Abstract
N-terminal proteolysis of huntingtin is thought to be an important mediator of HD pathogenesis. The formation of short N-terminal fragments of huntingtin (cp-1/cp-2, cp-A/cp-B) has been demonstrated in cells and in vivo. We previously mapped the cp-2 cleavage site by mass spectrometry to position Arg167 of huntingtin. The proteolytic enzymes generating short N-terminal fragments of huntingtin remain unknown. To search for such proteases, we conducted a genome-wide screen using an RNA-silencing approach and an assay for huntingtin proteolysis based on the detection of cp-1 and cp-2 fragments by Western blotting. The primary screen was carried out in HEK293 cells, and the secondary screen was carried out in neuronal HT22 cells, transfected in both cases with a construct encoding the N-terminal 511 amino acids of mutant huntingtin. For additional validation of the hits, we employed a complementary assay for proteolysis of huntingtin involving overexpression of individual proteases with huntingtin in two cell lines. The screen identified 11 enzymes, with two major candidates to carry out the cp-2 cleavage, bleomycin hydrolase (BLMH) and cathepsin Z, which are both cysteine proteases of a papain-like structure. Knockdown of either protease reduced cp-2 cleavage, and ameliorated mutant huntingtin induced toxicity, whereas their overexpression increased the cp-2 cleavage. Both proteases partially co-localized with Htt in the cytoplasm and within or in association with early and late endosomes, with some nuclear co-localization observed for cathepsin Z. BLMH and cathepsin Z are expressed in the brain and have been associated previously with neurodegeneration. Our findings further validate the cysteine protease family, and BLMH and cathepsin Z in particular, as potential novel targets for HD therapeutics.
Keywords: Cell Death, Huntington Disease, Neurodegeneration, Protein Degradation, Proteolytic Enzymes
Introduction
Huntington disease (HD)3 is caused by an expansion of the CAG repeat coding for polyglutamine (polyQ) within the HD gene product huntingtin (Htt) (1, 2). It is not clear how mutant Htt causes neuronal cell death, but evidence is accumulating that N-terminal fragments of mutant Htt are important mediators of pathogenesis. N-terminal fragments of Htt are found in human postmortem brain (3–8). In cell model experiments, shorter N-terminal fragments of Htt are generally more toxic to cells than longer fragments, and mouse HD models expressing shorter fragments usually develop more severe pathology than the full-length Htt models (6, 9–19). The proteolytic pathway for mutant Htt may include several cleavage events mediated by different enzymes involved in the generation of toxic fragments of various lengths. These enzymes may represent potential novel therapeutic targets for HD.
One example of such an enzyme is caspase-6, which cleaves mutant Htt at residue 586 from its N terminus and generates a toxic fragment; YAC128 mice expressing Htt with an altered caspase-6 site have a substantially ameliorated HD phenotype (20), whereas transgenic mice expressing a caspase-6 fragment with an expanded polyQ repeat develop an HD phenotype.4 Htt can be also cleaved by other caspases and calpains within the same region (10, 20–29), though their contribution to pathogenesis is less clear.
Htt is further cleaved into shorter N-terminal fragments through sequential or independent pathways, details of which are starting to emerge (8, 13, 30–32). Because the specific size of the Htt fragment may determine its pathogenic properties (15, 16, 20, 31, 33), it is important to map the exact location of the cleavage sites and to identify the proteases involved in specific proteolytic steps.
Cleavage of Htt near its N terminus produces short and potentially toxic fragments with expanded polyQ (within approximately the N-terminal 200 amino acids, numbering using a normal polyQ length of 23). Two short N-terminal fragments designated cp-A and cp-B had previously been observed in NG108–15 (neuroblastoma-glioma) cells with proteasomes inhibition, and the cp-A site was mapped to residues 105–114 (13). In several cell models, we consistently detect two similar fragments. We designated them as cp-1 and cp-2 because they appear similar to cp-A and cp-B, however, in our studies, neither of these fragments was mapped to the 105–114 region (32).
Using mass spectrometry, we previously mapped the cp-2 cleavage site to position Arg 167 of Htt. Furthermore, alterations of this cleavage site resulted in a decrease in toxicity in neuronal cells (31). Short fragments of mutant Htt also accumulate in mouse models of HD, in correlation with the disease progression (15, 18, 34).4 It is not known what proteolytic enzymes generate any of these short N-terminal fragments and in which subcellular compartment this cleavage occurs.
To search for the proteases involved in the N-terminal cleavage of Htt, we have employed an RNA-silencing screening approach using the entire collection from ON-TARGETplus siRNA library to all known human proteases (586 genes), and we have established an assay for Htt proteolysis, based on the detection of cp-2/cp-1 fragments by Western blotting. The primary screen was carried out in HEK293 cells, and the secondary screen was carried out in neuronal HT22 cells, transfected with the mutant truncated Htt (the N-terminal first 511 amino acids). As the result of the above screen, we found that two cysteine proteases of papain-like structure, bleomycin hydrolase (BLMH) and cathepsin Z, are likely candidates to carry out the cp-2 cleavage of Htt in HEK293 and HT22 cells. Both BLMH and cathepsin Z are expressed in brain and have been associated previously with neurodegeneration. We found that siRNA-mediated knockdown of either protease reduced the cp-2 cleavage and ameliorated mutant Htt-induced toxicity, whereas co-transfection of HEK293 and HT22 cells with the Htt and BLMH or cathepsin Z constructs increased the cp-2 cleavage. Both enzymes partially co-localized with Htt in the cytoplasm and at early and late endosomes. If validated in vivo, BLMH and cathepsin Z may develop into novel therapeutic targets for HD.
EXPERIMENTAL PROCEDURES
Plasmids and Mutagenesis
Truncated Htt expression constructs N511–8Q/82Q were generated as described previously (31) from the full-length Htt constructs (HD-FL-23/82Q) by an introduction of a stop codon after amino acid 511 of Htt. The length of 511 amino acids was chosen because it is comparable with but shorter than predicted caspase and calpain cleavage fragments, so that the construct would not be cleaved by one of these enzymes. The stop codons and the RLQL (Δ167–170) deletion used in this study were introduced by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) according to the manufacturer's protocol. The Htt-N511–52Q construct was produced from N511–82Q by random contractions of polyQ repeat in bacterial cells. Htt N586–20Q/128Q plasmids were a kind gift from Michael Hayden and were described previously (20, 31). The expression plasmids encoding BLMH, cathepsin Z, and expression clones of 18 other proteases that were tested in the screen were purchased from ATCC.
Cell Culture and Transfection
Human embryonic kidney (HEK)293FT cells (Invitrogen) and mouse hippocampal HT22 cells were maintained as described previously (31). All cells were transfected with constructs encoding Htt or with selected proteases using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
siRNA Screen and Western Blotting
For primary screening, we used the ON-TARGETplus library containing 586 siRNA pools to all human proteases (Thermo Scientific/Dharmacon). HEK293 cells were plated in antibiotic-free medium in 24-well plates at density 106 cells/well (two duplicate wells per each siRNA pool). 24 h later, cells were transfected with a mixture of the Htt-N511–52Q plasmid and individual siRNA pool (or control pool) at a final concentration of 75 nm using Lipofectamine 2000 (Invitrogen). The optimized transfection mixture per 1 well contained 0.5 μg of plasmid DNA, 30 pmol of siRNA (1.5 μl of 20 μm stock solution), and 3.7 μl of Lipofectamine 2000 in OptiMEM medium (Invitrogen). For Western blotting analysis, HEK293 cells were lysed 48 h after transfection in M-PER buffer (Pierce) with protease inhibitors (Protease Inhibitor Mixture III, Calbiochem), lysates were fractionated on NuPAGE 4–12% Bis-Tris polyacrylamide gels (Invitrogen), transferred to nitrocellulose membranes, and probed with antibodies to FLAG (M2, Sigma). Immunoblots were developed with peroxidase-conjugated secondary antibodies (Amersham Biosciences) and enhanced chemiluminescence (ECL-Plus detection reagent, Amersham Biosciences). The bands corresponding to N511, cp-1, and cp-2 fragments of Htt were quantitated using Molecular Imager Gel Doc XR System and Quantity One software (Bio-Rad). For a secondary screen in HT22 cells, we have used ON-TARGETplus siRNA pools to selected mouse proteases (Thermo Scientific/Dharmacon). Transfection and lysate preparation was carried out as mentioned above. NaCl was added to final concentration of 150 mm, and Htt-FLAG fusion proteins were immunoprecipitated using anti-FLAG M2 affinity gel (Sigma) followed by fractionation on NuPAGE 4–12% Bis-Tris polyacrylamide gels (Invitrogen), and detection with an antibody to Htt exon 1.
In Vitro Cleavage Assay
HEK293 cells were transfected as above with either normal (N511–8Q) or expanded (N511–52Q) Htt, or the fragment of Htt with deleted cp-2 site (N511–52Q-RLQL). Cell lysates were prepared 24 h post transfection in 150 mm NaCl, 50 mm Tris, pH 7.5, 5 mm EDTA, 50 mm MgCl2 and 0.5% Triton X-100 without protease inhibitors. For BLMH, cleavage lysates (50 μg of total protein) were incubated for 1 h at 37 °C with equal volumes of recombinant human BLMH (0.8 μg/reaction; R&D Systems) prepared in the assay buffer according to manufacturer's protocol (50 mm HEPES, 5 mm EDTA, 10 mm DTT, pH 7.0). For cathepsin Z cleavage, the recombinant human cathepsin Z (R&D Systems) was first activated for 5 min at room temperature in the activation buffer (25 mm sodium acetate, pH 3.5). Cell lysates (50 μg of total protein) were incubated for 1 h at 37 °C with equal volumes of the enzyme (0.4 or 0.6 μg/reaction) prepared in the assay buffer according to manufacturer's protocol (25 mm sodium acetate, pH 3.5, 5 mm DTT). The final pH of the cathepsin Z cleavage reaction was 6. The reactions were stopped by the addition of SDS sample buffer, and the proteins were analyzed by Western blotting as described above.
Antibodies
The rabbit polyclonal antibody to Htt (Htt 1–17 (against residues 1–17)) was described previously (35); the goat polyclonal antibody, prepared against the N-terminal Htt exon-1 fragment also was described previously (36); mouse monoclonal 1–82 antibody to Htt (against residues 1–82 of Htt) is from Millipore; antibody to the neoepitope Arg167 of Htt was described previously (31); mouse antibody to β-actin was from Sigma; mouse monoclonal antibody to BLMH was from Santa Cruz Biotechnology; goat polyclonal antibody to cathepsin Z was from R&D Systems; mouse monoclonal antibody to LAMP1 was from Abcam; mouse monoclonal antibody to GM130 was from BD Transduction Laboratories; rabbit polyclonal antibodies to LC3B, calreticulin, and EEA1 and rabbit monoclonal antibodies to COX IV, Rab 5, Rab 7, and Rab 11 were from Cell Signaling Technology; rabbit polyclonal antibody to active caspase-3 was from Promega.
Immunofluorescence
HT22 cells, expressing indicated constructs, were fixed (24 h after transfection) with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 (Sigma) for 10 min, blocked in 10% normal donkey serum (Sigma) for 30 min, and incubated (1 h, room temperature or overnight at 4 °C) with the following antibodies. For Htt and BLMH co-localization studies, we used a polyclonal antibody to Htt epitope 1–17 and mouse monoclonal antibody to BLMH, followed by donkey anti-rabbit Alexa Fluor 555 and donkey anti-mouse Alexa Fluor 488. For Htt and cathepsin Z co-localization, we used rabbit polyclonal antibody to Htt epitope 1–17 and goat polyclonal antibody to cathepsin Z, followed by donkey anti-rabbit Alexa Fluor 555 or donkey anti-goat Alexa Fluor 488. For Htt localization, we used mouse monoclonal 1–82 antibody to Htt and rabbit antibodies to organelles, followed by donkey anti-mouse Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 555. For BLMH localization, we used mouse monoclonal antibody to BLMH and rabbit antibodies to organelles, followed by donkey anti-rabbit Alexa Fluor 555 and donkey anti-mouse Alexa Fluor 488. For cathepsin Z localization studies, we used goat polyclonal antibody to cathepsin Z and mouse monoclonal antibody to LAMP1 or mouse antibody to GM130, followed by donkey anti-goat Alexa Fluor 555 and donkey anti-mouse Alexa Fluor 488 or rabbit antibodies to organelles with donkey anti-goat Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 555. Nuclei were stained with DAPI (Vector Laboratories). For triple staining of Htt, BLMH, and cellular organelles, we used goat polyclonal antibody to exon 1 of Htt, mouse monoclonal antibody to BLMH, and rabbit antibodies to organelles, followed by donkey anti-goat Alexa Fluor 350, donkey anti-mouse Alexa Fluor 488, and donkey anti-rabbit Alexa Fluor 555. For triple staining of Htt, cathepsin Z, and cellular organelles, we used mouse monoclonal 1–82 antibody to Htt, goat polyclonal antibody to cathepsin Z, and rabbit antibodies to organelles, followed by goat anti-mouse Alexa Fluor 350, donkey anti-goat Alexa Fluor 488, and donkey anti-rabbit Alexa Fluor 555. All Alexa Fluor antibodies were from Invitrogen. Confocal microscopy was performed using a Zeiss Axiovert 200 inverted microscope with 510-Meta confocal module and a 100× objective.
Caspase-3 Activation Cytotoxicity Assay
HT22 cells were plated in chamber slides and co-transfected with indicated Htt constructs and siRNA pools to BLMH or cathepsin Z as described above. 24 h post-transfection, immunofluorescence of Htt and active caspase-3 was performed as described above. Htt expression was detected using a monoclonal antibody to residues 1–82 of Htt (followed by anti-mouse Alexa Fluor 350), active caspase-3 was detected using anti-active caspase-3 antibody (followed by anti-rabbit Cy3). Microscopy was performed on Zeiss Axiovert 200 m microscope equipped with a 40× objective. In total, >200–400 transfected cells were counted per condition, and the number of cells expressing both Htt and active caspase-3 were presented as a percentage of the total number of Htt-positive cells.
RESULTS
Screen for Proteases Generating Short N-terminal Fragments of Htt
To search for enzymes that cleave Htt near its N terminus, we conducted a genome-wide screen using an RNA-silencing approach with the entire collection from the ON-TARGETplus siRNA library to all known human proteases. This library is composed of 586 siRNAs pools (four duplexes each) and is designed to reduce possible off-target effects. We developed a direct assay for Htt proteolysis based on the detection of cp-1 and cp-2 fragments, generated from a portion of expanded Htt (N511–52Q, Fig. 1A). The N511 fragment of Htt is comparable in length with Htt caspase fragments but does not contain any known caspase cleavage sites, which facilitated the focus of the screen on the proteolytic enzymes generating cp-1 and cp-2 fragments. The assay was based on our earlier studies in HEK293 and stable PC12 (pheochromocytoma) cells demonstrating the formation of two closely migrating N-terminal fragments (cp-1 and cp-2) of Htt in both cell systems (31, 32). In the primary screen, we used highly transfectable HEK293 cells, which allowed a reliable detection of both cp-1 and cp-2 fragments with an antibody to FLAG without proteasome inhibition.
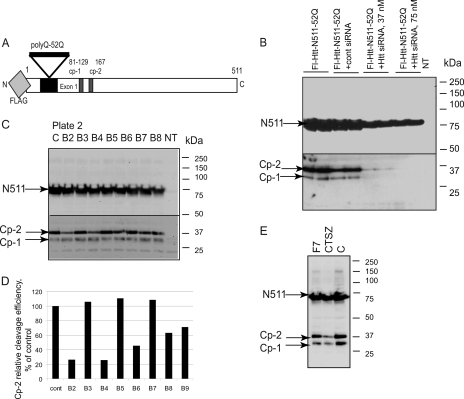
FIGURE 1.
Screen for proteases involved in the N-terminal cleavage of Htt. A, Htt construct used in the screen, comprising the first 511 amino acids of Htt with 52 polyQ and the N-terminal FLAG tag for detection. B, optimization of the transfection conditions with siRNA to Htt. HEK293 cells were transfected with indicated constructs and siRNAs. NT, nontransfected cells. C, example of a direct assay for Htt proteolysis. HEK293 cells were co-transfected with Htt N511–52Q and siRNA pools to proteases; the cp-1/cp-2 fragments were detected by Western blot using an antibody to FLAG. Lanes B2–B8, siRNA pools of indicated wells of plate two of the ON-TARGETplus library were co-transfected; lane C, co-transfection with nontargeting siRNA pool. NT, non-transfected cells. D, quantitation of the cp-2 cleavage efficiency upon co-transfection with protease siRNAs compared with nontargeting siRNA (cont), presented as a ratio (%) of the cp-2 bands to the corresponding N511 bands, shown in C. E, example of retesting of confirmed hits with lower concentration (25 nm) of siRNA.
Co-transfection conditions were first optimized for maximum knockdown efficiency with minimal cell toxicity using siRNA to Htt as a control (Fig. 1B). Based on these experiments, the final siRNA concentration of 75 nm, was chosen for the primary screen, which was carried out as follows; each of 586 siRNA pools was co-transfected with Htt-N511–52Q in HEK293 cells, in duplicate, in 24-well format; cell lysates were prepared 48 h later, and Htt proteins were detected using an antibody to FLAG epitope (Fig. 1 C). The change in cp-1/cp-2 cleavage efficiency induced by the knockdown of a specific protease was estimated by measuring the ratio of the cp-1 and cp-2 bands to the corresponding N511 bands (detected by Western blot, Fig. 1, C and D). Each ratio was normalized to the control cleavage efficiency of Htt co-transfected with the nontargeting siRNA. This control was included with every experimental set to reduce the assay variability due to variations in transfection efficiency and protein transfer.
At the next step, siRNA pools, which reduced the levels of cp-1 or cp-2 fragments (71 initial hits), were retested using the same assay (Fig. 2), resulting in 48 confirmed hits, which consistently decreased the production of either fragment at least 50% compared with control. These confirmed hits were rescreened using a lower concentration of siRNAs to reduce possible off-target effects. An example of this assay is shown on Fig. 1E, which demonstrates decreased cp1 and cp-2 upon cathepsin Z knockdown.
FIGURE 2.
The flow-chart of screening steps.
We have selected 20 proteases (12 highly ranked hits plus eight potentially relevant confirmed hits, Fig. 2) for a secondary assay in neuronal HT22 cells. Because these cells have lower transfection efficiency than HEK293 cells, there were lower levels of cp-1 and cp-2 in these cells, so immunoprecipitation with an antibody to FLAG tag was used for fragment detection. We employed two complementary approaches: co-transfection of Htt with siRNA pools to confirm the decrease in cp-1/cp-2 levels and co-transfection of Htt with expression constructs encoding selected proteases, expecting an increase in the fragments. Fig. 3 demonstrates both approaches. We observed some decline in the cp-1/cp-2 fragments upon cathepsin Z, and more so upon BLMH and elastase 2A (ELA2A) knockdown (Fig. 3A), and an increase in the fragments was induced by overexpression of cathepsin Z and BLMH (Fig. 3B). We also found some increase in the production of cp-1 upon co-expression with thrombin (F2) and calpain 10 (CAPN10), but these findings were not pursued any further in this study.
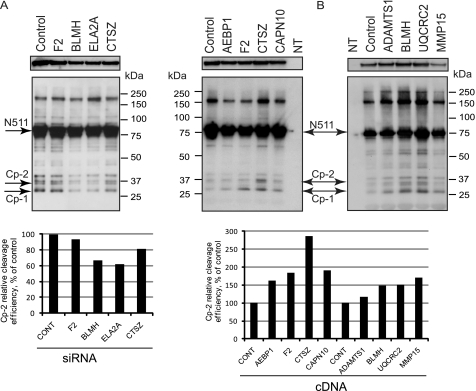
FIGURE 3.
Confirmation of the highly ranked hits from the primary assay in neuronal HT22 cells. A, HT22 cells were co-transfected with Htt-N511–52Q and either nontargeting siRNA pool (Control) or siRNAs to indicated proteases (F2, thrombin; ELA2A, elastase 2A; CTSZ, cathepsin Z; CONT, control). B, HT22 cells were co-transfected with Htt-N511–52Q and either pcDNA vector (Control) or cDNAs to indicated proteases (AEBP1, adipocyte enhancer binding protein; CAPN10, calpain 10; ADAMTS1, ADAM metallopeptidase with thrombospondin type 1; UQCRC2, ubiquitinol-cytochrome C reductase core protein 2; MMP15, matrix metallopeptidase 15; NT, nontransfected cells). In A and B, Htt proteins were immunoprecipitated with an antibody to FLAG, fractionated on SDS-PAGE, and detected with an antibody to exon 1 of Htt. The low exposure of the N511 band (top panels) demonstrates equal expression of Htt. The quantification of the cp-2 cleavage efficiency upon co-transfection with protease siRNAs (A) or cDNAs (B), presented as a ratio (%) of the cp-2 bands to the corresponding N511 bands, normalized to control, is shown below.
Upon completion of all the above steps of the screen, we have identified 11 enzymes, potentially involved in the cleavage of Htt yielding cp-1 and cp-2 fragments (Table 1). The five top proteases listed in Table 1 belong to the cysteine protease family with a papain-like active site structure, three are serine endopeptidases of chymotrypsin family, and two are metallopeptidases. The enzymes with papain-like active center (calpains and cathepsins) have been implicated previously in Htt proteolysis. Therefore, we have focused on two of these, BLMH and cathepsin Z, for further functional studies.
TABLE 1.
Protease identified in the siRNA screen
| Gene symbol | Name | Clan/family | Catalytic activity |
|---|---|---|---|
| BLMH | Bleomycin hydrolase | CA/C1 | Cysteine amino-/carboxy-/endopeptidase |
| CTSZ | Cathepsin Z (P, X) (Alternative names Cathepsin P, X) | CA/C1 | Cysteine carboxypeptidase |
| CTSB | Cathepsin B | CA/C1 | Cysteine endopeptidase |
| CTSH | Cathepsin H | CA/C1 | Cysteine endopeptidase |
| CALP10 | Calpain 10 | CA/C2 | Cysteine endopeptidase |
| F2 | Thrombin | PA/S1 | Serine endopeptidase |
| ELA2A | Elastase 2A | PA/S1 | Serine endopeptidase |
| PROZ | Protein Z | PA/S1 | Serine endopeptidase |
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 | MA/M11 | Metalloendopeptidase, zinc binding |
| UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | ME/M16 | Metalloendopeptidase, zinc binding |
| RBP3 | Retinol binding protein 2 | Unknown | Unknown |
Co-transfection of Htt with BLMH or Cathepsin Z Increases cp-2 Cleavage in HEK293 and HT22 Cells
To quantify an increase in normal and expanded Htt cleavage, induced by either protease, we conducted further experiments in HEK293 and HT22 cells (Fig. 4). We found that cleavage of both normal and expanded Htt was dramatically induced in HEK293 cells upon co-transfection with either BLMH or cathepsin Z expression plasmids together with Htt constructs. This increase was specific to the cp-2 fragment, whereas the levels of cp-1 were almost unchanged (Fig. 4, A and B). Furthermore, we did not observe an increase in fragments derived from the construct with deleted cp-2 site (N511–52Q-RLQL). In addition, the increased fragment was detected with specific antibody to Arg167 neoepitope (Fig. 4C). The elevated levels of BLMH and cathepsin Z upon co-transfection are shown on Fig. 4D. Notably co-transfection of both proteases has an additive effect on cp-2 cleavage of Htt in HEK293 cells (Fig. 4E). Thus, it appears that both BLMH and cathepsin Z are involved in a specific cleavage step at cp-2 site of Htt in HEK293 cells. Similar experiments were conducted in neuronal HT22 cells, with a dramatic increase in cp-2 fragment (Fig. 4F). Both cell lines were able to process cathepsin Z to its mature-active form, migrating slightly faster than the proform (Fig. 4, D and G).
FIGURE 4.
Co-expression of Htt with BLMH or cathepsin Z increases cp-2 cleavage in HEK293 and HT22 cells. HEK293 (A–E) or HT22 (F and G) cells were co-transfected with indicated Htt constructs and either BLMH or/and cathepsin Z expression plasmids. Htt proteins were detected by Western blotting with an antibody to exon 1 of Htt (A, E, and F). The cp-2 fragments were detected with a specific antibody to Arg167 neoepitope (C). Blots shown in A, E, and F were reprobed with an antibody to actin for loading control. The cp-2 fragment bands are indicated by arrows. The lines shown between gel samples indicate that some of the samples obtained from the same experiment were run on different parts of the same gel or on parallel gels at the same time. B, quantification of the cp-2 cleavage efficiency in HEK293 cells, calculated as a ratio (%) of the cp-2, total Htt (a sum of cp-2 and uncleaved N511 bands) shown in A (upper panel). The lower panel shows another representation of the same data (shown in A), demonstrating an increase in cp-2 cleavage with BLMH and cathepsin Z (CTSZ). Quantification of the cp-2 cleavage efficiency for Fig. 4E, calculated as above, is shown below the Western blot. The elevated levels of BLMH and cathepsin Z upon co-transfection in HEK293 cells (D) or in HT22 cells (G) are shown.
BLMH and Cathepsin Z Can Cleave Htt in Vitro
To establish whether BLMH and cathepsin Z can directly cleave Htt, we carried out the in vitro cleavage reactions using recombinant proteases.
Cell lysates were prepared 24 h post transfection from HEK293 cells expressing either normal (N511–8Q) or expanded (N511–52Q) Htt, or the fragment of Htt with deleted cp-2 site (N511–52Q-RLQL). At this time point, we observed little or no accumulation of the short Htt fragments in the cell culture (Fig. 5, control groups). An increase in both cp-1 and cp-2 cleavage products was detected upon incubation of the lysates with recombinant BLMH (Fig. 5A). Notably, there was no build up in the fragments produced from N511–52Q-RLQL construct with deleted cp-2 site. Incubation with increasing amounts of cathepsin Z at slightly acidic pH 6.0 also led to an increase in the levels of cp-1/cp-2-like fragments; however, cleavage of Htt by this protease seem to be less specific and produce multiple Htt fragments of various length (Fig. 5B). Again, less induction of the cleavage was observed with the construct lacking cp-2 site (N511–52Q-RLQL).
FIGURE 5.
BLMH and cathepsin Z can cleave Htt in vitro. A and B, cell lysates from HEK293 cells transfected with the indicated Htt constructs were prepared 24 h post transfection and incubated without (Control) or with indicated amounts of recombinant BLMH (A) or cathepsin Z (CTSZ; B). Htt proteins were detected by Western blotting with an antibody to exon 1 of Htt. Recombinant BLMH and cathepsin Z detected with specific antibodies are shown. Blots shown in A and B were re-probed with an antibody to actin as a loading control. The cp-2 fragment bands are indicated by arrows. The lines shown between gel samples in D indicate that the samples obtained from the same experiment were run on different parallel gels at the same time. C and D, quantification of the cp-2 cleavage efficiency in HEK293 cells, calculated as a ratio (%) of the cp-2 and total Htt (a sum of cp-2 and uncleaved N511 bands), shown in A and B. Lower panels show another representation of the same data (shown in A and B), demonstrating an increase in cp-2 cleavage with BLMH and cathepsin Z.
BLMH and Cathepsin Z Partially Co-localize with Htt in Cytoplasm and at Early and Late Endosomes of HT22 Cells
To investigate whether Htt co-localizes with either protease in cells, we used immunofluorescent confocal microscopy. HT22 cells, co-transfected with normal or expanded Htt-N511 constructs and each protease, were co-stained with antibodies to Htt and BLMH or cathepsin Z. We found that both BLMH and cathepsin Z partially co-localized with normal or expanded Htt in living cells (Fig. 6 and data not shown). To pinpoint the subcellular compartment where Htt and the above enzymes might co-localize, we used an antibody panel to organelle markers. Htt have been previously found to associate with multiple membranous organelles, including clathrin-coated and not coated endosomal vesicles (35, 37–40). Consistent with this, we found that transfected Htt partially co-localizes with early (EEA1 and Rab 5) and late (Rab 7) endosomal markers (supplemental Fig. S1A and data not shown).
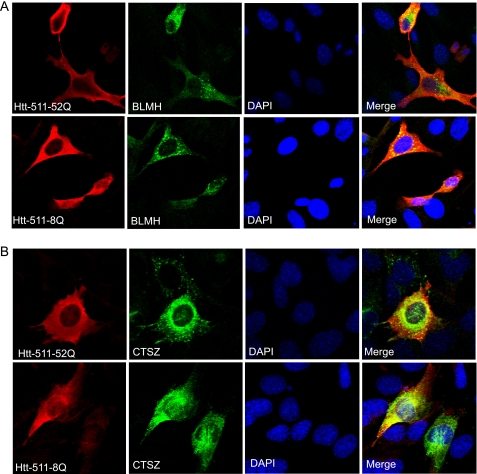
FIGURE 6.
BLMH and cathepsin Z co-localize with Htt in neuronal HT22 cells. HT22 cells were co-transfected with indicated Htt constructs and either BLMH (A) or cathepsin Z (CTSZ; B) expression constructs. Confocal immunofluorescent detection of Htt with an antibody to residues 1–17 (Alexa Fluor 555) is shown in red; detection of BLMH and cathepsin Z with specific antibodies is shown in green (Alexa Fluor 488). The nuclear staining (DAPI) is shown in blue. Yellow staining in merged images demonstrates partial co-localization.
BLMH, a cytoplasmic protease, also co-localized with EEA1, Rab5, Rab7, and recycling endosomal marker Rab11, but not with the endoplasmic reticulum marker calreticulin. Co-localization of endogenous BLMH with Rab7 was also detected. (supplemental Fig. S1B and data not shown). Cathepsins are primarily lysosomal proteases involved in terminal protein degradation. Consistent with this, we found cathepsin Z mostly co-localizing with lysosomal marker LAMP1 (supplemental Fig. S1C). However, confocal microscopy revealed a substantial overlap of transfected and endogenous cathepsin Z expression with early and late endosomal markers as well (supplemental Fig. S1C). Interestingly, confocal microscopy revealed some nuclear localization of transfected cathepsin Z in HT22 cells (Fig. 6B). No co-localization of Htt or either protease with mitochondrial marker COX IV or with LC3- positive puncta was detected (data not shown).
Because we found Htt, BLMH, and cathepsin Z expression in the proximity of endosomes, a triple staining using antibodies to Htt (blue), to either protease (green), and to endosomal markers (red) was performed with the goal to establish whether Htt and either enzyme co-localize at the endosomes (supplemental Fig. S1, D and E). Partial co-localization of Htt with either BLMH or cathepsin Z and early and late endosomal markers is evidenced by the appearance of white dots in the cytoplasm of HT22 cells. Co-localization of Htt with either protease elsewhere in the cytoplasm was also detected. These data are consistent with Htt proteolysis by BLMH and cathepsin Z occurring in the cytoplasm and within or in association with early and late endosomes; cleavage in the nucleus (by cathepsin Z) may also occur.
Knockdown of BLMH or Cathepsin Z Ameliorates Mutant Htt Toxicity in HT22 Cells
We have shown previously that alteration of the cp-2 site within Htt N511–52Q results in a decrease in Htt toxicity in HT22 cells (31). Thus, the knockdown of BLMH and cathepsin Z, enzymes involved in generating toxic cp-2 fragment, might diminish the pathogenic properties of expanded Htt. To test this hypothesis, we co-transfected HT22 cells with either normal (N586–20Q) or polyQ expanded (N586–128Q) Htt constructs together with siRNA pools to BLMH or cathepsin Z (Fig. 7A). In this assay, we used Htt constructs, encoding the pathogenic caspase-6 fragment of Htt with a longer glutamine stretch (N586–128Q) to achieve more cell toxicity as demonstrated by caspase-3 activation. We showed that BLMH and cathepsin Z are expressed and efficiently knocked down (∼60%) in this cell line (Fig. 7B). Caspase-3 activation was evaluated 48 h later on a cell-by-cell basis by co-staining of the cells with antibodies to active caspase-3 and Htt (Fig. 7C). To evaluate the effect of the protease knockdown on mutant Htt-mediated toxicity, we calculated the percentage of cells with activated caspase-3 among transfected cells co-stained with an Htt-specific antibody. At least 200–300 cells were counted for each condition in one experiment, and the experiment was repeated three times. As shown in Fig. 7D, siRNA-mediated knockdown of either BLMH or cathepsin Z resulted in significant reduction of expanded Htt toxicity in neuronal HT22 cells.
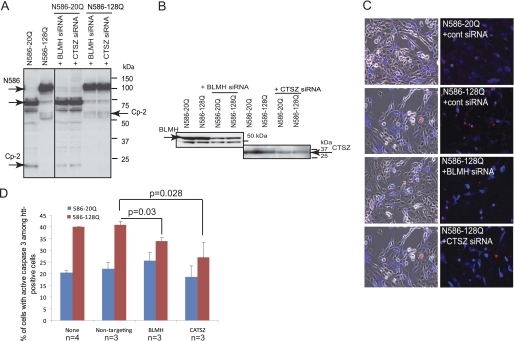
FIGURE 7.
Knockdown of BLMH or cathepsin Z ameliorates mutant Htt toxicity in HT22 cells. A, HT22 cells were co-transfected with either normal (N586–20Q) or polyQ expanded (N586–128Q) Htt constructs and siRNA pools to BLMH or cathepsin Z (CTSZ). A line shown between gel samples indicate that the samples obtained from the same experiment were run on different parallel gels at the same time. B, endogenous BLMH and cathepsin Z expression in HT22 cells and their knockdown detected with protease-specific antibodies. C, immunofluorescent detection of either normal (N586–20Q) or mutant (N586–128Q) Htt with an antibody to residues 1–82 is shown in blue (Alexa Fluor 350), and active caspase-3 is shown in red (Cy3). D, the graph shows percentage of cells with activated caspase-3 among transfected cells stained with an Htt-specific antibody. At least 200–300 cells were counted for each condition in one experiment. The experiment was repeated three or four times (n = 3 or n = 4; p = 0.03 nontargeting siRNA versus BLMH siRNA with Htt-N586–128Q; p = 0.028 nontargeting siRNA versus cathepsin Z siRNA with Htt-N586–128Q). cont, control.
DISCUSSION
The nature of proteolytic enzymes generating the short N-terminal fragments of Htt observed in many systems remains unknown. In this study, we have carried out a systematic siRNA screen of all known human proteases to determine which enzymes are capable of cleaving mutant Htt to form short N-terminal fragments. Our initial screens identified 11 enzymes. Further functional studies in two different cell lines resulted in two major candidates, BLMH and cathepsin Z. We have demonstrated that transfection of cDNAs and inhibition by siRNAs to BLMH and cathepsin Z had opposite effects, with the siRNAs inhibiting cleavage and the cDNAs enhancing cleavage of Htt. We found that both of these enzymes could generate the cp-2 fragment, specifically detected with our neoepitope antibody to the cp-2 site ending at position 167. In addition, we showed that the incubation of the cell lysates with recombinant enzymes induces Htt proteolysis at cp-1/cp-2 sites. By contrast, there was little or no increase in fragments when using the construct with a deleted cp-2 site (N511–52Q-RLQL). Both enzymes were found to at least partially co-localize with Htt in the cytoplasm and within or in association with early and late endosomes, with some possible nuclear co-localization observed for cathepsin Z. Finally, our functional assay indicates that cleavage by these enzymes at this position has a role in mutant Htt cell toxicity.
The primary screen was based on our earlier studies in HEK293 and stable PC12 cells, where we demonstrated the formation of two closely migrating N-terminal fragments generated from full-length Htt in both cell systems (32). These fragments, which we call cp-1 and cp-2, may be different from the previously described cp-A and cp-B (5) because they can be detected without proteasome inhibition, and they are not affected by a deletion of residues 105–114 of Htt. More recently, we have characterized cp-1 and cp-2 fragments derived from truncated Htt, comparable with length to caspase-6 fragment (N586), which may be a pathogenic intermediate. The cp-2 fragment was mapped by mass spectrometry to position Arg167 of Htt (31).
A number of features of our experimental design facilitate the identification of specific cleavage of Htt. For the primary screen, we developed a Western-based assay performed in highly transfectable HEK293 cells. These cells were transiently transfected with a construct expressing the first 511 amino acids of Htt with 52Q and an N-terminal FLAG tag for reliable detection of cp-1/cp-2 fragments. Although these two fragments can also be easily detected with antibodies to the N terminus or exon 1 of Htt (Fig. 4), Htt-specific antibodies may also detect possible endogenous fragments formed from the full-length Htt with normal repeat length, which would complicate the interpretation of results. Therefore, we chose to use the FLAG antibody for the screen. Because the Htt-N511 construct does not contain cleavage sites previously mapped to caspases (residues 513–586), it may be cleaved into short N-terminal fragments via caspase-independent pathways (32). Thus, our assay was specifically designed to search for the enzymes involved in the production of the cp-1 and cp-2 fragments. The assays were all conducted in the absence of proteasome inhibition to provide a physiological environment. Although our initial screen was conducted in HEK293 cells, the follow-up experiments were carried out in both HEK293 and HT22 neuronal cells.
There are several limitations of our approach. For technical reasons, screens and assays were carried out in cell lines rather than primary neurons as biochemical studies are very difficult in primary neurons. However, we did use the HT22 neuronal cell line, which is suitable for functional studies such as Htt toxicity (31). Although incubation of the cell lysates containing Htt with recombinant enzymes induced Htt proteolysis at cp-1/cp-2 sites, we were unable to demonstrate a direct proteolysis of in vitro-translated Htt by BLMH and cathepsin Z. It is possible that additional activation of recombinant BLMH and cathepsin Z by cofactors or proteases present in the cell lysate is required. Another possibility is that certain conformation of Htt protein acquired through its interaction with adaptors or other proteins may be necessary for the above proteolysis to take place, which cannot be achieved in the cell-free system.
A recent report by the Ellerby/Hughes research team describes a similar screen for proteases capable of yielding short N-terminal fragments of Htt (41). However, there are a number of important differences in the experimental design between these two studies, explaining different (possibly complementary) findings. The initial screen in the cited study was designed to examine the generation of the smallest N-terminal fragment produced from full-length Htt, corresponding to the range of cp-1/cp-2 (cp-A/cp-B), within the N-terminal 200 amino acids. The screen identified several members of the matrix metalloprotease family. One enzyme, MMP10, was capable of cleaving Htt in vitro yielding a fragment of 402 amino acids, which does not correspond to the fragment sought in the initial screen. Although Htt was found to be a substrate for MMP10, the above screen fell short of identifying enzymes that cleave Htt further toward the N terminus, in the region of the cp-1/cp-2-like fragments. It is conceivable, however, that MMP10 initially cleaves full-length Htt, which is followed by subsequent proteolysis carried out by other proteases (e.g. BLMH and cathepsin Z), yielding cp-1/cp-2 size fragments. We did find two members of the matrix metalloprotease family, MMP2 and MMP15, among our hits from the primary screen, performed with 75 nm siRNA (Fig. 2). However, these enzymes did not affect the cleavage in the next screening step using lower concentration of siRNA and were not confirmed in neuronal cells. In the above report, an even higher concentration of siRNAs (136 nm) was used throughout the study, which raises concerns about possible off-target effects. There are some other findings in the described study that are difficult to interpret; co-localization of MMP10 and Htt was observed predominantly in dead cells, so the physiological relevance is unclear; in an HD Drosophila model, many different MMPs showed some degree of amelioration of the HD phenotype, rendering specificity uncertain; the effects that proteasome inhibitors might have on Htt proteolysis were not defined. Finally, MMPs are secretory pathway enzymes, which are known to process extracellular matrix and cytokines, which may modulate toxicity. Thus, the mechanism of their action on mutant Htt toxicity may be indirect.
Both BLMH and cathepsin Z belong to the cysteine protease family with a papain-like active site structure. The proteases of this family have been implicated previously in Htt proteolysis; calpain-mediated cleavage of Htt (at residues 469 and 536) may be linked to mutant Htt toxicity (10, 21–23, 25). Our previous findings indicate that calpain may be also involved in degradation of cp-1/cp-2 fragments as calpain inhibitors caused increased fragment accumulation in PC12 cells (32). Cathepsins have been also found to contribute to Htt degradation and accumulation of stable N-terminal fragments in clonal striatal cells (42). Our new findings that BLMH and cathepsin Z mediate Htt proteolysis and toxicity further validate this protease family, and BLMH and cathepsin Z in particular, as potential novel targets for HD therapeutics. The other members of this family (and the other proteases identified in our screen) have not been further explored in the current study and might merit additional investigation.
BLMH was first identified by its ability to inactivate the anti-cancer drug bleomycin (43). It is a self-compartmentalizing neutral protease that forms a hexameric barrel structure in the cytoplasm with six active sites embedded in the channel, somewhat similar to the proteasome (44, 45). BLMH substrate specificity is diverse and is controlled by the position of its own C terminus in the active site, as opposed to the specific sequence in the substrate (45). The enzyme acts as a carboxypeptidase on its own C terminus to convert itself into an aminopeptidase; however, its intrinsic endopeptidase activity was also demonstrated (46). Acting as an endopeptidase, it can cleave amyloid β peptide at positions His14 and Phe19 (47). Thus, BLMH has been implicated in Alzheimer disease in the regulation of processing of amyloid precursor protein (48) with increased BLMH reactivity found in senile plaque in the human brain (49). BLMH is widely expressed in neurons in all brain regions (50). Because there are no highly specific BLMH inhibitors, we have not attempted further pharmacological experiments.
Cathepsin Z is also a member of the cysteine protease family with a papain-like structure (51). It is widely expressed via housekeeping-like promoter (52), including all areas of brain, predominantly in glia, but also in neurons (53). Cathepsin Z is a carboxypeptidase (54) with broad substrate specificity, which excludes proline in the vicinity of the cleavage site (55). Whether it can act as an endopeptidase has not been studied in detail. Similar to other cathepsins, cathepsin Z is primarily a lysosomal enzyme, but it has also been found in the cytoplasm (53). Cathepsins are known to be mainly involved in terminal protein degradation. However, recently distinct cathepsins have been suggested to have specific physiological functions. One example is a related protease cathepsin L; an isoform devoid of signal peptide was found in the nucleus, where it regulates cell cycle progression (56). Of potential interest, we have found some endogenous or transfected cathepsin Z reactivity in the nucleus of HT22 cells (Fig. 6), though interpretation should be cautious until this can be studied with more antibodies. Cathepsin Z can be activated by other proteases, including cathepsin L. This enzyme was found to be up-regulated in glial cells of degenerating brain regions in a mouse model of amyotrophic lateral sclerosis and associated with plaques in amyloid precursor protein/PS1 mice and AD postmortem material (53).
These two proteases that we implicate in the cleavage of Htt have not been extensively studied, so the manner by which they might act on Htt is uncertain. It is conceivable that they could act in tandem, with BLMH acting as an endopeptidase and cathepsin Z following as a carboxypepsidase to trim amino acids back to position Arg167, which is preceded by proline. This is consistent with our observation that co-transfection of both proteases has an additive effect on the cp-2 cleavage. However, it is possible that other proteolytic enzymes may be also involved at this step because cathepsin Z does not favor proline in the penultimate position (P2). This mechanism is compatible with our previous study where we found that the cp-2 fragment, ending at Arg167, appears to be one component of Htt aggregates with other cp-1/cp-2-like fragments of similar size present as well (31). Another possibility is that these two enzymes act on Htt independently, even in different subcellular compartments and at different time points during Htt protein metabolic of functional pathway. Both BLMH and cathepsin Z exhibit mostly positional specificity less dependent on the substrate sequence. Our in vitro cleavage experiments showed that cathepsin Z can cleave Htt at multiple sites (Fig. 5). Therefore, the role of BLMH and cathepsin Z in Htt proteolysis could also result from the enzyme and substrate co-localization in a specific cellular environment and may include other molecules acting as modulators or adaptors. This may explain the N-terminal cleavage variability observed in different systems.
Our immunofluorescence studies demonstrate that Htt co-localizes with both BLMH and cathepsin Z in the cytoplasm and at the early and late endosomes. Because the methods used in this study do not allow establishing whether such co-localization occurs inside the endosomes or on their outer membrane facing cytosol or both, there are several possibilities for interaction.
Htt has been shown to associate with acidic phospholipids (39) and it is palmitoylated at cysteine 214, by which it may attach to the cytoplasmic face of intracellular membranes (57). Co-localization of Htt with various membranous vesicles is well established and is consistent with the Htt role in vesicular trafficking and with the function of Htt-interacting partners (35, 38–40, 58, 59). Htt modulates brain-derived neurotrophic factor vesicular transport along microtubules via its interaction with HAP1 (Htt-associated protein-1)/p150Glued dynactin complex, which is altered in HD (60, 61). HAP40 interaction with Htt has been shown to affect Rab5-mediated endosomal motility. The latter is impaired in HD cells due to up-regulation of HAP40 (62). Htt is important for activation of Rab11-GTPase, and mutant Htt impairs vesicle formation from recycling endosomes by interfering with Rab11 activity (63, 64). Future studies will determine whether the cleavage of Htt by BLMH and cathepsin Z occurs on the outer surface of early and late endosomes and whether it is linked to Htt function in vesicular transport and/or disruption of these processes in HD conditions.
Htt was previously found within endosomal-lysosomal-like organelles in clonal striatal cells, and the endosomal-lysosomal system was suggested to be a major path for Htt degradation and proteolysis (65). Our data suggest that Htt N-terminal proteolysis can occur within the endosomes or at the close proximity to endosomes in the cytoplasm. However, the possibility that this cleavage occurs elsewhere in the cytoplasm or nucleus (by cathepsin Z) cannot be excluded. Cleavage of Htt within the cytoplasm would be in agreement with most current models of HD pathogenesis. It is also conceivable that Htt proteolysis in the secretory pathway could be relevant to HD. Recent studies have shown that polyglutamine peptide aggregates can penetrate the cell membrane and enter the cytosolic compartment (66), and it was suggested that the prion-like transmission of protein aggregates between cells may play a role in neurodegenerative diseases (67).
In summary, although there are a number of limitations to our approach, we provide convergent evidence from several different kinds of experiments suggesting that cleavage of Htt at the cp-2 site by BLMH and cathepsin Z could be involved in HD pathogenesis. In particular, our data indicate that these enzymes may be involved in promotion of mutant Htt-induced cell toxicity. To more conclusively demonstrate a role for these cleavage events in vivo, it would be useful to conduct genetic cross experiments between knock-outs of BLMH or cathepsin Z and HD models expressing either the caspase-6 fragment or full-length Htt. BLMH knockouts demonstrate astrogliosis and behavioral changes (68), whereas cathepsin Z-null mice have normal phenotype, possibly due to functional compensation by cathepsin family members. These crosses would be interesting future experiments to test in vivo the role of BLMH and cathepsin Z in cleavage of Htt and in HD pathogenesis. The studies we report here provide the initial evidence that these enzymes contribute to mutant Htt proteolysis and toxicity in cells and may be potential therapeutic targets for HD.
Acknowledgment
We thank Michael Hayden (University of British Columbia, Vancouver) for providing us with Htt expression constructs.
This work was supported, in whole or in part, by NINDS, National Institutes of Health, Grant 16375. This work was also supported by HDSA Coalition for the Cure and the High Q Foundation CHDI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
E. Waldron, T. Ratovitski, X. Wang, M. Jiang, W. Duan, S. Mori, D. Borchelt, D. Swing, L. Tessarollo, and C. Ross, manuscript in preparation.
- HD
- Huntington disease
- polyQ
- polyglutamine
- BLMH
- bleomycin hydrolase
- MMP
- matrix metallopeptidase.
REFERENCES
- 1. Huntington Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Ross C. A., Margolis R. L., Rosenblatt A., Ranen N. G., Becher M. W., Aylward E. (1997) Medicine 76, 305–338 [DOI] [PubMed] [Google Scholar]
- 3. Becher M. W., Kotzuk J. A., Sharp A. H., Davies S. W., Bates G. P., Price D. L., Ross C. A. (1998) Neurobiol. Dis. 4, 387–397 [DOI] [PubMed] [Google Scholar]
- 4. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 5. Mende-Mueller L. M., Toneff T., Hwang S. R., Chesselet M. F., Hook V. Y. (2001) J. Neurosci. 21, 1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schilling G., Klevytska A., Tebbenkamp A. T., Juenemann K., Cooper J., Gonzales V., Slunt H., Poirer M., Ross C. A., Borchelt D. R. (2007) J. Neuropathol. Exp. Neurol. 66, 313–320 [DOI] [PubMed] [Google Scholar]
- 7. Vonsattel J. P. (2008) Acta. Neuropathol. 115, 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ross C. A., Tabrizi S. J. (2011) Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 9. Cooper J. K., Schilling G., Peters M. F., Herring W. J., Sharp A. H., Kaminsky Z., Masone J., Khan F. A., Delanoy M., Borchelt D. R., Dawson V. L., Dawson T. M., Ross C. A. (1998) Hum. Mol. Genet. 7, 783–790 [DOI] [PubMed] [Google Scholar]
- 10. Gafni J., Ellerby L. M. (2002) J. Neurosci. 22, 4842–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi S., Morita H., Bennett K. M., Tanaka Y., Engelender S., Peters M. F., Cooper J. K., Wood J. D., Sawa A., Ross C. A. (2003) Neuroreport. 14, 565–568 [DOI] [PubMed] [Google Scholar]
- 12. Kim M., Lee H. S., LaForet G., McIntyre C., Martin E. J., Chang P., Kim T. W., Williams M., Reddy P. H., Tagle D., Boyce F. M., Won L., Heller A., Aronin N., DiFiglia M. (1999) J. Neurosci. 19, 964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lunkes A., Lindenberg K. S., Ben-Haïem L., Weber C., Devys D., Landwehrmeyer G. B., Mandel J. L., Trottier Y. (2002) Mol. Cell. 10, 259–269 [DOI] [PubMed] [Google Scholar]
- 14. Lunkes A., Mandel J. L. (1998) Hum. Mol. Genet. 7, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 15. Rubinsztein D. C. (2002) Trends Genet. 18, 202–209 [DOI] [PubMed] [Google Scholar]
- 16. Schilling G., Becher M. W., Sharp A. H., Jinnah H. A., Duan K., Kotzuk J. A., Slunt H. H., Ratovitski T., Cooper J. K., Jenkins N. A., Copeland N. G., Price D. L., Ross C. A., Borchelt D. R. (1999) Hum. Mol. Genet. 8, 397–407 [DOI] [PubMed] [Google Scholar]
- 17. Sun B., Fan W., Balciunas A., Cooper J. K., Bitan G., Steavenson S., Denis P. E., Young Y., Adler B., Daugherty L., Manoukian R., Elliott G., Shen W., Talvenheimo J., Teplow D. B., Haniu M., Haldankar R., Wypych J., Ross C. A., Citron M., Richards W. G. (2002) Neurobiol. Dis. 11, 111–122 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka Y., Igarashi S., Nakamura M., Gafni J., Torcassi C., Schilling G., Crippen D., Wood J. D., Sawa A., Jenkins N. A., Copeland N. G., Borchelt D. R., Ross C. A., Ellerby L. M. (2006) Neurobiol. Dis. 21, 381–391 [DOI] [PubMed] [Google Scholar]
- 19. Zhou H., Cao F., Wang Z., Yu Z. X., Nguyen H. P., Evans J., Li S. H., Li X. J. (2003) J. Cell. Biol. 163, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 21. Bizat N., Hermel J. M., Boyer F., Jacquard C., Créminon C., Ouary S., Escartin C., Hantraye P., Kajewski S., Brouillet E. (2003) J. Neurosci. 23, 5020–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gafni J., Hermel E., Young J. E., Wellington C. L., Hayden M. R., Ellerby L. M. (2004) J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- 23. Goffredo D., Rigamonti D., Tartari M., De Micheli A., Verderio C., Matteoli M., Zuccato C., Cattaneo E. (2002) J. Biol. Chem. 277, 39594–39598 [DOI] [PubMed] [Google Scholar]
- 24. Hermel E., Gafni J., Propp S. S., Leavitt B. R., Wellington C. L., Young J. E., Hackam A. S., Logvinova A. V., Peel A. L., Chen S. F., Hook V., Singaraja R., Krajewski S., Goldsmith P. C., Ellerby H. M., Hayden M. R., Bredesen D. E., Ellerby L. M. (2004) Cell. Death. Differ. 11, 424–438 [DOI] [PubMed] [Google Scholar]
- 25. Kim M., Roh J. K., Yoon B. W., Kang L., Kim Y. J., Aronin N., DiFiglia M. (2003) Exp. Neurol. 183, 109–115 [DOI] [PubMed] [Google Scholar]
- 26. Sawa A., Nagata E., Sutcliffe S., Dulloor P., Cascio M. B., Ozeki Y., Roy S., Ross C. A., Snyder S. H. (2005) Neurobiol. Dis. 20, 267–274 [DOI] [PubMed] [Google Scholar]
- 27. Wellington C. L., Ellerby L. M., Gutekunst C. A., Rogers D., Warby S., Graham R. K., Loubser O., van Raamsdonk J., Singaraja R., Yang Y. Z., Gafni J., Bredesen D., Hersch S. M., Leavitt B. R., Roy S., Nicholson D. W., Hayden M. R. (2002) J. Neurosci. 22, 7862–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wellington C. L., Ellerby L. M., Hackam A. S., Margolis R. L., Trifiro M. A., Singaraja R., McCutcheon K., Salvesen G. S., Propp S. S., Bromm M., Rowland K. J., Zhang T., Rasper D., Roy S., Thornberry N., Pinsky L., Kakizuka A., Ross C. A., Nicholson D. W., Bredesen D. E., Hayden M. R. (1998) J. Biol. Chem. 273, 9158–9167 [DOI] [PubMed] [Google Scholar]
- 29. Wellington C. L., Singaraja R., Ellerby L., Savill J., Roy S., Leavitt B., Cattaneo E., Hackam A., Sharp A., Thornberry N., Nicholson D. W., Bredesen D. E., Hayden M. R. (2000) J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- 30. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Hou Z., Liang Y., Poirier M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., Ross C. A. (2009) J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ratovitski T., Nakamura M., D'Ambola J., Chighladze E., Liang Y., Wang W., Graham R., Hayden M. R., Borchelt D. R., Hirschhorn R. R., Ross C. A. (2007) Cell. Cycle. 6, 2970–2981 [DOI] [PubMed] [Google Scholar]
- 33. Slow E. J., Graham R. K., Osmand A. P., Devon R. S., Lu G., Deng Y., Pearson J., Vaid K., Bissada N., Wetzel R., Leavitt B. R., Hayden M. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11402–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L. M., Trottier Y., Richards W. G., Osmand A., Paganetti P., Bates G. P. (2010) J. Biol. Chem. 285, 8808–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharp A. H., Loev S. J., Schilling G., Li S. H., Li X. J., Bao J., Wagster M. V., Kotzuk J. A., Steiner J. P., Lo A., et al. (1995) Neuron 14, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 36. Peters M. F., Ross C. A. (2001) J. Biol. Chem. 276, 3188–3194 [DOI] [PubMed] [Google Scholar]
- 37. Atwal R. S., Xia J., Pinchev D., Taylor J., Epand R. M., Truant R. (2007) Hum. Mol. Genet. 16, 2600–2615 [DOI] [PubMed] [Google Scholar]
- 38. DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J. P., Carraway R., Reeves S. A., et al. (1995) Neuron 14, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 39. Kegel K. B., Sapp E., Yoder J., Cuiffo B., Sobin L., Kim Y. J., Qin Z. H., Hayden M. R., Aronin N., Scott D. L., Isenberg G., Goldmann W. H., DiFiglia M. (2005) J. Biol. Chem. 280, 36464–36473 [DOI] [PubMed] [Google Scholar]
- 40. Velier J., Kim M., Schwarz C., Kim T. W., Sapp E., Chase K., Aronin N., DiFiglia M. (1998) Exp. Neurol. 152, 34–40 [DOI] [PubMed] [Google Scholar]
- 41. Miller J. P., Holcomb J., Al-Ramahi I., de Haro M., Gafni J., Zhang N., Kim E., Sanhueza M., Torcassi C., Kwak S., Botas J., Hughes R. E., Ellerby L. M. (2010) Neuron 67, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim Y. J., Sapp E., Cuiffo B. G., Sobin L., Yoder J., Kegel K. B., Qin Z. H., Detloff P., Aronin N., DiFiglia M. (2006) Neurobiol. Dis. 22, 346–356 [DOI] [PubMed] [Google Scholar]
- 43. Akiyama S., Ikezaki K., Kuramochi H., Takahashi K., Kuwano M. (1981) Biochem. Biophys. Res. Commun. 101, 55–60 [DOI] [PubMed] [Google Scholar]
- 44. O'Farrell P. A., Gonzalez F., Zheng W., Johnston S. A., Joshua-Tor L. (1999) Structure 7, 619–627 [DOI] [PubMed] [Google Scholar]
- 45. Zheng W., Johnston S. A., Joshua-Tor L. (1998) Cell 93, 103–109 [DOI] [PubMed] [Google Scholar]
- 46. Koldamova R. P., Lefterov I. M., Gadjeva V. G., Lazo J. S. (1998) Biochemistry 37, 2282–2290 [DOI] [PubMed] [Google Scholar]
- 47. Kajiya A., Kaji H., Isobe T., Takeda A. (2006) Protein. Pept. Lett. 13, 119–123 [DOI] [PubMed] [Google Scholar]
- 48. Lefterov I. M., Koldamova R. P., Lazo J. S. (2000) FASEB. J. 14, 1837–1847 [DOI] [PubMed] [Google Scholar]
- 49. Namba Y., Ouchi Y., Takeda A., Ueki A., Ikeda K. (1999) Brain. Res. 830, 200–202 [DOI] [PubMed] [Google Scholar]
- 50. Kamata Y., Itoh Y., Kajiya A., Karasawa S., Sakatani C., Takekoshi S., Osamura R. Y., Takeda A. (2007) J. Biochem. 141, 69–76 [DOI] [PubMed] [Google Scholar]
- 51. Nägler D. K., Ménard R. (1998) FEBS Lett. 434, 135–139 [DOI] [PubMed] [Google Scholar]
- 52. Deussing J., von Olshausen I., Peters C. (2000) Biochim. Biophys. Acta. 1491, 93–106 [DOI] [PubMed] [Google Scholar]
- 53. Wendt W., Zhu X. R., Lübbert H., Stichel C. C. (2007) Exp. Neurol. 204, 525–540 [DOI] [PubMed] [Google Scholar]
- 54. Nägler D. K., Zhang R., Tam W., Sulea T., Purisima E. O., Ménard R. (1999) Biochemistry 38, 12648–12654 [DOI] [PubMed] [Google Scholar]
- 55. Devanathan G., Turnbull J. L., Ziomek E., Purisima E. O., Ménard R., Sulea T. (2005) Biochem. Biophys. Res. Commun. 329, 445–452 [DOI] [PubMed] [Google Scholar]
- 56. Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. (2004) Mol. Cell. 14, 207–219 [DOI] [PubMed] [Google Scholar]
- 57. Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R. (2006) Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harjes P., Wanker E. E. (2003) Trends Biochem. Sci. 28, 425–433 [DOI] [PubMed] [Google Scholar]
- 59. Truant R., Atwal R., Burtnik A. (2006) Biochem. Cell. Biol. 84, 912–917 [DOI] [PubMed] [Google Scholar]
- 60. Engelender S., Sharp A. H., Colomer V., Tokito M. K., Lanahan A., Worley P., Holzbaur E. L., Ross C. A. (1997) Hum. Mol. Genet. 6, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 61. Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., Saudou F. (2004) Cell 118, 127–138 [DOI] [PubMed] [Google Scholar]
- 62. Pal A., Severin F., Lommer B., Shevchenko A., Zerial M. (2006) J. Cell. Biol. 172, 605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li X., Sapp E., Valencia A., Kegel K. B., Qin Z. H., Alexander J., Masso N., Reeves P., Ritch J. J., Zeitlin S., Aronin N., Difiglia M. (2008) Neuroreport 19, 1643–1647 [DOI] [PubMed] [Google Scholar]
- 64. Li X., Standley C., Sapp E., Valencia A., Qin Z. H., Kegel K. B., Yoder J., Comer-Tierney L. A., Esteves M., Chase K., Alexander J., Masso N., Sobin L., Bellve K., Tuft R., Lifshitz L., Fogarty K., Aronin N., DiFiglia M. (2009) Mol. Cell. Biol. 29, 6106–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kegel K. B., Kim M., Sapp E., McIntyre C., Castaño J. G., Aronin N., DiFiglia M. (2000) J. Neurosci. 20, 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ren P. H., Lauckner J. E., Kachirskaia I., Heuser J. E., Melki R., Kopito R. R. (2009) Nat. Cell. Biol. 11, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brundin P., Melki R., Kopito R. (2010) Nat. Rev. Mol. Cell. Biol. 11, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Montoya S. E., Thiels E., Card J. P., Lazo J. S. (2007) Neuroscience 146, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]