Abstract
In multicellular organisms, cells are interconnected by cell adhesion molecules. Nectins are immunoglobulin (Ig)-like cell adhesion molecules that mediate homotypic and heterotypic cell-cell adhesion, playing key roles in tissue organization. To mediate cell-cell adhesion, nectin molecules dimerize in cis on the surface of the same cell, followed by trans-dimerization of the cis-dimers between the neighboring cells. Previous cell biological studies deduced that the first Ig-like domain of nectin and the second Ig-like domain are involved in trans-dimerization and cis-dimerization, respectively. However, to understand better the steps involved in nectin adhesion, the structural basis for the dimerization of nectin must be determined. In this study, we determined the first crystal structure of the entire extracellular region of nectin-1. In the crystal, nectin-1 formed a V-shaped homophilic dimer through the first Ig-like domain. Structure-based site-directed mutagenesis of the first Ig-like domain identified four essential residues that are involved in the homophilic dimerization. Upon mutating the four residues, nectin-1 significantly decreased cis-dimerization on the surface of cultured cells and abolished the homophilic and heterophilic adhesion activities. These results indicate that, in contrast with the previous notion, our structure represents a cis-dimer. Thus, our findings clearly reveal the structural basis for the cis-dimerization of nectins through the first Ig-like domains.
Keywords: Cell Adhesion, Cell Junctions, Cell-Cell Interaction, Protein Structure, X-ray Crystallography
Introduction
In multicellular organisms, cells are interconnected by cell-cell junctions, which are involved in embryogenesis, tissue organization, and tissue homeostasis. Cell adhesion molecules (CAMs)3 are the main adhesive components of the cell-cell junctions (1–3). CAMs trans-interact with the CAMs on the neighboring cells via their extracellular regions. CAMs assemble into a ladder-like configuration, thereby converting their trans-interactions into physiological cell-cell adhesion (1–3). Disruption of cell-cell adhesion causes various serious diseases, including carcinogenesis and cancer metastasis. Therefore, it is important to understand how CAMs organize cell-cell adhesion. Of the CAMs identified to date, numerous studies have focused on cadherins because they are the main CAMs in adherens junctions (1, 3, 4). The extracellular regions of cadherins homophilically interact in trans with each other (1, 3, 4), mediating homotypic cell-cell adhesion involved in embryogenesis, tissue morphogenesis, and neural development (1, 3, 4). Crystal structures of the extracellular regions of cadherins have provided structural insights into the trans-interaction of cadherins. However, different studies have proposed different models for the minimum essential adhesive units underlying the conversion of the trans-interactions into cadherin adhesion (5, 6). Currently, the molecular mechanisms that enable CAMs to assemble and densely pack into the cell-cell junctions remain poorly understood.
Nectins are immunoglobulin (Ig)-like CAMs that mediate both homotypic and heterotypic cell-cell adhesion. Nectins comprise a family with four members (2, 7–9), and the primary sequence analysis has predicted that each member of the nectin family contains an extracellular region with three Ig-like domains, a single transmembrane region, and a cytoplasmic tail region (2, 9). The cytoplasmic tail region is responsible for binding to the PDZ domain of afadin, an actin-filament-binding protein (10). Nectins homophilically interact in cis on the same cell surface and homophilically and heterophilically interact in trans between neighboring cells, thereby mediating homotypic cell-cell adhesion as well as heterotypic one (2, 7–9). Of the combinations of trans-interactions, the trans-interaction between nectins-1 and -3 is the strongest (11). Indeed, the trans-interaction between nectins-1 and -3 plays important roles in various aspects of the central nervous system, including axon guidance and synaptogenesis (2, 7–9). It has been deduced from previous cell biological and biochemical studies that the first Ig-like domain of nectin is necessary for the formation of trans-dimers, but not for cis-dimers, whereas the second Ig-like domain contributes to the formation of cis-dimers (12–14). The function of the third Ig-like domain is currently unknown. It has been also deduced from intermolecular force microscopy measurement that nectins can adhere through another state, in addition to the adhesion mediated by interactions between the first Ig-like domains (15). In this novel adhesion state, the first, second, and third Ig-like domains interact in trans with the third, second, and first Ig-like domains of another nectin molecule, respectively, in a zipper-like fashion. However, the adhesive strength induced by this state is weaker than that mediated by the interaction between the first Ig-like domains. To understand nectin adhesion better, the structural basis for the interactions between all of the extracellular domains of nectins must be determined.
Evidence is accumulating that the formation of adherens junctions is guided by nectins (2, 7–9). Nectins first form the initial cell-cell contact between two neighboring cells and encircle the cells in a belt-like fashion, along with assembly of peripheral actin filament bundles. Following the formation of the nectin-based cell-cell adhesions, cadherin is recruited to the nectin-based adhesion sites, eventually forming strong cell-cell adhesions. In addition to the cooperative role of nectins with cadherins, nectins can organize cell-cell adhesion, particularly heterotypic adhesion, in a cadherin-independent manner (2, 7–9, 16, 17). During early development of the vertebrate central nervous system, nectins-1 and -3 but not cadherins are expressed in commissural axons and floor plate cells, respectively (17). The trans-interaction between nectins-1 and -3 dominantly organizes the heterotypic cell-cell adhesion between commissural axons and floor plate cells and regulates changes in the trajectory of commissural axons (17). Nectins also play a dominant role in germ cell differentiation in the testis. In the latter half of spermatogenesis, spermatids form prominent heterotypic cell-cell junctions with Sertoli cells called Sertoli cell-spermatid junctions, where the existence of the cadherin system has been questionable (16). Nectins-2 and -3 reside in Sertoli cells and spermatids, respectively, and the trans-interaction between nectins-2 and -3 regulates the organization of the Sertoli cell-spermatid junctions (16). Therefore, structural insight into nectin-mediated cell adhesion is required to understand the molecular anatomy of cell-cell adhesion better. Mutations in the nectin gene are also associated with some disorders whose pathologies are characterized by cleft lip/palate, syndactyly, mental retardation, and ectodermal dysplasia (18–21). In addition, nectins regulate many other cellular activities such as movement, proliferation, survival, differentiation, polarization, and viral entry, in cooperation with other CAMs and cell surface membrane receptors (2, 7–9, 22). Thus, understanding the architecture of nectin adhesion is particularly important.
Here, we show the crystal structure of the entire extracellular region of nectin-1. In the crystal, nectin-1 forms a pseudo 2-fold symmetric and a V-shaped homophilic dimer through polar interactions between the first Ig-like domains. Structure-based site-directed mutagenesis studies of the first Ig-like domain identified four residues that are essential for the homophilic dimerization. Upon mutating the four residues, nectin-1 significantly decreased cis-dimerization on the surface of the cultured cells and lost homoptypic adhesion as well as heterotypic adhesion with nectin-3. Therefore, these results reveal that nectin cis-dimerizes through the first Ig-like domain. Based on our findings, we discuss the role of the cis-dimer of nectin as an adhesive unit in the formation of nectin-based cell-cell junctions. To our knowledge, this is the first report of the crystal structure of this member of the nectin family.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
cDNAs corresponding to the extracellular region of human nectin-1α (residues 30–335) (nectin-1-EC) and mouse nectin-1α mutant (residues 30–335, T63A, Q64A, E125A, and N133A) (nectin-1-EC-4mut) were subcloned into the BamHI and NotI sites of the prokaryotic expression vector pET21b(+) (Novagen), which encodes a C-terminal His6 tag. The proteins were expressed in Escherichia coli strain BL21 (DE3) (Novagen) as inclusion bodies. The inclusion bodies were dissolved in a buffer containing 50 mm MES-NaOH, pH 6.0, 8 m urea, 1 mm EDTA, 1 mm DTT. The samples were then refolded by 300-fold dilution into refolding buffer containing 500 mm l-arginine, 100 mm Tris-HCl, pH 9.0, 2 mm oxidized glutathione, and 1 mm reduced glutathione, followed by incubation at 4 °C for 48 h. The samples were subjected to size-exclusion chromatography on a Hiload 16/60 Superdex 200 column (GE Healthcare). The fractions indicative of the oligomeric form of nectin-1-EC proteins were dialyzed against 20 mm MES-NaOH, pH 6.0, to precipitate almost misfolded proteins, and subjected to a HiTrap SP HP column (GE Healthcare) followed by a Mono Q column (GE Healthcare). Overall, ∼0.5 mg of the refolded nectin-1-EC and ∼0.1 mg of the refolded nectin-1-EC-4mut were purified from 0.1 g of inclusion bodies.
To express the native nectin-1-EC protein, cDNA corresponding to the signal peptide and the extracellular region of mouse nectin-1α (residues 1–335) was subcloned into mammalian expression vector pcDNA3.1 with a C-terminal extension consisting of a TEV cleavage site and a His8 tag followed by a myc tag and a His6 tag, and transfected into HEK293 cells with Effectene (Qiagen). Cells stably expressing and secreting nectin-1-EC with the C-terminal His8-myc-His6 tag (nectin-1-EC-myc) were selected by culturing the cells in the presence of 500 μg/ml G418. The native nectin-1-EC-myc was purified with nickel-agarose (Qiagen) from the conditioned medium in accordance with the manufacturer's instruction. The extracellular region of mouse nectin-3α fused to an Fc fragment of the human IgG (Nef-3) was prepared as described previously (23, 24).
Size-exclusion Chromatographic Analysis
For the binding of nectin-1-EC to Nef-3, 1.1 μm nectin-1-EC, 1.1 μm Nef-3, and a mixture containing 1.1 μm each nectin-1-EC and Nef-3 were incubated in a buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm NaCl at 4 °C for 60 min. The samples (50 μl) were subjected to a Superdex 200 PC 3.2/30 (GE Healthcare) column and fractionated (100 μl/fraction). For analysis of the homophilic dimerization, nectin-1-EC and nectin-1-EC-4mut were concentrated to 1.5 mg/ml, and filtered using an Ultrafree-MC filter (Millipore). Then, 200 μl of the protein solutions was subjected to a Superdex 200 10/300 column (GE Healthcare) with 200 μl.
Crystallization and Data Collection
Crystals were observed in drops containing equal volumes of nectin-1-EC solution (5 mg/ml in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 6% 1,6-hexanediol) and precipitant solution (50 mm citric acid, 50 mm Bis-Tris propane and 1–3% PEG3350) at 23 °C. To improve the quality of crystal diffraction, the crystals were dehydrated using 25% PEG300 solution. A heavy atom derivative was obtained by soaking the crystals overnight in this buffer also containing 1 mm K2PtBr4. The diffraction datasets for native and derivative proteins were collected on a BL44XU at the SPring-8 synchrotron facility at 100 K using wavelengths of 0.9000 Å and 1.07195 Å, respectively. All datasets were processed and scaled with the HKL-2000 suite (25).
Structural Determination
The structure of nectin-1-EC was solved by single-wavelength anomalous dispersion with HKL2MAP interface (26). Model development and refinement were performed with COOT (27) and phenix.refine (28). The final model contains residues 36–336 of chain A, 33–336 of chain B, as well as 37 water molecules, one 1,6-hexanediol, and two citric acid molecules. The stereochemical quality of the final model was checked with MolProbity (29). This analysis showed no residues in disallowed regions (96.4% in favored regions and 3.6% in allowed regions) in the Ramachandran plot. Homophilic cis-dimerization was analyzed using PISA (30) and Contact (31). All structural figures were prepared using PyMOL (32).
Analytical Ultracentrifugation
Sedimentation velocity measurements were performed using a Beckman-Coulter Optima XL-I analytical ultracentrifuge (Beckman Coulter). 360 μl of protein solution at concentrations of 1.00, 0.67, and 0.33 mg/ml for nectin-1-EC and nectin-1-EC-4mut were first centrifuged at 3,000 rpm (700 × g) for 5 min to stabilize its temperature, followed by centrifugation at 50,000 rpm (201,600 × g). Absorbance data at 280 nm was measured for 5 h at 5-min intervals. All measurements were carried out at a constant temperature of 4 °C with a radial increment of 0.003 cm in the continuous scanning mode. Sedimentation velocity data were analyzed using the program SedAnal (33). For sedimentation equilibrium experiments on nectin-1-EC and nectin-1-EC-4mut, the protein solutions were centrifuged at concentrations of 1.20, 0.56, and 0.24 mg/ml. The rotor speeds were 12,000 rpm (11,612 × g) and 15,000 rpm (18,144 × g) for nectin-1-EC, and 15,000 rpm (18,144 × g) and 18,000 rpm (26,127 × g) for nectin-1-EC-4mut. The equilibrium concentration profiles were recorded by monitoring absorbance at 280 nm across the centrifugation cell with a radial increment of 0.001 cm in the continuous scanning mode. All measurements were carried out at a constant temperature of 4 °C. Sedimentation equilibrium data were analyzed using Beckman analytical software (34).
Establishment of Stable Transformants
cDNA encoding a full-length mouse nectin-1α mutant (T63A, Q64A, E125A, and N133A) (nectin-1-4mut) was subcloned into the pCAGIpuro vector (14) and the pPGKIH vector (12). L cells were transfected with pCAGIpuro-nectin-1-4mut or pPGKIH-nectin-1-4mut with Effectene (Qiagen). The cells were cultured for 1 day, replated, and selected by culturing in the presence of 5 μg/ml puromycin (Sigma) or 500 μg/ml hygromycin (InvivoGen), respectively, as described previously (12). For a trans-interaction-deficient mutant, cDNA for the full-length mouse nectin-1α mutant (F129L) was used instead of nectin-1-4mut.
Immunofluorescence Microscopy
The cells cultured on coverslips for 2–3 days were fixed in 1% formaldehyde, and blocked in PBS containing 1% bovine serum albumin (BSA) for 60 min without permeabilization. The samples were incubated with a rat anti-nectin-1 mAb raised against the extracellular region of nectin-1 (35), followed by incubation with an Alexa Fluor 488-labeled anti-rat IgG secondary Ab (Invitrogen) for 30 min. The specimens were analyzed by confocal laser scanning microscope as described previously (12, 36).
Chemical Cross-linking
Chemical cross-linking was performed as described previously (12, 13, 22) with a slight modification. Briefly, to obtain a single-cell suspension, the cells were incubated in 0.03125% trypsin and 1 mm EDTA at 37 °C for 5 min and dispersed by gentle pipetting. The cell suspension (1 × 106 cells/ml) was then incubated with PBS containing 1 mm bis-(sulfosuccinimidyl) suberate (BS3) (Pierce). After incubation at 14 °C for 15 min, the reaction was stopped by adding 10 mm Tris-HCl, pH 7.5. The cells were washed with PBS and counted to confirm that there was no aggregation in the cell suspension. The cells were then lysed and subjected to immunoblotting with a rabbit anti-nectin-1 pAb raised against the cytoplasmic region of nectin-1 (22, 36, 37).
Cell Aggregation Assay
The cell aggregation assay was performed as described previously (12, 13, 22, 23). Briefly, a single-cell suspension (1 × 106 cells/ml) was obtained as described above, and suspended in Hanks' balanced salt solution. The cells were then placed in 12-well plates precoated with BSA and rotated on a gyratory shaker at 37 °C for the indicated times. Aggregation was stopped with the addition of 2% glutaraldehyde. The extent of aggregation of cells is represented by the ratio of the total number of particles at time t of incubation (Nt) to the initial number of particles (No). In some instances, nectin-3-L cells were prelabeled with DiI (Invitrogen) as described previously (38).
RESULTS
Crystallization of the Extracellular Region of Nectin-1α
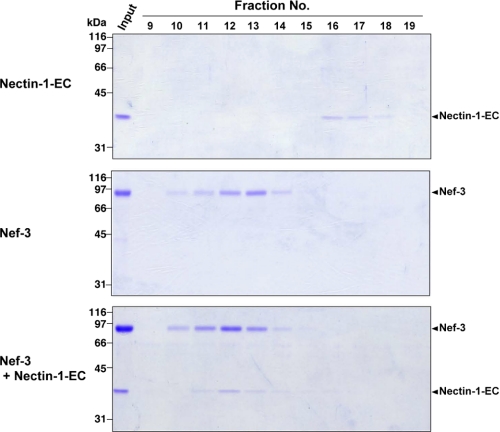
To gain insight into the structure of nectin, we first attempted to crystallize the entire extracellular region of nectin-1α (nectin-1-EC) expressed in HEK293 cells. Despite the high purity of the glycosylated nectin-1-EC proteins, no crystals were obtained, suggesting that glycosylation presumably inhibits the crystallization. Therefore, we next prepared the nonglycosylated form of nectin-1-EC using E. coli. Because nectin-1-EC formed inclusion bodies in E. coli, it was extracted with urea and refolded with rapid dilution method (see “Experimental Procedures”). The activity of the purified nectin-1-EC proteins was validated by its binding to the extracellular region of nectin-3α. Nectin-1-EC, the extracellular region of nectin-3α fused to an Fc fragment of the human IgG (Nef-3), or a mixture of nectin-1-EC and Nef-3 was subjected to size-exclusion chromatography followed by SDS-PAGE. Nectin-1-EC alone and Nef-3 alone were eluted in fractions 16–17 and 10–14, respectively (Fig. 1). On the other hand, in the mixture of nectin-1-EC and Nef-3, nectin-1-EC was co-eluted with Nef-3 in fractions 10–14. We also carried out similar assays using native nectin-1-EC expressed in HEK293 cells instead of the refolded one and observed that the native nectin-1-EC was also co-eluted with Nef-3 (supplemental Fig. 1, A and B). These results indicate that, similar to native nectin-1-EC, refolded nectin-1-EC has a potency to bind to nectin-3, allowing us to assume that the nectin-1-EC proteins are renatured. Therefore, we proceeded to crystallize the nectin-1-EC proteins and successfully obtained a sufficient number of crystals enough for high resolution analysis.
FIGURE 1.
Binding of nectin-1-EC to Nef-3. Nectin-1-EC proteins expressed in E. coli, Nef-3, and a mixture of the nectin-1-EC proteins and Nef-3 were subjected to a Superdex 200 column, respectively. The resulting fractions were subjected to SDS-PAGE followed by staining with Coomassie Brilliant Blue.
Crystal Structure of Nectin-1-EC
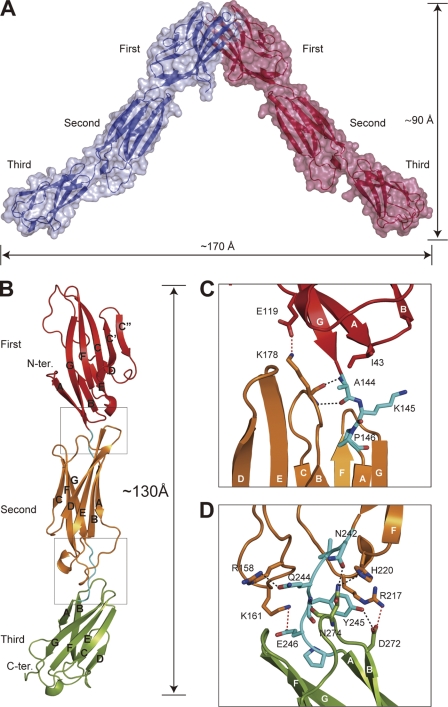
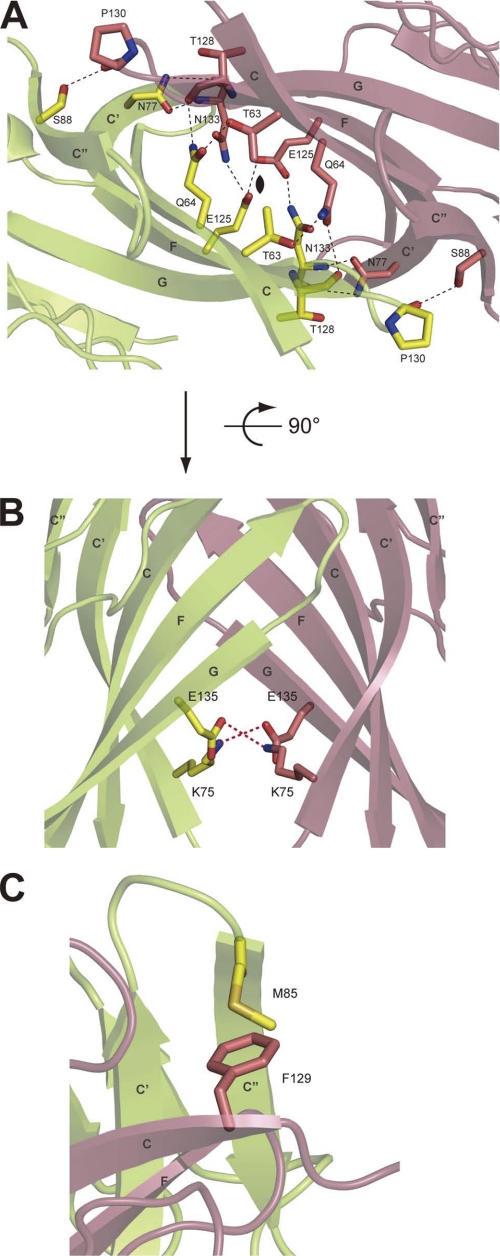
The obtained crystal structure of nectin-1-EC is shown in Fig. 2. The final model has been refined to Rwork and Rfree of 19.0% and 22.6%, respectively, and comprises residues 36–336 of chain A and 33–336 of chain B (Table 1). The crystallographic asymmetric unit was composed of two nectin-1-EC molecules, each of which had a similar structure and exhibited a root mean square deviation of 1.2 Å as calculated in COOT with the SSM algorithm (27). Primary sequence analyses of nectins have predicted that each member of the nectin family has an extracellular region containing three Ig-like domains (2,9). The first Ig-like domain has been deduced to belong to the V-set domain, an Ig-like domain resembling the variable domain of immunoglobulin, whose structure is composed of two β-sheets with nine β-strands (39). Consistent with this prediction, the crystal structure of the nectin-1-EC molecule displays three tandem Ig-like domains, termed the first, second and third Ig-like domains (Fig. 2A). Two nectin-1-EC molecules assemble into a V-shaped homophilic dimer through the first Ig-like domains. In this dimer, the distance between the third Ig-like domains is ∼170 Å, and the distance from the first Ig-like domain to a line connecting the third Ig-like domains is ∼90 Å as shown in Fig. 2A. The first Ig-like domain contains two β-sheets with nine β-strands. The nine β-strands are denoted, from the N terminus, as A, B, C, C′, C", D, E, F, and G (Fig. 2B), and the amino acid position of each strand is shown in supplemental Fig. 2. A disulfide bond between Cys51 and Cys124 and a salt bridge between Arg96 and Asp118 connect the two β-sheets (strands BED and AGFCC′C"). In addition, the first Ig-like domain shares high sequence identity (27.8%) and structural similarity (root mean square deviation = 3.0 Å) with the V-set domain of nectin-like molecule (Necl)-5 (also known as CD155), an Ig-like CAM (40). These results support the notion that the first Ig-like domain is a V-set domain. On the other hand, both the second and third Ig-like domains were classified as C2-set Ig-like domains because of the absence of strands C′ and C" (39). Although the frameworks of these C2-set domains are very similar (root mean square deviation = 2.2 Å), the third Ig-like domain has much shorter strands D and E and a flipped CD loop. The dimeric interface, which buries a solvent-accessible area of ∼1668 Å2 and is most extensive during crystal packing (the other contact area is less than ∼1332 Å2), comprises the β-strands GFCC′C" and the FG loop. The side chain atoms of Thr63, Gln64, Asn77, Ser88, Glu125, and Asn133, and the main chain atoms of Thr128, Pro130, and Asn133 from the A and B molecules form thirteen hydrogen bonds in the dimeric interface (supplemental Table 1). In addition, the dimeric interface involves two salt bridges (Lys75–Glu135) and three van der Waals interactions between the side chains of Thr63 and Thr63 and between those of Met85 and Phe129 (Fig. 3).
FIGURE 2.
Crystal structure of nectin-1-EC and the interdomain interface at 2.8 Å. A, solvent-accessible surface and ribbon diagram of the V-shaped nectin-1-EC dimer. Each molecule is colored differently. The Ig-like domains (first, second, and third) are labeled. B, ribbon diagram of nectin-1-EC. The first, second, and third Ig-like domains, and the short linker regions between the Ig-like domains are shown in red, orange, green, and cyan, respectively. The β-strands are labeled with capital letters in alphabetical order starting from the N terminus of each Ig-like domain, according to the immunoglobulin convention. C and D, close-up views of boxed interdomain interfaces. The residues involved in domain-domain interactions are drawn in stick representation. Oxygen and nitrogen molecules are colored red and blue, respectively. Hydrogen bonds and salt bridges are drawn as dotted black and red lines, respectively.
TABLE 1.
Data collection and refinement statistics of human nectin-1α
| Collection and statistics | Extracellular region native | Extracellular region K2PtBr4 |
|---|---|---|
| Data collection | ||
| Space group | P213 | P213 |
| Cell dimensions | ||
| a, b, c (Å) | 164.9, 164.9, 164.9 | 166.3, 166.3, 166.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50-2.8 (2.9-2.8)a | 50-3.75 (3.88-3.75)a |
| Rmerge | 6.8 (35.8) | 9.6 (47.0) |
| I/σI | 19.4 (4.3) | 20.5 (4.3) |
| Completeness (%) | 97.9 (96.8) | 100.0 (100.0) |
| Redundancy | 3.8 (3.9) | 6.3 (6.5) |
| Refinement | ||
| Resolution (Å) | 35.99-2.80 | |
| No. reflections | 34,698 | |
| Rwork/Rfree | 0.190/0.226 | |
| No. atoms | ||
| Protein | 4,646 | |
| Ligand/ion | 34 | |
| Water | 37 | |
| B-factors | ||
| Protein | 80.19 | |
| Ligand/ion | 99.11 | |
| Water | 61.36 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.010 | |
| Bond angles (°) | 1.247 | |
a Highest resolution shell is shown in parentheses.
FIGURE 3.
Homophilic dimer interface of nectin-1-EC. A, ribbon diagram of the nectin-1-EC dimer interface viewed along a pseudo 2-fold axis. The residues that form hydrogen bonds are drawn in stick representation. Each molecule is colored differently. Oxygen and nitrogen molecules are colored red and blue, respectively. Hydrogen bonds are drawn as dotted black lines. B, a 90° rotated view of A. The residues that formed salt bridges are drawn in stick representation. C, van der Waals interactions between the side chains of Met85 and Phe129. Phe129 is a key residue for trans-interaction.
The first, second, and third Ig-like domains were configured into a rod-like shape with a length of ∼130 Å in length (Fig. 2B). In this region, the long axis of the second Ig-like domain was inclined at ∼15° and ∼40° to the first and third Ig-like domains respectively. The first and second Ig-like domains were connected by the linker (residues 144–146), an EF loop in the first Ig-like domain, and a BC loop in the second Ig-like domain, and were stabilized by the two hydrogen bonds between the main chain atoms of Ala144 and Lys178, one salt bridge (Glu119–Lys178), and one van der Waals interaction between the side chains of Ile43 and Ala144 (Fig. 2C). The second and third Ig-like domains were connected by the linker region (residues 242–247), AB and EF loops in the second Ig-like domain, and a BC loop in the third Ig-like domain, and were stabilized by the hydrogen bonds among the linker residues Asp242, Gln244, and Tyr245, three salt bridges (Glu246–Lys161, Asp272–Arg325, and Asp272–Arg217), and the van der Waals interaction between the side chains of Arg217 and Tyr245 (Fig. 2D). These interactions contributed to stabilize the orientations of each domain of nectin-1-EC (supplemental Table 2).
Involvement of the Hydrogen Bonds between the First Ig-like Domains in Homophilic Dimerization of Nectin-1-EC
We next examined the role of the hydrogen bonds between the first Ig-like domains in the homophilic dimerization of nectin-1. To impair the hydrogen bond network, we mutated four residues that formed the hydrogen bonds in the dimeric interface (Thr63, Gln64, Glu125, and Asn133) to alanines, and the mutant protein (nectin-1-EC-4mut) was prepared in the same manner as nectin-1-EC. Nectin-1-EC and nectin-1-EC-4mut were subjected to analytical unltracentrifugation, and the sedimentation equilibrium data were analyzed with the program SedAnal (33) and Beckman analytical software (34). The sedimentation coefficient for nectin-1-EC increased from ∼2.6 up to ∼2.9 in a concentration-dependent manner, whereas nectin-1-EC-4mut sedimented as a single species with S ∼2.5 at all of the tested concentrations (Fig. 4A). The sedimentation equilibrium data showed that nectin-1-EC existed in a monomer-dimer equilibrium at lower concentrations (absorbance at 280 nm < ∼1.1) but in higher order oligomeric states at higher concentrations (absorbance at 280 nm > ∼1.1) (supplemental Fig. 3A). Therefore, we used absorbance data up to 1.1 (absorbance at 280 nm) for the global-fit calculation to avoid an effect of this higher order oligomeric state. In contrast, nectin-1-EC-4mut existed in a monomer-dimer equilibrium across the concentration range tested (supplemental Fig. 3B). The Kd values for the monomer-dimer equilibrium of nectin-1-EC and nectin-1-EC-4mut were ∼3 μm and 275 μm, respectively, based on the sedimentation data (supplemental Fig. 3, C and D). These results indicate that the mutations weaken the homophilic dimerization. Next, nectin-1-EC and nectin-1-EC-4mut were subjected to the size-exclusion chromatography using a calibrated Superdex 200 10/300 GL column. Nectin-1-EC was eluted as a single peak at an elution volume of 14.4 ml, corresponding to a globular protein with an apparent molecular mass of ∼63 kDa (Fig. 4B). Nectin-1-EC-4mut eluted at an elution volume of 15.1 ml, with an apparent molecular mass of ∼51 kDa. These results further support the notion that these mutations weaken the homophilic dimerization. Taken together, these results indicate that the hydrogen bond network between the first Ig-like domains is essential for the homophilic dimerization of nectin-1.
FIGURE 4.
Decreased dimerization of nectin-1-EC by mutations that impair hydrogen bond formation. A, g(s*) analysis of nectin-1-EC and nectin-1-EC-4mut by sedimentation velocity. The protein concentration ranges from 0.33 to 1.00 mg/ml. The curves are normalized to a total area of unity. B, size-exclusion chromatographs of nectin-1-EC and nectin-1-EC-4mut.
Role of the Hydrogen Bonds in cis-Dimerization of Nectin-1 in Cultured Cells
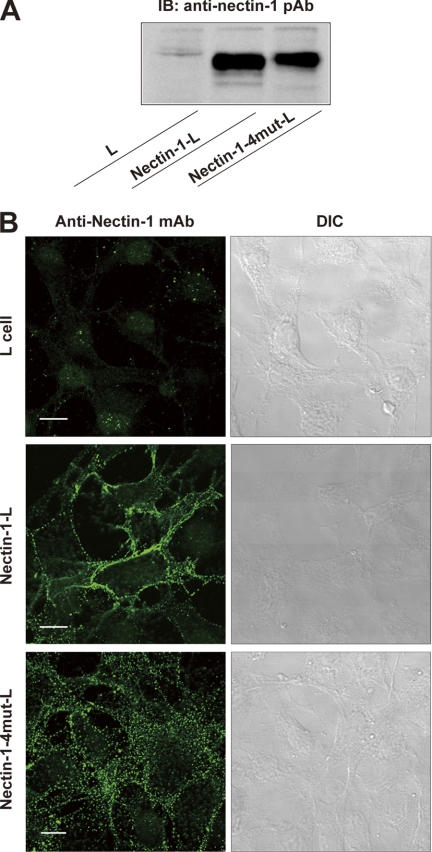
To determine the roles of the homophilic dimerization occurring via the first Ig-like domains in the cell adhesion activity of nectin-1, we established L fibroblasts stably expressing full-length nectin-1 (nectin-1-L cells) or the full-length nectin-1 alanine mutant (nectin-1-4mut-L cells). Immunoblotting of the cell lysates with the anti-nectin-1 pAb showed that the expression level of mutant nectin-1 in nectin-1-4mut-L cells was similar to that of wild-type nectin-1 in nectin-1-L cells (Fig. 5A). We immunostained the nonpermeabilized cells with an anti-nectin-1 mAb raised against the extracellular region of nectin-1. Consistent with findings of earlier studies (12, 22, 36, 37), the extracellular immunoreactive signals for nectin-1 were localized predominantly to the cell-cell junctions in the nectin-1-L cells (Fig. 5B). Extracellular immunoreactive signals for mutant nectin-1 were also detected on the cell surfaces of nectin-1-4mut-L cells, indicating that impairment of the hydrogen bonds does not affect cell surface trafficking of the nectin-1 molecules. However, mutant nectin-1 was not concentrated at the cell-cell junctions, but was instead broadly distributed across the entire cell surfaces. These results suggest that the hydrogen bonds might be involved in the formation of the nectin-1-based cell-cell junctions, which will be discussed in more detail below.
FIGURE 5.
Characterization of L fibroblast stably expressing the nectin-1 mutant where the hydrogen bonds in the dimer interface are impaired. A, expression level of mutant nectin-1. Cell lysates were prepared from parental L cells, nectin-1-L cells, and nectin-1-4mut-L cells and subjected to SDS-PAGE followed by immunoblotting with the anti-nectin-1 pAb. B, cell surface localization of mutant nectin-1. Parental L cells, nectin-1-L cells, and nectin-1-4mut-L cells were fixed in formalin. Without permeabilization, the cells were immunostained with the anti-nectin-1 mAb raised against the extracellular region of nectin-1. DIC, differential interference contrast. Scale bars, 10 μm.
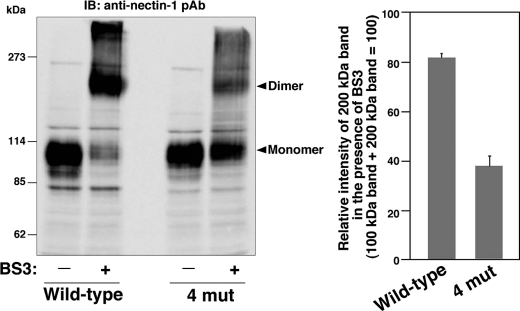
We next examined whether impairment of the hydrogen bonds affected cis-dimerization of nectin-1. Nectin-1-L cells and nectin-1-4mut-L cells were dissociated to single-cell suspensions and incubated with or without a cell surface chemical cross-linker, BS3, followed by immunoblotting with the anti-nectin-1 pAb. In the absence of BS3, both cell types showed immunoreactive bands at about 100 kDa that were indicative of monomeric nectin-1 molecules (Fig. 6). Consistent with earlier studies (12, 22), in the presence of BS3, intensity of the 100-kDa immunoreactive band corresponding to monomeric nectin-1 was decreased in nectin-1-L cells; instead, these cells showed an additional band with a molecular mass of about 200 kDa corresponding to the cis-dimer of nectin-1. In contrast, in the presence of BS3, the nectin-1-4mut-L cells showed significant decrease of the immunoreactive band corresponding to the cis-dimers, and instead showed increase of the immunoreactive band corresponding to monomeric nectin-1. Similar results were also obtained when we used three independent clones of nectin-1-4mut-L cells (data not shown).
FIGURE 6.
Decreased cis-dimerization of nectin-1 by impairment of hydrogen bond formation. Nectin-1-L cells and nectin-1-4mut-L cells were dissociated to single-cell suspensions and incubated with or without a membrane impermeable cross-linker, BS3, for 15 min at 14 °C. The cells were lysed with the SDS sample buffer and subjected to SDS-PAGE followed by immunoblotting with the anti-nectin-1 pAb. The result shown is representative of three independent experiments. Quantification of the relative intensities of the 200-kDa bands in the presence of BS3 is shown in the right panel.
We also confirmed that the 200-kDa immunoreactive band represented the cis-dimer of nectin-1 by using a point mutant of nectin-1 that abolished trans-interaction but not cis-dimerization. It has been reported that substituting phenylalanine at position 136 with leucine abolishes trans-interaction but not cis-dimerization of nectin-2 (12). Based on this report, we generated a point mutant of nectin-1 (nectin-1-F129L) in which phenylalanine at position 129 (equivalent to Phe136 in nectin-2) was substituted with leucine, and confirmed that the L cells stably expressing nectin-1-F129L (nectin-1-F129L-L cells) were unable to trans-interact in the cell aggregation assay (supplemental Fig. 4). Then, nectin-1-L cells and nectin-1-F129L-L cells were subjected to the chemical cross-linking assay. In the absence of BS3, both cell types showed the 100-kDa immunoreactive bands corresponding to monomeric nectin-1 (supplemental Fig. 5). In both cell types, in the presence of BS3, the intensity of the 100-kDa band decreased whereas the band with a molecular mass of ∼200 kDa became prominent. These results indicate that trans-interaction is not involved in the formation of the 200-kDa immunoreactive band and confirm that this band represents the cis-dimer of nectin-1.
Collectively, these results indicate that impairing the formation of hydrogen bonds greatly reduces the cis-dimerization of nectin-1. Taken together, our present results indicate that the solved crystal structure of the nectin-1 dimer, which is formed by the interactions between the first Ig-like domains, reflects a cis-dimer.
Abolishment of Cell Adhesion Activities of Nectin-1 by Impairment of the Hydrogen Bonds
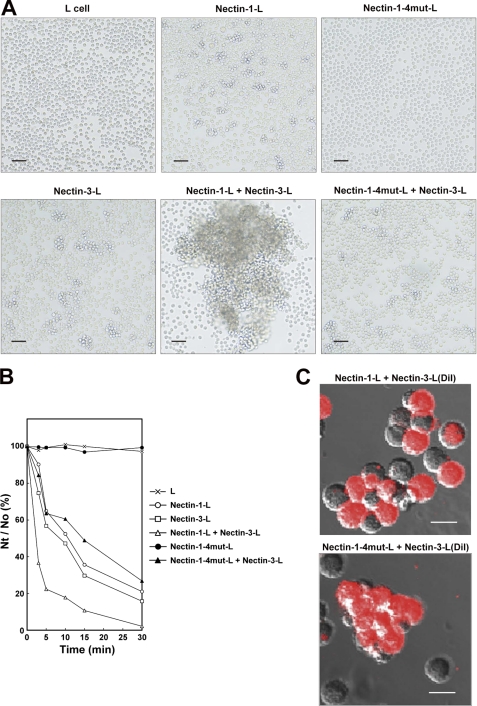
cis-Dimerization of nectin is thought to be a prerequisite for trans-interaction followed by the formation of the cell-cell junctions (2, 9). Therefore, the impaired cis-dimerization of mutant nectin-1 would be expected to lead to the loss of the cell adhesion capability. In agreement with this notion, the localization of mutant nectin-1, as shown in Fig. 5B, suggests the loss of the nectin-1-based cell-cell junctions. To confirm further that the crystal structure reflects a cis-dimer and that the impaired cis-dimerization led to the loss of the cell adhesion capability, we performed cell aggregation assays. Nectin-1-L cells, nectin-1-4mut-L cells, and L cells were dissociated to single-cell suspensions as in Fig. 6 and gently agitated. Consistent with the findings of earlier studies (12, 22, 37), nectin-1-L cells, but not L cells, formed cell aggregates in a time-dependent manner (Fig. 7, A and B). In contrast, nectin-1-4mut-L cells did not aggregate. These results indicate that impairing the hydrogen bonds abolishes the homophilic cell adhesion activity of nectin-1. We next assessed the effect of these mutations on the heterophilic cell adhesion activity by agitating nectin-1-L cells or nectin-1-4mut-L cells with dissociated L cells stably expressing full-length nectin-3 (nectin-3-L cells). Consistent with the previous reports showing that nectin-1 preferentially trans-interacts with nectin-3 (11, 14, 23, 35), the nectin-1-L cells and nectin-3-L cells formed large cell aggregates in a time-dependent manner. DiI-labeling of nectin-3-L cells revealed that the large aggregates were mosaic arrangements of nectin-1-L cells and nectin-3-L cells (Fig. 7C), indicating heterophilic cell adhesion activity of nectin-1 and nectin-3. In contrast, the mixture of nectin-1-4mut-L cells and nectin-3-L cells formed moderately sized cell aggregates, similar to those formed by nectin-3-L cells alone. The DiI-labeling also showed that the cell aggregates in the mixture of nectin-1-4mut-L cells and nectin-3-L cells were only composed of nectin-3-L cells. Therefore, impairing the hydrogen bonds abolishes the heterophilic cell adhesion activity of nectin-1. Similar results were also obtained when we used three independent clones of nectin-1-4mut-L cells (data not shown). Taken together, these results indicate that the cis-dimerization of nectin-1 through the first Ig-like domains is a prerequisite for both homophilic and heterophilic cell adhesion activities.
FIGURE 7.
Abolishment of cell adhesion activities of nectin-1 by impairment of hydrogen bond formation. A, cell aggregation assay. Parental L cells, nectin-1-L cells, nectin-1-4mut-L cells, and nectin-3-L cells were dissociated to single-cell suspensions. Then, the indicated combinations of cells were agitated for 5 min and fixed in glutaraldehyde. Scale bars, 100 μm. The results shown are representative of three independent experiments. B, quantification of time-dependent cell aggregation. The cell aggregation assays were carried out for the indicated times. The extent of cell aggregation is represented as the ratio of the total number of particles at time t of incubation (Nt) to the initial number of particles (No). The results shown are representative of three independent experiments. C, cell aggregation assay using DiI-labeled nectin-3-L cells. Nectin-3-L cells prelabeled with DiI, nectin-1-L cells, and nectin-1-4mut-L cells were dissociated to single-cell suspensions. The indicated combinations of cells were agitated for 3–5 min and immediately observed under a fluorescence microscope. Scale bars, 20 μm. The results shown are representative of three independent experiments.
DISCUSSION
In this study, we have provided the first crystal structure of the entire extracellular region of nectin-1, a member of the nectin family of proteins, and showed that nectin-1 forms the cis-dimer by interactions between the first Ig-like domains. We also found that the first Ig-like domain-mediated cis-dimerization is a prerequisite for the cell adhesion activity of nectin-1 (Fig. 7), indicating that cis-dimerization precedes trans-interaction.
In contrast to our crystallographic data, a previous cell biological study that used several deletion mutants of nectin, suggested that the second Ig-like domain is involved in cis-interaction (13). How can we explain these differing results? In general, CAMs have been postulated to cluster in a lateral direction to convert the intrinsic binding activity of their extracellular regions into physiological cell-cell adhesion (1). Therefore, one possibility is that the second Ig-like domain of nectin may be involved in the lateral clustering of the cis-dimer. In this case, the previous result that deletion of the second Ig-like domain abolishes the cell adhesion activity of nectin (13) is in good agreement with the importance of the lateral clustering. In addition to cis-dimerization, the first Ig-like domain is also involved in trans-interaction (12–14). The previous site-directed mutational analysis of nectin (12) and the results obtained in this study (supplemental Figs. 4 and 5) indicate that Phe129 in the first Ig-like domain is critical for trans-interaction but not for cis-dimerization. Our structure of nectin-1 indicates that Phe129 is located on the top of the dimeric interface and is solvent-accessible (Fig. 3C and supplemental Fig. 6). The trans-interaction is likely to be formed by the two cis-dimers in a head-to-head manner around the common pseudo 2-fold axis. Collectively, these results suggest a possible molecular mechanism to explain how nectins built cell-cell junctions. Nectins first form cis-dimers through the first Ig-like domains on the surface of the same cell, which is followed by lateral clustering of the cis-dimers through the second Ig-like domains. Then, the lateral clusters of the cis-dimers undergo trans-interactions between the neighboring cells through the first Ig-like domains, resulting in the formation of nectin-based cell-cell junctions. The x-ray structures of the first Ig-like domain of Necl-1 and the first-second Ig-like domains of Necl-5 were recently reported (40, 41). In the crystals, these Necls formed the homophilic dimers via the AGFCC′C" β-sheets in the first Ig-like domains in the same manner as cis-dimerization of nectin-1. Notably, it was presumed that the crystal structure of Necl-1 revealed the homophilic trans-interaction at synapses. However, expecting that the mechanisms by which nectin-1 and Necl-1 that form the cell-cell junctions are similar, our findings may also lead to persuasive proposal that the Necl-1 dimer reflects a cis-dimer.
Our crystal structure revealed that nectin-1 possesses two short linkers of three and five residues that connect the first and second Ig-like domains, and the second and third Ig-like domains, respectively. In the interdomain interface between the first and second Ig-like domains, one van der Waals interaction between the side chains of Ile43 and Ala144 and one salt bridge (Glu119–Lys178) are interconnected by two hydrogen bonds between the main chain atoms of Ala144 and Lys178 (supplemental Fig. 7A). In the interdomain interface of the second and third Ig-like domains, the two hydrogen bonds of His220–Asn274 and Asn242–Asn274 and one van der Waals interaction between the side chains of Arg217 and Tyr245 are interconnected by two hydrogen bonds between the main chain atoms of Tyr245 and Asn274. These interactions define the relative orientations of the first and second, and the second and third Ig-like domains, resulting in the rod-like shape of nectin-1. Sequence alignments of the extracellular regions of human nectins show that all of the residues located at equivalent positions in each nectin are probably capable of participating in these interactions, except for an absence of the salt bridges in the interdomain interface between the first and second Ig-like domains in nectins-3 and -4 (supplemental Fig. 2). Therefore, all of the human nectins likely adopt the similar orientations of the first and second, and the second and third Ig-like domains, resulting in a rod-like shape.
The residues mediating the homophilic cis-dimer formation of nectin-1 are not conserved among the members of the nectin family, except for three residues, namely Thr128–Pro130 (supplemental Fig. 2). Although the main chain atoms of Thr128 and Pro130 contribute to the nectin-1-EC dimer interface by forming hydrogen bonds with the side chain atoms of Gln64 and Ser88, respectively, the side chains of Thr128 and Pro130 do not directly participate in this dimer interface. Therefore, human nectins are expected to form homophilic dimer via different interactions through the AGFCC′C" β-sheets. Given that the association angle of the first Ig-like domains determines the height of the homophilic cis-dimer, any difference in the interaction between the AGFCC′C" β-sheets is likely to define the height of the cis-dimer for each nectin (supplemental Fig. 7B).
Evidence is accumulating that specific nectins regulate cell-cell adhesion between specific cell types in vivo (2, 7–9). Synapses (special junctions that form between the axons and dendrites of neurons) are sites of neurotransmission. Two types of junctions have been identified: synaptic junctions and puncta adherentia junctions (PAJs) (42). Synaptic junctions function as neurotransmission sites, whereas PAJs are regarded as mechanical adhesion sites between axon terminals and their targets. At the PAJs in the synapses between the mossy fiber terminals and dendrites of pyramidal cells in the CA3 region of the hippocampus, nectins-1 and -3 are asymmetrically localized on the presynaptic and postsynaptic sides, respectively (35). The heterophilic trans-interaction between nectins-1 and -3 regulates the organization of the PAJs (35). A heterophilic trans-interaction between nectins-2 and -3 dominantly organizes the heterotypic cell-cell adhesion that is essential for germ cell differentiation in the testis. During germ cell differentiation, spermatogenic cells are embraced and cultivated by Sertoli cells during spermatogenesis (43). In the latter half of spermatogenesis, spermatids form prominent heterotypic cell-cell junctions with Sertoli cells (44), where the existence of the cadherin system has been questionable. Nectins-2 and -3 are specifically expressed in Sertoli cells and spermatids, respectively, and the heterophilic trans-interaction between nectins-2 and -3 dominantly organizes the Sertoli cell-spermatid junctions (16). The unique height of the cis-dimer of each nectin, as mentioned above, might determine the distance between the neighboring cells in these particular cell-cell junctions.
In addition to its role in the cell adhesion, nectin-1 serves as an entry receptor for herpes simplex virus (HSV) by binding with the virus envelope glycoprotein D (gD) (22, 45, 46). cis-Dimerization of nectin-1 through the first Ig-like domain also provides structural features involved in contacting gD. Although the structural details of the nectin-1-gD complex are unknown, several mutagenesis studies have shown that the nectin-1 mutant Q76A/N77A/M85A severely reduced the HSV entry activity (47, 48). These residues are located on the top and the side regions of the dimeric interface in the first Ig-like domain of nectin-1 and are solvent-accessible (supplemental Fig. 6). Soluble gD ectodomains are monomeric (49) and inhibit cell-cell adhesion activity (trans-interaction) but not cis-dimerization of nectin-1 (22, 50). Therefore, the cis-dimer of nectin-1 may have two contact surfaces for binding to the gD monomer on the upper sides of the first Ig-like domains. Further structural studies are required to investigate this in more detail.
Acknowledgments
We thank Dr. Kazumasa Sakurai and Miyo Sakai for the ultracentrifugation experiments. We also thank Dr. Junichi Takagi (Osaka University, Osaka, Japan) for kindly providing the mammalian expression vector encoding the C-terminal His8-myc-His6 tag.
This work was supported by grants-in-aid for the National Project on Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7 and Tables 1 and 2.
The atomic coordinates and structure factors (code 3ALP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CAM
- cell adhesion molecule
- Bis-Tris propane
- 1,3-bis(tris(hydroxymethyl)methylamino)propane
- BS3
- bis-(sulfosuccinimidyl) suberate
- gD
- glycoprotein D
- pAb
- polyclonal antibody
- PAJ
- puncta adherentia junction.
REFERENCES
- 1. Yap A. S., Brieher W. M., Gumbiner B. M. (1997) Annu. Rev. Cell Dev. Biol. 13, 119–146 [DOI] [PubMed] [Google Scholar]
- 2. Takai Y., Ikeda W., Ogita H., Rikitake Y. (2008) Annu. Rev. Cell Dev. Biol. 24, 309–342 [DOI] [PubMed] [Google Scholar]
- 3. Harris T. J., Tepass U. (2010) Nat. Rev. Mol. Cell Biol. 11, 502–514 [DOI] [PubMed] [Google Scholar]
- 4. Takeichi M. (1995) Curr. Opin. Cell Biol. 7, 619–627 [DOI] [PubMed] [Google Scholar]
- 5. Shapiro L., Weis W. I. (2009) Cold Spring Harbor Perspect. Biol. 1, a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koch A. W., Pokutta S., Lustig A., Engel J. (1997) Biochemistry 36, 7697–7705 [DOI] [PubMed] [Google Scholar]
- 7. Sakisaka T., Takai Y. (2004) Curr. Opin. Cell Biol. 16, 513–521 [DOI] [PubMed] [Google Scholar]
- 8. Sakisaka T., Ikeda W., Ogita H., Fujita N., Takai Y. (2007) Curr. Opin. Cell Biol. 19, 593–602 [DOI] [PubMed] [Google Scholar]
- 9. Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nat. Rev. Mol. Cell Biol. 9, 603–615 [DOI] [PubMed] [Google Scholar]
- 10. Mandai K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., Matsuda Y., Tsukita S., Takai Y. (1997) J. Cell Biol. 139, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikeda W., Kakunaga S., Itoh S., Shingai T., Takekuni K., Satoh K., Inoue Y., Hamaguchi A., Morimoto K., Takeuchi M., Imai T., Takai Y. (2003) J. Biol. Chem. 278, 28167–28172 [DOI] [PubMed] [Google Scholar]
- 12. Miyahara M., Nakanishi H., Takahashi K., Satoh-Horikawa K., Tachibana K., Takai Y. (2000) J. Biol. Chem. 275, 613–618 [DOI] [PubMed] [Google Scholar]
- 13. Momose Y., Honda T., Inagaki M., Shimizu K., Irie K., Nakanishi H., Takai Y. (2002) Biochem. Biophys. Res. Commun. 293, 45–49 [DOI] [PubMed] [Google Scholar]
- 14. Yasumi M., Shimizu K., Honda T., Takeuchi M., Takai Y. (2003) Biochem. Biophys. Res. Commun. 302, 61–66 [DOI] [PubMed] [Google Scholar]
- 15. Tsukasaki Y., Kitamura K., Shimizu K., Iwane A. H., Takai Y., Yanagida T. (2007) J. Mol. Biol. 367, 996–1006 [DOI] [PubMed] [Google Scholar]
- 16. Ozaki-Kuroda K., Nakanishi H., Ohta H., Tanaka H., Kurihara H., Mueller S., Irie K., Ikeda W., Sakai T., Wimmer E., Nishimune Y., Takai Y. (2002) Curr. Biol. 12, 1145–1150 [DOI] [PubMed] [Google Scholar]
- 17. Okabe N., Shimizu K., Ozaki-Kuroda K., Nakanishi H., Morimoto K., Takeuchi M., Katsumaru H., Murakami F., Takai Y. (2004) Dev. Biol. 273, 244–256 [DOI] [PubMed] [Google Scholar]
- 18. Suzuki K., Hu D., Bustos T., Zlotogora J., Richieri-Costa A., Helms J. A., Spritz R. A. (2000) Nat. Genet. 25, 427–430 [DOI] [PubMed] [Google Scholar]
- 19. Sözen M. A., Suzuki K., Tolarova M. M., Bustos T., Fernández Iglesias J. E., Spritz R. A. (2001) Nat. Genet. 29, 141–142 [DOI] [PubMed] [Google Scholar]
- 20. Avila J. R., Jezewski P. A., Vieira A. R., Orioli I. M., Castilla E. E., Christensen K., Daack-Hirsch S., Romitti P. A., Murray J. C. (2006) Am. J. Med. Genet. A 140, 2562–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scapoli L., Palmieri A., Martinelli M., Vaccari C., Marchesini J., Pezzetti F., Baciliero U., Padula E., Carinci P., Carinci F. (2006) Ann. Hum. Genet. 70, 410–413 [DOI] [PubMed] [Google Scholar]
- 22. Sakisaka T., Taniguchi T., Nakanishi H., Takahashi K., Miyahara M., Ikeda W., Yokoyama S., Peng Y. F., Yamanishi K., Takai Y. (2001) J. Virol. 75, 4734–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Honda T., Shimizu K., Kawakatsu T., Yasumi M., Shingai T., Fukuhara A., Ozaki-Kuroda K., Irie K., Nakanishi H., Takai Y. (2003) Genes Cells 8, 51–63 [DOI] [PubMed] [Google Scholar]
- 24. Honda T., Shimizu K., Fukuhara A., Irie K., Takai Y. (2003) Biochem. Biophys. Res. Commun. 306, 104–109 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Macromol. Crystallogr. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 31. Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2002) PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 33. Stafford W. F., Sherwood P. J. (2004) Biophys. Chem. 108, 231–243 [DOI] [PubMed] [Google Scholar]
- 34. McRorie D. K., Voelker P. J. (1993) Self-associating Systems in the Analytical Ultracentrifuge, Beckman Instruments, Palo Alto, CA [Google Scholar]
- 35. Mizoguchi A., Nakanishi H., Kimura K., Matsubara K., Ozaki-Kuroda K., Katata T., Honda T., Kiyohara Y., Heo K., Higashi M., Tsutsumi T., Sonoda S., Ide C., Takai Y. (2002) J. Cell Biol. 156, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Satoh-Horikawa K., Nakanishi H., Takahashi K., Miyahara M., Nishimura M., Tachibana K., Mizoguchi A., Takai Y. (2000) J. Biol. Chem. 275, 10291–10299 [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K., Nakanishi H., Miyahara M., Mandai K., Satoh K., Satoh A., Nishioka H., Aoki J., Nomoto A., Mizoguchi A., Takai Y. (1999) J. Cell Biol. 145, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Honig M. G., Hume R. I. (1986) J. Cell Biol. 103, 171–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bork P., Holm L., Sander C. (1994) J. Mol. Biol. 242, 309–320 [DOI] [PubMed] [Google Scholar]
- 40. Zhang P., Mueller S., Morais M. C., Bator C. M., Bowman V. D., Hafenstein S., Wimmer E., Rossmann M. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18284–18289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong X., Xu F., Gong Y., Gao J., Lin P., Chen T., Peng Y., Qiang B., Yuan J., Peng X., Rao Z. (2006) J. Biol. Chem. 281, 10610–10617 [DOI] [PubMed] [Google Scholar]
- 42. Spacek J., Lieberman A. R. (1974) J. Anat. 117, 487–516 [PMC free article] [PubMed] [Google Scholar]
- 43. Russell L. D., Ettlin R., ShinhaHikim A. P., Clegg E. D. (1990) Histological and Histopathological Evaluation of the Testis, Cache River Press, Vienna [Google Scholar]
- 44. Cheng C. Y., Mruk D. D. (2002) Physiol. Rev. 82, 825–874 [DOI] [PubMed] [Google Scholar]
- 45. Lopez M., Eberlé F., Mattei M. G., Gabert J., Birg F., Bardin F., Maroc C., Dubreuil P. (1995) Gene 155, 261–265 [DOI] [PubMed] [Google Scholar]
- 46. Geraghty R. J., Krummenacher C., Cohen G. H., Eisenberg R. J., Spear P. G. (1998) Science 280, 1618–1620 [DOI] [PubMed] [Google Scholar]
- 47. Martinez W. M., Spear P. G. (2002) J. Virol. 76, 7255–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Struyf F., Martinez W. M., Spear P. G. (2002) J. Virol. 76, 12940–12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krummenacher C., Supekar V. M., Whitbeck J. C., Lazear E., Connolly S. A., Eisenberg R. J., Cohen G. H., Wiley D. C., Carfí A. (2005) EMBO J. 24, 4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krummenacher C., Baribaud I., Sanzo J. F., Cohen G. H., Eisenberg R. J. (2002) J. Virol. 76, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]