit (PerkinElmer Life Sciences). Viral titer (TCID50) was determined on the Rev-dependent GFP indicator cell, Rev-CEM (24, 25). For infection of resting CD4 T cells, unless specified, for most replication assays, 103.5 to 104.5 TCID50 units of HIV-1 were used to infect 106 cells.
Measurement of CXCR4 Receptor Internalization and Recycling
A half million cells were incubated with 1 μg of FITC-labeled anti-CXCR4 antibody (R&D Systems) in 0.5 ml of PBS + 0.1% BSA at 37 °C for 20 to 90 min, washed with cold PBS + 0.1% BSA and then resuspended in cold PBS + 0.1% BSA for flow cytometry analysis. To measure antibody-induced receptor internalization, 1 million cells were incubated with 1 μg of biotin-labeled anti-CXCR4 antibody (R&D Systems) at 4 °C for 2 h, washed with cold PBS + 0.1% BSA, and then incubated at 37 °C for 5 to 60 min. Cells were washed with acidic medium (PRMI 1640 (pH 4.0)) for 3 min twice, followed by washing with cold PBS + 0.1% BSA, and subsequently lysed in 100 μl SDS lysis buffer for Western blotting to detect intracellular CXCR4 with peroxidase-labeled streptavidin (KPL).
Chemotaxis Assay
A half million cells were resuspended into 100 μl RPMI 1640 medium and then added to the upper chamber of a 24-well transwell plate (Corning). The lower chamber was filled with 600 μl of medium premixed with SDF-1. The plate was incubated at 37 °C for 2 h, and then the upper chamber was removed and cells in the lower chamber were counted.
Viral Entry Assays
Viral entry assays were performed as described previously (26). We also used a Nef-luciferase-based entry assay. Viruses containing Nef-luciferase were produced as described previously (27). For entry assays, cells (1 × 106) were infected with 200 ng of Nef-luciferase containing viruses at 37 °C for 2 h, and then washed three times with medium. Cells were resuspended in 0.1 ml of luciferase assay buffer (Promega), and luciferase activity was measured in live cells using a GloMax-Multi detection system (Promega).
FITC-Phalloidin Staining of F-actin and Flow Cytometry
F-actin staining using FITC-labeled phalloidin (Sigma) was carried out as described previously (9). Cells were resuspended in 1% paraformaldehyde and analyzed on a FACSCalibur (BD Biosciences).
Quantitative PCR
Viral DNA and 2-LTR circles in shRNA lentiviral vector transduced cells were carried out as described previously (9) using the following primers and probes: HIV-Env-5F, HIV-Env-3R, and Env-Probe for HIV-1 DNA; and MH536, MH535, and MH603 for viral 2-LTR circles.
Western Blotting to Detect LIMK and PAK activation
Blots were incubated with rabbit polyclonal antibodies specific for either phospho-LIMK1/2, phospho-PAK1/2, phospho-PAK1, phospho-PAK2, or Phospho-PAK4 (1:1000 dilution) (Cell Signaling Technology, Inc.) for 1 h or overnight at 4 °C. The blots were washed and then incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies (KPL) (1:5000) for 1 h and then developed with SuperSignal West Femto maximum sensitivity substrate (Pierce). The same blots were also stripped and reprobed with antibodies against LIMK1, PAK1, PAK2, PAK4 (Cell Signaling Technology, Inc.), or GAPDH (Abcam). On some occasions, Western blotting was also performed using infrared imaging (Odyssey infrared imager, Li-cor Biosciences) with IRDye goat anti-rabbit 680 or 800cw antibodies (Li-cor Biosciences).
Assays to Measure Rac1, cdc42, and Rho Activation
GST pull-down assays for active RhoA, Rac1, and Cdc42 were performed as recommended by the manufacturer (Pierce). Briefly, cells were lysed in lysis/binding/wash buffer, and then the supernatants were transferred to a glutathione column with either GST-human Pak1-pzi-binding domain (for Rac1 and Cdc42) or GST-human Rhotekin (for RhoA) attached. As a positive control, a portion of the cell lysate was incubated with GTPγS. As a negative control, a portion of the cell lysate was also incubated with GDP. The columns were washed, eluted with SDS sample buffer, and then immunoblotted with either anti-Rac1, anti-Cdc42, or anti-RhoA mouse monoclonal antibodies (Pierce).
SiRNA and shRNA Knockdown of LIMK1 in Human CD4 T Cells
Plasmids (pLKO.1-puro) carrying LIMK1 shRNAs and a non-targeting shRNA (NTC) were purchased from Sigma. Viral particles were assembled as suggested by the manufacturer and used to transduce primary human CD4 T cells that had been prestimulated with anti-CD3/CD28 beads (2 beads per cell) for 12 h. After 12–24 h, the beads were removed and cells were selected and grown in fresh medium containing 1 μg/ml puromycin. CEM-SS cells were directly transduced with the shRNA lentiviral particles and selected and grown in puromycin-containing medium to maintain the knockdown phenotype.
Additional experimental procedures can be found under supplemental experimental procedures.
RESULTS
HIV-1 Mediates Early Actin Polymerization and LIMK Activation
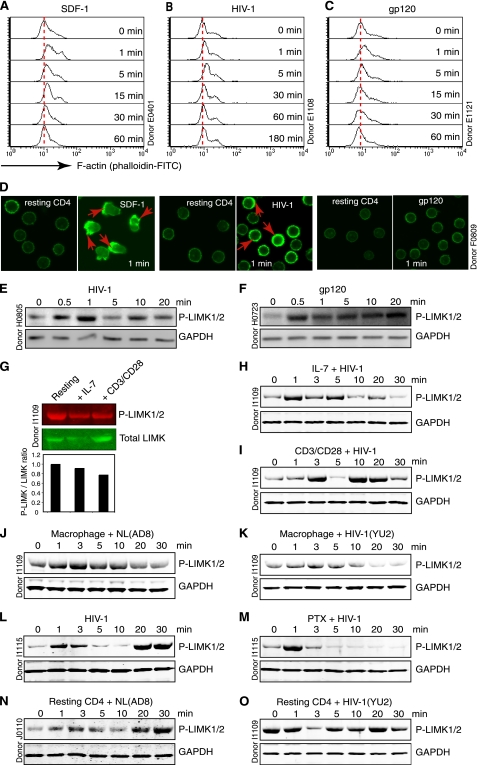
Binding of HIV-1 to resting T cells triggers a rapid, transient actin polymerization, a process resembling the SDF-1-mediated chemotactic response (Fig. 1, A–D and supplemental Fig. 1A). In SDF-1-treated human Jurkat and peripheral blood T cells, this rapid actin polymerization is associated with a concomitant, transient LIMK1 activation within 1 min (20). LIMK1 is the kinase responsible for the serine 3 phosphorylation of cofilin (28, 29), an actin-binding protein directly depolymerizing and severing actin filaments (30, 31). Overexpression of LIMK1 has been shown to promote accumulation of F-actin, whereas expression of a dominant negative form of LIMK1 resulted in a loss of F-actin (28, 29), suggesting that LIMK1 is a direct modulator of actin polymerization. To investigate whether HIV-1 infection of human resting CD4 T cells resulted in a similar LIMK activation, we chose to monitor the phosphorylation of LIMK. Blood resting CD4 T cells were purified by two rounds of negative depletion, rested overnight, and then infected with HIV-1 and immunoblotted for phospho-LIMK with an antibody specific for phosphorylation of LIMK1 at Thr-508 and LIMK2 at Thr-505. These two isoforms of LIMKs are genetic duplicates and are structurally and functionally similar, with LIMK1 expressed mostly in neurons and cells of the hematopoietic lineage (32, 33). We found that LIMK was transiently activated (Fig. 1E and supplemental Fig. 1B) and that gp120 alone was sufficient to trigger this activation (Fig. 1F).
FIGURE 1.
HIV-mediated early actin polymerization and LIMK activation. A–D, actin polymerization triggered by stimulation with SDF-1 (50 ng/ml) (A), HIV-1NL4–3 (800 ng p24) (B), or gp120 (10 nm) (C). F-actin was stained with FITC-phalloidin and analyzed with flow cytometry or confocal microscopy (D). E and F, resting CD4 T cells treated with HIV-1 (100 ng) (E) or gp120 (50 nm) (F) and then analyzed by Western blotting for LIMK activation using an anti-phospho-LIMK antibody. The same blots were also probed with an anti-human GAPDH antibody for loading control. G–I, resting CD4 T cells were stimulated with anti-CD3/CD28 beads or culture in IL-7 for 4 days. LIMK activation was measured by Western blotting using an anti-phospho-LIMK or anti-LIMK antibody followed by IRDye goat anti-rabbit 680 or 800cw antibodies for infrared imaging. Density of bands was scored as the ratio of phospho-LIMK to total LIMK relative to untreated resting T cells (G). The pretreated cells were also treated with HIV-1 (100 ng) for measuring LIMK activation (H and I). J and K, human monocyte-derived macrophages were treated with HIV-1, NL(AD8) (200 ng) or YU2 (100 ng) and LIMK activation was measured. L and M, resting CD4 T cells were either untreated (L) or pretreated with pertussis toxin (PTX) (100 ng/ml) for 1 h (M) and then treated with HIV-1 (100 ng) to measure LIMK activation. N and O, resting CD4 T cells were treated with HIV-1, NL(AD8) (200 ng), or YU2 (100 ng) to measure LIMK activation.
Given that productive viral replication occurs in active T cells and in cells exposed to cytokines (34), we examined LIMK activation in these cells following HIV infection. Blood CD4 T cells were either preactivated with CD3/CD28 stimulation or cultured in IL-7 for 4 days. Both treatments did not increase LIMK phosphorylation (Fig. 1G). However, when infected with HIV, these cells demonstrated a quick LIMK activation (1–3 min), followed by LIMK dephosphorylation and then reactivation at later times (Fig. 1, H and I). Similar LIMK activation was also observed in the infection of human primary macrophages with the CCR5-utilizing (R5) HIV-1, NL(AD8), and YU2 (Fig. 1, J and K). These results suggest that activation of LIMK is rather a general mechanism occurring early in HIV infection of both primary resting/active CD4 T cells and macrophages.
We also examined whether LIMK activation is mediated through CXCR4 and the pertussis toxin-sensitive Gαi signaling because cofilin has been shown to be activated through CXCR4 and Gαi (9). Surprisingly, although pertussis toxin completely shut down late LIMK activation (at 20–30 min), it did not inhibit the early activation of LIMK (at 1–3 min) (Fig. 1, L and M), suggesting that the signals may be transduced from Gαi as well as other Gα subunits such as Gαq (18). Alternatively, HIV may use both CD4 and CXCR4 to activate LIMK. It has been shown that HIV-mediated CD4 signaling can lead to the phosphorylation of cofilin (7), suggesting possible involvement of CD4 in LIMK activation. Because a majority of resting CD4 T cells express high levels of CD4 and CXCR4 but undetectable levels of CCR5, we treated resting CD4 T cells with the R5 virus, NL(AD8), and YU2 to determine LIMK activation in the presence of CD4 and absence of CCR5. As shown in Fig. 1, N and O, we observed modulation of LIMK activities by both NL(AD8) and YU2, demonstrating that in addition to CXCR4, CD4 may also be used by the virus to trigger LIMK activation. This receptor redundancy in triggering LIMK activity suggest that LIMK activation is likely a conserved pathway critical for the initiation of viral infection.
HIV-1-mediated LIMK Activation Is through the Rack-PAK-LIMK Pathway
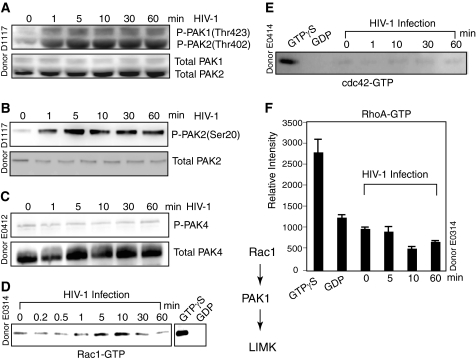
The Rho family of GTPases such as Rho, Rac, and Cdc42 are well known regulators of actin dynamics, and LIMK is normally activated through phosphorylation by ROCK and PAK, which are downstream effectors of Rho and Rac/Cdc42, respectively. We next examined the PAK family proteins because both PAK1 and PAK4 have been shown to phosphorylate and regulate the activity of LIMK (35, 36). Resting CD4 T cells were treated with HIV-1 and immunoblotted with an antibody to the phosphorylated forms of both PAK1 and PAK2 at Thr-423 and Thr-402, respectively. We detected phosphorylation of both PAK1 and PAK2 following HIV-1 treatment of resting T cells (Fig. 2A). PAK2 activation is less studied and is not known to be directly involved in LIMK phosphorylation. Thus, we repeated the Western blotting using a second antibody to detect PAK2 phosphorylation at a separate site, serine 20, and confirmed that PAK2 was indeed phosphorylated in response to HIV-1 infection (Fig. 2B) or gp120 treatment (data not shown). Because both PAK1 and PAK2 share a number of substrates and can be activated by the small GTPase Rac and Cdc42, we could not exclude the possibility that PAK2 may also contribute to the phosphorylation of LIMK in HIV-1-infected resting CD4 T cells (37). Active PAK4 also interacts specifically with LIMK1 and stimulates LIMK1 activation (36), but we did not detect PAK4 phosphorylation in resting CD4 T cells at any time point following HIV infection (Fig. 2C).
FIGURE 2.
HIV-1-mediated activation of the Rack-PAK-LIMK pathway. A–C, resting CD4 T cells were stimulated with HIV-1 and analyzed by Western blotting for the activation of PAKs using an anti-phospho-PAK1/2 antibody (A), an anti-phospho-PAK2 antibody (B), and an anti-phospho-PAK4 antibody (C). The same blots were stripped and reprobed with antibodies against total PAK1/2, PAK2, and PAK4. Activation of Rac1 (D), cdc42 (E), or RhoA (F) was also analyzed by pulling down cell lysates through GST-PAK1 (D and E) or GST-Rhotekin (F) columns and immunoblotting for active Rac1, cdc42, or RhoA. As controls, uninfected cell lysates were preincubated with GTPγS or GDP and similarly pulled down and immunoblotted.
We further examined upstream of LIMK and PAK activation by investigating whether HIV-1 infection leads to the Rho family GTPase activation. When Rac1 and Cdc42 are in their activated GTP-bound state, they can bind to PAK1, disrupting an intramolecular interaction between the regulatory and catalytic domains of PAK1 and leading to PAK1 activation. Resting CD4 T cells were infected with HIV-1 for a time course. Cell lysates were harvested and used to pull down active Rho GTPases through a GST-PAK1 column. The samples were then eluted and immunoblotted for active Rac1 or Cdc42. As shown in Fig. 2D, we observed HIV-1-induced transient Rac-1 activation that begins at 1 min, peaks at 10 min, and then returns to the level found in uninfected resting CD4 T cells. Interestingly, HIV-1 infection of resting CD4 T cells resulted in no detectable activation of Cdc42 (Fig. 2E). We also examined the activation of RhoA using GST-human Rhotekin to pull down active RhoA but detected no RhoA activation in resting T cells (Fig. 2F). Taken together, our data indicate that the Rac-PAK-LIMK pathway is briefly activated by HIV-1 at an early time in the infection of resting CD4 T cells. Our results are consistent with previous findings that Rac1, but not Cdc42 nor Rho, mediates SDF-1-induced LIMK1 activation in human T cells (20). Rac1 is also required for HIV envelope-mediated syncytium formation, indicating that Rac1 may be activated by gp120 to trigger actin polymerization to mediate cell-cell (13) and virus-cell fusion (8).
Okadaic Acid Mediates Transient LIMK Activation, Promoting HIV Infection
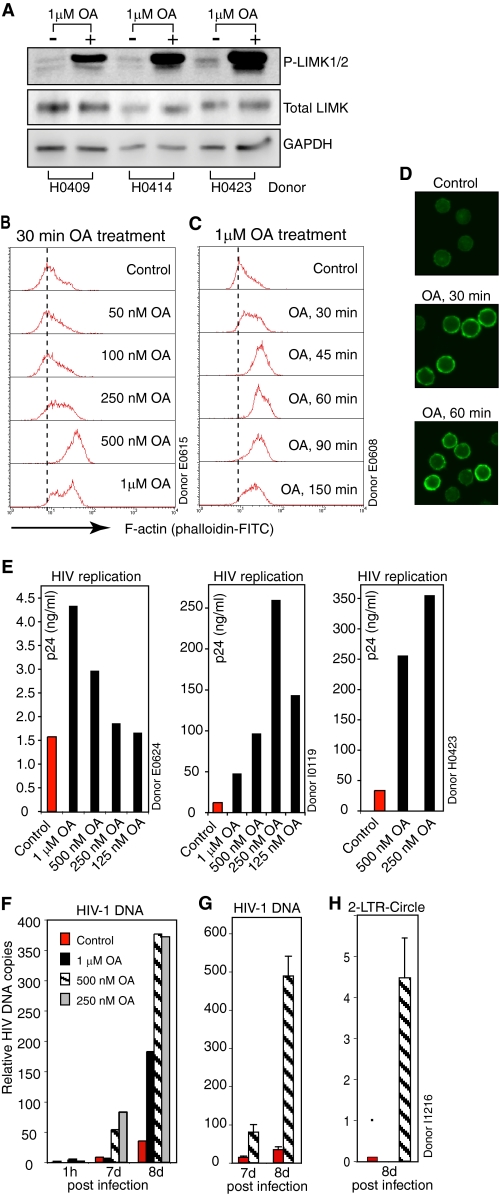
To confirm the role of LIMK in HIV-1 infection of CD4 T cells, we sought to use pharmacological agents to modulate LIMK activity. Currently, there is no specific LIMK inhibitor or activator identified. During a drug screening, we discovered that okadaic acid (OA), an inhibitor of PP1 and PP2A, triggered dramatic LIMK activation in resting CD4 T cells (Fig. 3A). This result suggested that LIMK is possibly dephosphorylated by PP1 or PP2A phosphatases, whereas a protein phosphatase 3 (formerly PP2B) inhibitor, FK-506, did not result in LIMK activation (data not shown). This OA-mediated LIMK activation is concomitant with a drastic polymerization of the cortical actin filaments (Fig. 3, B–D, and supplemental Fig. 2, A and B), consistent with the role of LIMK in regulating actin polymerization. This LIMK activation is also correlated with an OA-mediated enhancement of HIV-1 latent infection of resting T cells (Fig. 3E and supplemental Fig. 2, C and D). The enhancement was further confirmed by real-time PCR quantification of viral DNA and 2-LTR circles in OA-treated resting T cells (Fig. 3, F–H), indicating that transient LIMK activation and actin polymerization positively affect HIV-1 infection.
FIGURE 3.
Okadaic acid mediates LIMK activation, promoting actin polymerization and HIV-1 latent infection of resting CD4 T cells. A, resting T cells from three donors were treated with okadaic acid (OA) for 30 min and then analyzed for LIMK activation by Western blotting using an anti-phospho-LIMK antibody. The same blot was stripped and reprobed with antibodies against total LIMK or human GAPDH for loading controls. B–D, OA treatment of resting CD4 T cells triggers dosage- and time-dependent cortical actin polymerization. Cells were treated with OA at different dosages (B) or for different time periods (C) and then stained with FITC-phalloidin for analysis of F-actin by flow cytometry (B and C) or confocal microscopy (D). E, OA-mediated enhancement of HIV-1 latent infection of T cells. Resting CD4 T cells were briefly treated with OA for 10 min, infected with HIV-1 for 50 min, washed, incubated for 5 days in the absence of OA, and then activated with anti-CD3/CD28 magnetic beads. Shown is viral replication at day 7 (donor E0624) or 8 (donors I0119 and H0423) postinfection. F–H, resting CD4 T cells from another donor were similarly OA-treated and infected as in E. Syntheses of total HIV DNA (F and G) and 2-LTR circles (H) following infection were also measured with real-time PCR as described under supplemental experimental procedures. G, shown is the PCR triplicate of viral DNA in cells untreated or treated with 500 nm of OA.
LIMK1 regulates cortical actin polymerization and CXCR4 receptor cycling in CD4 T cells
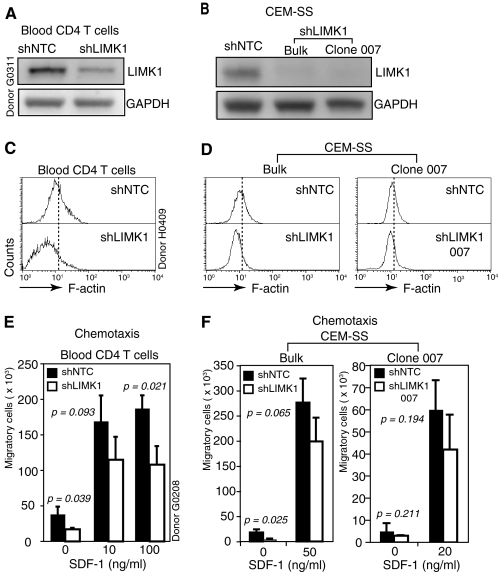
To specifically address the involvement of LIMK1 in actin polymerization and HIV-1 infection of CD4 T cells, we suppressed LIMK1 expression in transformed and primary human CD4 T cells using small interfering RNA (siRNA) or short hairpin RNA (shRNA) that transcribes siRNA to specifically target LIMK1 (Fig. 4A and supplemental Fig. 3, A–E). We achieved successful knockdown of LIMK1 in primary CD4 T cells in multiple donors tested (data not shown). Consistently, the LIMK1 knockdown cells from all donors demonstrated a decrease of F-actin (Fig. 4C and supplemental Fig. 3G). Nevertheless, the degree of F-actin loss in LIMK1 knockdown CD4 T cells varied from donor to donor, and the phenotype also diminished quickly after reaching its peak level (supplemental Fig. 3G). This transient knockdown phenotype plus donor variations limited our ability to conduct extensive phenotypic characterization of the knockdown cells. We decided to repeat the LIMK1 knockdown in a stable human CD4 T cell line, CEM-SS, so that donor variations could be eliminated. We were able to effectively suppress LIMK1 using shRNA in CEM-SS cells (Fig. 3B and supplemental Fig. 3, B and E). Again, the LIMK1 knockdown cells had a measurable loss of F-actin, consistent with the LIMK knockdown results in primary CD4 T cells (Fig. 4D and supplemental Fig. 3F). These results demonstrated that LIMK1 directly regulates actin polymerization both in primary and in transformed CD4 T cells. The phenotype of F-actin decrease in the LIMK1 knockdown CEM-SS cells was also transient, diminishing within a day or so after reaching its peak level (data not shown). We noticed that cells with LIMK1 knockdown appeared to grow slowly, and indeed a growth curve analysis confirmed that the knockdown cells had a significantly slower growth rate (supplemental Fig. 3I), suggesting that inhibition of LIMK1 and F-actin polymerization affects the cell cycle. This result appears to be consistent with recent findings demonstrating that LIMK1 is involved in control of cytokinesis of mammalian cells (38–40). This result also indicated that within the knockdown population, cells with minimal or mild LIMK1 knockdown would quickly outgrow cells with efficient LIMK1 knockdown, resulting in the loss of the knockdown phenotype. A common strategy to stabilize the shRNA knockdown phenotype would be to clone individual knockdown cells. Thus, we performed cell cloning of the LIMK1 knockdown CEM-SS cells. Clones carrying stable LIMK1 knockdown were selected, and F-actin staining confirmed that they all carried the expected decrease in F-actin as seen in the bulk LIMK knockdown cell populations. These cells have been cultured for extended period of time, and the knockdown and F-actin phenotypes were stably maintained (Fig. 4, B and D, and supplemental Fig. 3, K and L).
FIGURE 4.
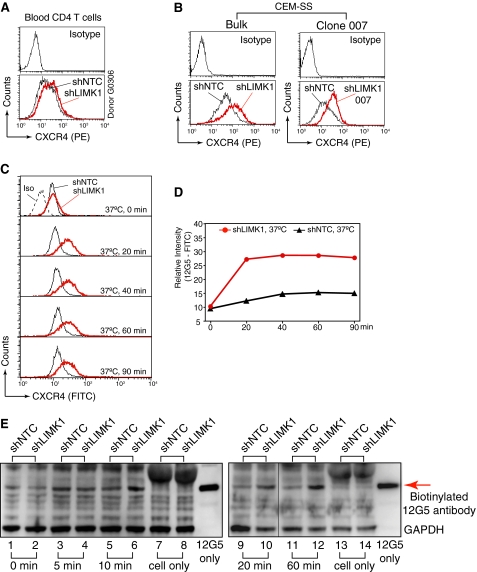
LIMK1 regulates actin polymerization and CD4 T cell chemotaxis. A and B, shRNA-mediated LIMK1 knockdown in primary blood CD4 T cells (A), CEM-SS T cells (B, bulk cell populations (Bulk)), and a derived clone (B, Clone 007). shNTC is the control shRNA against no human gene. Cells carrying stable LIMK1 knockdown or shNTC were selected in puromycin and analyzed by Western blotting using an anti-LIMK antibody. The same blots were stripped and reprobed with antibodies against human GAPDH for loading controls. C and D, LIMK1 knockdown decreases F-actin in primary CD4 T cells (C), CEM-SS T cells (D, Bulk), and clone 007 (D). The decreases of F-actin in the knockdown cells were measured by FITC-phalloidin staining and flow cytometry. E and F, effects of LIMK1 knockdown on T cell mobility in primary blood CD4 T cells (E), CEM-SS T cells (F, Bulk), and clone 007 (F). The effects in cell mobility were assayed for SDF-1-induced chemotaxis as described under “Experimental Procedures.” Data were analyzed with Student's t test on the means. The resulting p value is shown.
Actin polymerization is the driving force for T cell chemotactic migration following SDF-1 binding to CXCR4. We tested the effect of decreased F-actin on SDF-1-induced chemotaxis of the LIMK knockdown cells and observed moderate decreases in cell migration (Fig. 4, E and F), confirming a role of LIMK1 and F-actin in T cell chemotaxis (20).
We next investigated surface CXCR4 receptor expression on LIMK knockdown cells because decreased chemotactic response may result from lower surface CXCR4. Surprisingly, we detected a marked increase rather than decrease of CXCR4 on these knockdown cells (Fig. 5, A and B, and supplemental Fig. 3, H and L). This increase did not result from a general up-regulation of all surface receptors. Staining of CD4 in the LIMK knockdown CEM-SS cells detected lower surface CD4 (supplemental Fig. 4), a contrast to CXCR4 up-regulation. We also repeated the LIMK1 knockdown experiment in primary CD4 T cells to confirm that the CXCR4 receptor modulation was original to primary CD4 T cells and observed identical, transient up-regulation of the CXCR4 receptor (Fig. 5A) but not the down-modulation of the CD4 receptor (supplemental Fig. 4). We did not pursue the mechanism of CD4 down-modulation, as the phenotype was not observed in primary blood CD4 T cells and may be limited to certain transformed T cell lines.
FIGURE 5.
LIMK1 regulates CXCR4 receptor trafficking in CD4 T cells. A and B, LIMK1 knockdown up-regulates surface CXCR4 on primary CD4 T cells (A), CEM-SS T cells (B, Bulk), and clone 007 (B). The CXCR4 up-regulation was measured by surface staining with an anti-CXCR4 antibody (12G5). C and D, LIMK1 knockdown increases CXCR4 receptor internalization and recycling. Cells were incubated with FITC-labeled 12G5 at 37 °C for 20 to 90 min and then chilled on ice for 20 min, washed three times with cold PBS, and analyzed by flow cytometry. Binding of 12G5 to CXCR4, receptor endocytosis, and receptor recycling occur at 37 °C. E, the rate of CXCR4 endocytosis was further measured with biotin-labeled 12G5. Cells were incubated with biotin-labeled 12G5 at 4 °C for 2 h to permit sufficient binding to CXCR4. Cells were then washed with cold PBS. Subsequently, the temperature was shifted to 37 °C to induce 12G5-CXCR4 internalization. All cell surface 12G5 was stripped away by washing with cold acidic medium (pH 4.0). Cells were then immediately lysed and immunoblotted with streptavidin-labeled peroxidase to quantify the internalization of 12G5.
Given that the rate of receptor internalization and recycling can control its surface density, we further measured CXCR4 trafficking in one of the LIMK knockdown cells (LIMK-007). We used a well characterized monoclonal antibody, 12G5, (41), to track CXCR4 internalization and recycling. CXCR4 is internalized through endocytosis to endosome compartments, from where it could recycle to the cell surface after removal of the bound antibody (42, 43). Cells were incubated with FITC-labeled 12G5 at 37 °C for 20 to 90 min. As shown in Fig. 5, C and D, we observed a drastic enhancement of 12G5 accumulation in the LIMK1 knockdown cells, suggesting a greater increase in intracellular CXCR4. This drastic increase of 12G5 accumulation could result from an altered rate of either CXCR4 endocytosis or exocytosis. Thus, we developed a new assay based on a previous method (43) to measure CXCR4 internalization through endocytosis. In this assay, cells were incubated with biotin-labeled 12G5 at 4 °C for 2 h to permit sufficient binding to CXCR4. Subsequently, the temperature was shifted to 37 °C to induce 12G5-CXCR4 internalization. All cell surface 12G5 was stripped away by washing with a cold acidic buffer (pH 4.0) that can elute cell surface antibody as shown in a control experiment (supplemental Fig. 5). Cells were then immediately lysed and immunoblotted with streptavidin-labeled peroxidase to quantify the internalization of 12G5. As shown in Fig. 5E, we observed a marked increase of intracellular 12G5 in the LIMK1 knockdown cells, suggesting that LIMK knockdown caused an increase in the rate of CXCR4 internalization and endocytosis. These results imply that both the endocytosis and exocytosis must be increased in the LIMK knockdown cell because the surface display of CXCR4 was up-regulated. Our data are consistent with previous suggestions that the cortical actin is a barrier to cellular uptake and secretory processes (44). Thus, it is possible that the LIMK-mediated early cortical actin polymerization, a transient buildup of the cortical actin, may result in a temporary block to CXCR4 internalization that could potentially regulate viral entry and early postentry processes.
LIMK1 Regulates Viral Entry and DNA Synthesis
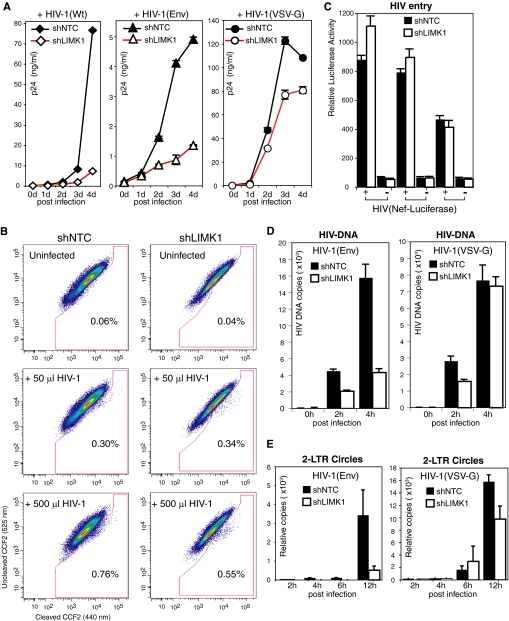
We further investigated the effect of LIMK1 knockdown on viral replication by infecting cells with HIV-1NL4–3 and observed significant inhibition of HIV replication during a 4-day time course (Fig. 6A). Because the cortical actin is also required for viral assembly (45), we chose to use a single-cycle replicating virus to study viral early process. For comparison, we identically infected the cells with a VSV-G pseudotyped HIV-1 that enters cells via endocytosis, thereby bypassing CD4/CXCR4 and the cortical actin. We observed much greater inhibition of HIV-1 replication by LIMK1 knockdown in comparison to that on the VSV-G-pseudotyped viral replication (Fig. 6A), suggesting that LIMK1 most likely affects viral infection through CD4/CXCR4 and/or the cortical actin. Thus, we measured the relevant viral processes, such as receptor-mediated entry and early reverse transcription. We did not find a significant difference in viral entry at low viral dosages using two different entry assays (26, 27) (Fig. 6, B and C, and supplemental Fig. 6, A and B). However, at high dosages, viral entry was slightly decreased in the LIMK1 knockdown cell (Fig. 6B and supplemental Fig. 6A), likely resulting from the use of saturating amounts of viruses that require extensive receptor clustering beyond the natural associate and close disposition of CD4 and CXCR4 (12, 46–48). It is possible that the LIMK1 knockdown decreases actin activities and thus affects receptor clustering and the entry of excess levels of the virus (8, 14). We next investigated early viral DNA synthesis following a low-dosage viral infection at which viral entry was not affected. We observed a significant decrease in the LIMK1 knockdown cell both at 2 and 4 h postinfection (Fig. 6D). In contrast, we did not observe a significant inhibition of viral DNA synthesis in the VSV-G pseudotyped HIV-1 infection (4 h), although there was a slight delay in viral DNA synthesis at an early time point (2 h) (Fig. 6D). This slight delay likely resulted from a slowdown of membrane scission of clathrin-coated pits in early endocytosis, which is known to be affected by actin dynamics (49). We also examined viral nuclear DNA using viral 2-LTR circles as a surrogate and observed diminished 2-LTR circles in the LIMK1 knockdown cells at both 6 and 12 h postinfection (Fig. 6E). In contrast, there was only a slight decrease in 2-LTR circles in the VSV-G pseudotyped HIV-1 infection at 12 h in the LIMK1 knockdown cells (Fig. 6E).
FIGURE 6.
LIMK1 regulates HIV-1 entry and DNA synthesis. A, CEM-SS T cells carrying stable LIMK1 knockdown (shLIMK1, clone 007) or the control (shNTC) were infected with HIV-1NL4–3 (Wt) or with single-cycle replication HIV-1 pseudotyped with either the HIV envelope (Env) or with VSV-G (equal p24, 200 ng per million cells) for 2 h. Cells were subsequently washed twice and cultured for 4 days. Viral replication was monitored by the release of extracellular p24. B and C, viral entry was measured with a Vpr-BlaM-based entry assay (B) or a Nef-luciferase-based entry assay. B, two dosages of HIV-1 (50 and 500 μl of HIV-1, 17.5 ng/μl of p24) were used to infect 1 million cells. C, 270 ng (p24) of HIV-1(Nef-luciferase) were used to infect 1 million cells. The syntheses of total HIV DNA (D) and 2-LTR circles (E) following infection (250 ng p24 per million cells) were also measured with real-time PCR as described under supplemental experimental procedures.
DISCUSSION
In this article, we demonstrate that at the earliest time of HIV infection, binding of the viral envelope to resting CD4 T cells triggers a quick, transient activation of LIMK and actin polymerization, which directly regulates CXCR4 receptor internalization and viral DNA synthesis. These results suggest that LIMK is a major regulator controlling the spatiotemporal dynamics of the cortical actin critical for the initiation of HIV-1 infection.
The observed impairment in viral DNA synthesis in LIMK knockdown cells is consistent with previous suggestions that actin filaments are required for the establishment of the viral reverse transcription complex (9, 15). It is also in accordance with our recent data showing that blocking actin polymerization with jasplakinolide (120 nm) or latrunculin A (2.5 μm) inhibits viral DNA synthesis and that promoting actin polymerization through siRNA knockdown of cofilin enhances viral DNA synthesis (9). Nevertheless, the exact molecular mechanism of how actin directly affects viral DNA synthesis remains to be resolved. It is remarkable that LIMK knockdown resulted in a several-fold reduction in viral replication (Fig. 6A), given that the knockdown only caused a moderate decrease in the cortical actin (Fig. 3D). The virus should have a sufficient amount of actin to interact with. Mechanistically, it is possible that actin polymerization may simply act as a driving force for viral uncoating and that decreased actin dynamics hinder this process. Alternatively, the cortical actin may function as an anchorage for the viral reverse transcription complex, and a decrease in the cortex density may result in less contact time. Certainly, multiple HIV proteins such as the viral nucleocapsid, the large subunit of the viral reverse transcriptase, the viral integrase, and Nef in the preintegration complex (PIC) are known to directly interact with actin (50–56), suggesting possible anchorage of PIC onto the cortical actin for efficient reverse transcription. Among these actin-interacting proteins, Nef has been known to affect viral DNA synthesis in cells without being directly involved in reverse transcription because Nef-defective virions display normal levels of endogenous reverse transcriptase activity (57–59), suggesting that the effect of Nef on viral DNA synthesis may involve a cellular cofactor such as actin or cofilin. Interestingly, in nef-expressing cells, the protein has also recently been shown to cause the phosphorylation of cofilin and the remodeling of the actin cytoskeleton (60). The direct involvement of F-actin in the viral transcriptional processes has been demonstrated previously in other viruses (61).
Our data suggest that LIMK is a main regulator hijacked by HIV to transiently take control of the cortical actin dynamics important for optimal reverse transcription. The surprising finding that LIMK1 also regulates CXCR4 receptor cycling suggests that the LIMK-triggered transient actin polymerization may temporarily block CXCR4 internalization to promote the formation of the CD4-CXCR4 fusion complex for entry. It may also prolong CXCR4 signaling that leads to the subsequent activation of cofilin to facilitate viral postentry migration (9) (Fig. 7). In chemotaxis, prolonged CXCR4 signaling at the leading edge could help to maintain directional cell migration toward the chemokine gradient.
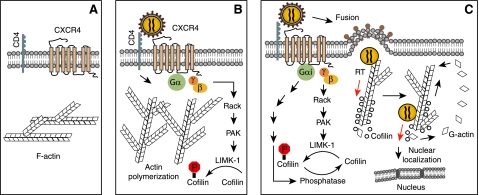
FIGURE 7.
Model of LIMK1 regulating actin dynamics and HIV-1 infection. A, in the absence of chemotactic stimulation, the cortical actin in resting CD4 T cells remains relatively static. B, binding of gp120 to CD4 and CXCR4 activates LIMK1, triggering actin polymerization that transiently blocks CXCR4 internalization to stabilize CD4/CXCR4 clustering and virus-receptor interaction. C, subsequent CXCR4 signaling through Gαi activates cofilin, leading to the depolymerization of cortical actin. Following entry, actin dynamics may also facilitate viral uncoating and permit the direct anchorage of the viral PIC onto the cortical actin for reverse transcription. Cofilin-mediated actin treadmilling also promotes intracellular migration of PIC across the actin cortex.
Our data also showed that HIV-triggered LIMK activation is mediated through the Rho family GTPase Rac1 and its downstream effector PAK1, in agreement with previous findings that Rac1 but not Cdc42 and Rho mediates SDF-1-induced LIMK1 activation in human T cells (20). Our results also agree with a previous report showing that HIV envelope-mediated cell-cell syncytium formation relies on Rac-1 but not on Cdc42 and Rho activation, suggesting possible activation of Rac1 by gp120 to trigger actin polymerization to mediate fusion (13).
The elucidation of LIMK-mediated actin polymerization and its role in HIV infection is instrumental to the understanding of viral pathogenesis and to the development of novel therapeutics. It is certain that increased actin activity, either through induction of transient actin polymerization or depolymerization, benefits HIV infection (9), enhancing viral entry and DNA synthesis by actin polymerization and promoting viral intracellular migration by actin treadmilling (9). Consistent with these in vitro observations, in a recent pilot study, we have found that in the resting CD4 T cells of HIV-infected patients, phosphorylation of an actin-depolymerizing factor, cofilin, is altered (62). LIMK is the kinase responsible for the serine 3 phosphorylation of cofilin. It remains to be determined whether the abnormal cofilin activation observed in HIV patients is the direct result of altered activities of LIMK or a cofilin phosphatase.
A temporal actin event is normally triggered by ligand binding and subsequent signal transduction. Although severe inhibition of actin dynamics is normally associated with severe cytotoxicity, drug-mediated prevention of viral disturbance of the actin cortex is possible. The surface receptors as well as upstream signaling molecules are viable targets for inhibition of virus-initiated actin activities. In addition, the viral PIC could also be directly targeted by preventing viral contact with the cortical actin.
Acknowledgments
We thank the George Mason University Student Health Center for blood donation; the National Institutes of Health AIDS Research and Reference Reagent Program for reagents; D. A. Stephany and K. L. Holmes of NIAID, National Institutes of Health for the fusion assay; J. Willett, L. Liotta, F. Kashanchi, and J. Kuhn for comments; and J. Guernsey for editorial assistance.
This work was supported, in whole or in part, by NIAID National Institutes of Health Grants AI069981 and 1R01AI081568 (to Y. W.). This work was also supported by the 2009 New York City to Washington, D. C. AIDS Ride (to Y. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6, experimental procedures, and references.
- LIMK
- LIM domain kinase
- LIM1
- LIM domain kinase 1
- R5
- CCR5-utilizing
- OA
- okadaic acid
- PIC
- preintegration complex
- NTC
- non-targeting control
- SDF-1
- stromal cell-derived factor-1.
REFERENCES
- 1. Gottlieb M. S., Schroff R., Schanker H. M., Weisman J. D., Fan P. T., Wolf R. A., Saxon A. (1981) N. Engl. J. Med. 305, 1425–1431 [DOI] [PubMed] [Google Scholar]
- 2. Stahl R. E., Friedman-Kien A., Dubin R., Marmor M., Zolla-Pazner S. (1982) Am. J. Med. 73, 171–178 [DOI] [PubMed] [Google Scholar]
- 3. Dalgleish A. G., Beverley P. C., Clapham P. R., Crawford D. H., Greaves M. F., Weiss R. A. (1984) Nature 312, 763–767 [DOI] [PubMed] [Google Scholar]
- 4. Feng Y., Broder C. C., Kennedy P. E., Berger E. A. (1996) Science 272, 872–877 [DOI] [PubMed] [Google Scholar]
- 5. Naghavi M. H., Valente S., Hatziioannou T., de Los Santos K., Wen Y., Mott C., Gundersen G. G., Goff S. P. (2007) EMBO J. 26, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrero-Villar M., Cabrero J. R., Gordón-Alonso M., Barroso-González J., Alvarez-Losada S., Muñoz-Fernández M. A., Sánchez-Madrid F., Valenzuela-Fernández A. (2009) J. Cell. Sci. 122, 103–113 [DOI] [PubMed] [Google Scholar]
- 7. Jimenez-Baranda S., Gomez-Mouton C., Rojas A., Martinez-Prats L., Mira E., Ana Lacalle R., Valencia A., Dimitrov D. S., Viola A., Delgado R., Martinez A. C., Manes S. (2007) Nat. Cell. Biol. 9, 838–846 [DOI] [PubMed] [Google Scholar]
- 8. Harmon B., Ratner L. (2008) J. Virol. 82, 9191–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoder A., Yu D., Dong L., Iyer S. R., Xu X., Kelly J., Liu J., Wang W., Vorster P. J., Agulto L., Stephany D. A., Cooper J. N., Marsh J. W., Wu Y. (2008) Cell 134, 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ploubidou A., Way M. (2001) Curr. Opin. Cell Biol. 13, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naghavi M. H., Goff S. P. (2007) Curr. Opin. Immunol. 19, 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ugolini S., Moulard M., Mondor I., Barois N., Demandolx D., Hoxie J., Brelot A., Alizon M., Davoust J., Sattentau Q. J. (1997) J. Immunol. 159, 3000–3008 [PubMed] [Google Scholar]
- 13. Pontow S. E., Heyden N. V., Wei S., Ratner L. (2004) J. Virol. 78, 7138–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iyengar S., Hildreth J. E., Schwartz D. H. (1998) J. Virol. 72, 5251–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bukrinskaya A., Brichacek B., Mann A., Stevenson M. (1998) J. Exp. Med. 188, 2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y., Belkina N. V., Shaw S. (2009) Sci. Signal. 2, pe23. [DOI] [PubMed] [Google Scholar]
- 17. Bukrinsky M. (2008) Retrovirology 5, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y., Yoder A. (2009) PLoS. Pathog. 5, e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balabanian K., Harriague J., Décrion C., Lagane B., Shorte S., Baleux F., Virelizier J. L., Arenzana-Seisdedos F., Chakrabarti L. A. (2004) J. Immunol. 173, 7150–7160 [DOI] [PubMed] [Google Scholar]
- 20. Nishita M., Aizawa H., Mizuno K. (2002) Mol. Cell. Biol. 22, 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Y., Marsh J. W. (2001) Science 293, 1503–1506 [DOI] [PubMed] [Google Scholar]
- 22. Kelly J., Beddall M. H., Yu D., Iyer S. R., Marsh J. W., Wu Y. (2008) Virology 372, 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu D., Wang W., Yoder A., Spear M., Wu Y. (2009) PLoS Pathog. 5, e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Y., Beddall M. H., Marsh J. W. (2007) Retrovirology 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Y., Beddall M. H., Marsh J. W. (2007) Curr. HIV Res. 5, 394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavrois M., De Noronha C., Greene W. C. (2002) Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 27. Saeed M. F., Kolokoltsov A. A., Davey R. A. (2006) J. Virol. Methods 135, 143–150 [DOI] [PubMed] [Google Scholar]
- 28. Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. (1998) Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 29. Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O., Caroni P. (1998) Nature 393, 805–809 [DOI] [PubMed] [Google Scholar]
- 30. Bamburg J. R., Harris H. E., Weeds A. G. (1980) FEBS Lett. 121, 178–182 [DOI] [PubMed] [Google Scholar]
- 31. Lappalainen P., Drubin D. G. (1997) Nature 388, 78–82 [DOI] [PubMed] [Google Scholar]
- 32. Bernard O., Burkitt V., Webb G. C., Bottema C. D., Nicholl J., Sutherland G. R., Matthew P. (1996) Genomics 35, 593–596 [DOI] [PubMed] [Google Scholar]
- 33. Ikebe C., Ohashi K., Fujimori T., Bernard O., Noda T., Robertson E. J., Mizuno K. (1997) Genomics 46, 504–508 [DOI] [PubMed] [Google Scholar]
- 34. Unutmaz D., KewalRamani V. N., Marmon S., Littman D. R. (1999) J. Exp. Med. 189, 1735–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. (1999) Nat. Cell. Biol. 1, 253–259 [DOI] [PubMed] [Google Scholar]
- 36. Dan C., Kelly A., Bernard O., Minden A. (2001) J. Biol. Chem. 276, 32115–32121 [DOI] [PubMed] [Google Scholar]
- 37. Misra U. K., Deedwania R., Pizzo S. V. (2005) J. Biol. Chem. 280, 26278–26286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Amano T., Kaji N., Ohashi K., Mizuno K. (2002) J. Biol. Chem. 277, 22093–22102 [DOI] [PubMed] [Google Scholar]
- 39. Davila M., Jhala D., Ghosh D., Grizzle W. E., Chakrabarti R. (2007) Mol. Cancer 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaji N., Muramoto A., Mizuno K. (2008) J. Biol. Chem. 283, 4983–4992 [DOI] [PubMed] [Google Scholar]
- 41. Endres M. J., Clapham P. R., Marsh M., Ahuja M., Turner J. D., McKnight A., Thomas J. F., Stoebenau-Haggarty B., Choe S., Vance P. J., Wells T. N., Power C. A., Sutterwala S. S., Doms R. W., Landau N. R., Hoxie J. A. (1996) Cell 87, 745–756 [DOI] [PubMed] [Google Scholar]
- 42. Signoret N., Rosenkilde M. M., Klasse P. J., Schwartz T. W., Malim M. H., Hoxie J. A., Marsh M. (1998) J. Cell Sci. 111, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 43. Signoret N., Oldridge J., Pelchen-Matthews A., Klasse P. J., Tran T., Brass L. F., Rosenkilde M. M., Schwartz T. W., Holmes W., Dallas W., Luther M. A., Wells T. N., Hoxie J. A., Marsh M. (1997) J. Cell Biol. 139, 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfeiffer J. R., Seagrave J. C., Davis B. H., Deanin G. G., Oliver J. M. (1985) J. Cell Biol. 101, 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jolly C., Mitar I., Sattentau Q. J. (2007) J. Virol. 81, 5547–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zaitseva M., Romantseva T., Manischewitz J., Wang J., Goucher D., Golding H. (2005) J. Leukocyte Biol. 78, 1306–1317 [DOI] [PubMed] [Google Scholar]
- 47. Xiao X., Wu L., Stantchev T. S., Feng Y. R., Ugolini S., Chen H., Shen Z., Riley J. L., Broder C. C., Sattentau Q. J., Dimitrov D. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7496–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singer II, Scott S., Kawka D. W., Chin J., Daugherty B. L., DeMartino J. A., DiSalvo J., Gould S. L., Lineberger J. E., Malkowitz L., Miller M. D., Mitnaul L., Siciliano S. J., Staruch M. J., Williams H. R., Zweerink H. J., Springer M. S. (2001) J. Virol. 75, 3779–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Merrifield C. J., Perrais D., Zenisek D. (2005) Cell 121, 593–606 [DOI] [PubMed] [Google Scholar]
- 50. Rey O., Canon J., Krogstad P. (1996) Virology 220, 530–534 [DOI] [PubMed] [Google Scholar]
- 51. Wilk T., Gowen B., Fuller S. D. (1999) J. Virol. 73, 1931–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hottiger M., Gramatikoff K., Georgiev O., Chaponnier C., Schaffner W., Hübscher U. (1995) Nucleic Acids Res. 23, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu B., Dai R., Tian C. J., Dawson L., Gorelick R., Yu X. F. (1999) J. Virol. 73, 2901–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turlure F., Devroe E., Silver P. A., Engelman A. (2004) Front. Biosci. 9, 3187–3208 [DOI] [PubMed] [Google Scholar]
- 55. Niederman T. M., Hastings W. R., Ratner L. (1993) Virology 197, 420–425 [DOI] [PubMed] [Google Scholar]
- 56. Fackler O. T., Kienzle N., Kremmer E., Boese A., Schramm B., Klimkait T., Kücherer C., Mueller-Lantzsch N. (1997) Eur. J. Biochem. 247, 843–851 [DOI] [PubMed] [Google Scholar]
- 57. Aiken C., Trono D. (1995) J. Virol. 69, 5048–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chowers M. Y., Pandori M. W., Spina C. A., Richman D. D., Guatelli J. C. (1995) Virology 212, 451–457 [DOI] [PubMed] [Google Scholar]
- 59. Schwartz O., Maréchal V., Danos O., Heard J. M. (1995) J. Virol. 69, 4053–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stolp B., Reichman-Fried M., Abraham L., Pan X., Giese S. I., Hannemann S., Goulimari P., Raz E., Grosse R., Fackler O. T. (2009) Cell Host Microbe 6, 174–186 [DOI] [PubMed] [Google Scholar]
- 61. De B. P., Burdsall A. L., Banerjee A. K. (1993) J. Biol. Chem. 268, 5703–5710 [PubMed] [Google Scholar]
- 62. Wu Y., Yoder A., Yu D., Wang W., Liu J., Barrett T., Wheeler D., Schlauch K. (2008) Retrovirology 5, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]