Abstract
Pathogenic bacteria acquire the essential element iron through specialized uptake pathways that are necessary in the iron-limiting environments of the host. Members of the Gram-negative Neisseriaceae and Pasteurellaceae families have adapted to acquire iron from the host iron binding glycoprotein, transferrin (Tf), through a receptor complex comprised of transferring-binding protein (Tbp) A and B. Because of the critical role they play in the host, these surface-exposed proteins are invariably present in clinical isolates and thus are considered prime vaccine targets. The specific interactions between TbpB and Tf are essential and ultimately might be exploited to create a broad-spectrum vaccine. In this study, we report the structure of TbpBs from two porcine pathogens, Actinobacillus pleuropneumoniae and suis. Paradoxically, despite a common Tf target, these swine related TbpBs show substantial sequence variation in their Tf-binding site. The TbpB structures, supported by docking simulations, surface plasmon resonance and hydrogen/deuterium exchange experiments with wild-type and mutant TbpBs, explain why there are structurally conserved elements within TbpB homologs despite major sequence variation that are required for binding Tf.
Keywords: Bacteria; Cell Surface Protein; Iron; Protein Structure; X-ray Crystallography; Bacterial Lipoprotein.; Bacterial Transferrin Receptor,; Iron Acquisition,; Vaccine Candidate; Membrane Protein
Introduction
While colonizing or infecting the host, bacterial pathogens utilize surface proteins to acquire nutrients and interact with host components to survive the antibacterial properties of the host milieu. For bacteria, the acquisition of the essential nutrient iron is of critical importance as vertebrates sequester free iron within the extracellular milieu using specific iron carrying glycoproteins, lactoferrin (Lf), and transferrin (Tf)3 (1).
To colonize, bacteria have selectively evolved with their host to express specific mechanisms for iron uptake. The pathogenic Gram-negative bacteria from the Neisseriaceae and Pasteurellaceae families have developed a host Tf-specific iron uptake system mediated by a bacterial receptor complex, the Tf-binding proteins A and B (TbpA, TbpB) (2). TbpA is a TonB-dependent outer-membrane protein, strictly essential for iron uptake during bacteria infection (3). TbpA binds both of the holo or apo forms of Tf (4), and it is proposed to serve as a channel for iron transport across the outer membrane (5). TbpB, the second component of the receptor complex, is a surface exposed lipoprotein that binds holo-Tf in a selective manner (4, 6). This enables the bacterial receptor to recruit iron more efficiently from the partially iron-saturated forms of Tf in vivo (7). TbpB is essential for colonization of the host (8), and depending on the strain and species, TbpB-defective bacteria present several different growth-impaired phenotypes. TbpBs display species specificity (9), binding only to their host Tf, while the human Tf-receptor has no species specificity retrieving Tf through endocytosis where the low pH of the endosome drives the iron delivery from Tf (10, 11).
Surface lipoproteins such as TbpB have been targeted for vaccine development as they elicit a strong immune response, and antibodies to specific surface lipoproteins are bactericidal (12–14). Because TbpB functions to specifically associate with the host protein Tf, it is likely that the site of interaction may harbor a conserved binding interface (15) that could be exploited to create a broad spectrum vaccine.
Recently the structure of the TbpB from the porcine pathogen Actinobacillus pleuropneumoniae together with functional binding studies has illustrated that the residues from the site of interaction with porcine Tf (pTf) do not appear to be conserved among porcine pathogens (16). The structure illustrates that TbpB is a bilobed protein requiring the N-lobe domain for Tf binding function. Both the N- and C-lobe adopt a similar overall fold; the C-lobe is the most conserved domain in sequence along all TbpB proteins, while N-lobes show paradoxically a less well-conserved sequence even between strains targeting the same host and thus the same Tf (16).
A. pleuropneumoniae along with Actinobacillus suis and Haemophilus parasuis are important pathogens in pigs that share a common mechanism for acquiring iron directly from the host Tf (17–19). This parallels the situation with human pathogens in which Neisseria meningitidis, Moraxella catarrhalis, and H. influenzae acquire iron from human Tf through a conserved, highly specific interaction common to these species (15). In the absence of structural information for TbpBs from human pathogens, a study of TbpBs from porcine pathogens provides the greatest opportunity to probe the details of this interaction and determine how a conserved interaction can be maintained in the face of considerable sequence diversity.
In this study, we describe crystal structures of two divergent TbpB receptors from the porcine pathogens A. suis (H57) and A. pleuropneumoniae (H87) and provide models of the pTf-TbpB complexes that are supported by hydrogen-deuterium exchange coupled to mass spectrometry (H/DX-MS) and binding experiments with site-directed mutant TbpBs.
EXPERIMENTAL PROCEDURES
Sequence Analysis of tbpB Genes
The tbpB genes from all of our clinical isolates of A. pleuropneumoniae and A. suis were amplified from whole colonies using a set of primers designed to bind ∼100 bp outside of the gene. These sequences were combined with all of the unique publicly available tbpB sequences, and a ClustalW2 multiple sequence alignment was generated. The alignment was manually inspected and edited, and a bootstrapped neighbor-joining unrooted phylogenetic tree was generated from this alignment using the program Geneious.
Expression and Purification of Recombinant TbpB
Protein was expressed and purified essentially as described previously (16). The genes encoding TbpB from A. suis strain H57 (AsH57) and A. pleuropneumoniae strain H87 (ApH87) were cloned in to an expression vector encoding an N-terminal maltose-binding protein fusion partner containing a polyhistidine tag and a TEV cleavage site preceding the region encoding the mature TbpB protein. The recombinant fusion proteins were isolated on a pTf-Sepharose column as described previously (16) or by Ni-NTA chromatography. After cleavage by TEV protease the TbpB proteins were purified by Q-Sepharose chromatography as described previously (16).
Protein Crystallization
Purified AsH57 and ApH87 TbpBs were screened against the JSCG+ suite commercial screen using hanging drop vapor diffusion at 10 mg/ml. Initial AsH57 TbpB crystals were observed in 0.2 m tri-potassium citrate pH 8.3 and 20% PEG 3350 then optimized with a 1:1 ratio sitting drop at 20 °C in a precipitant solution composed of 0.1 m Bis-Tris pH 5.5, 0.4 m lithium sulfate, 17% PEG 3350 and 20% glycerol yielding crystals in space group P21. ApH87 TbpBs were first observed in 0.16 m calcium acetate, 0.08 m sodium cacodylate pH 6.5, 14.4% PEG 8000, and 20% glycerol and optimized at 20 °C using 0.16 m sodium acetate, 0.08 m calcium cacodylate pH 6.5, 23% PEG 3350 and 15% glycerol to yield crystals in space group C2221. Unit cell dimensions and statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics for AsH57 and ApH87 TbpB
| AsH57 TbpB27–577 | ApH87 TbpB26–528 | |
|---|---|---|
| Data collection | ||
| Space group | P21 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 90.7, 74.5, 106.4 | 132.6, 151.3, 90.3 |
| a, b, g (°) | 90.0, 105.7, 90.0 | 90, 90, 90 |
| Resolution (Å) | 50-2.1 (2.17-2.1)a | 50-2.1 (2.17-2.1) |
| Rsym or Rmerge | 0.145 (0.50) | 0.093 (0.45) |
| I/σI | 15.7 (3.1) | 28.6 (4.4) |
| Completeness (%) | 99.7 (99.2) | 99.9 (99.8) |
| Redundancy | 3.9 (2.3) | 13.7 (10.1) |
| Refinement | ||
| Resolution (Å) | 44.76-2.1 | 47.14-2.1 |
| No. reflections | 75629 | 50535 |
| Rwork/Rfree | 0.17/0.21 | 0.16/0.20 |
| No. atoms | 9509 | 4595 |
| Protein | 8589 | 4033 |
| Glycerol | 30 | 90 |
| Cacodylic acid | 0 | 5 |
| Water | 890 | 498 |
| B-factors | 50.1 | 64.5 |
| Protein | 50.5 | 62.8 |
| Ligand/ion | 69.6 | 98.6 |
| Water | 44.8 | 71.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.006 |
| Bond angles (°) | 1.071 | 1.062 |
a Highest resolution shell is shown in parenthesis.
Structure Determination
Data were gathered on crystals frozen at 105 K on beamline 08ID-1 at the Canadian Light Source (CLS). Diffraction data sets of AsH57 TbpB and ApH87 TbpB were collected at a wavelength of 0.9795 Å using 1° oscillations with 360 and 300 images, respectively. Data were processed with XDS (20) to resolutions of 2.1 Å for ApH87 TbpB and AsH57 TbpB. The first structural model was obtained by molecular replacement based on ApH49 TbpB (PDB code 3HOL) using PHASER (21), yielding an initial Rwork/Rfree 0.29/0.32 and 0.34/038 for AsH57 and ApH87, respectively. Final models were generated following several rounds of model building and refinement using Coot (22) and Refmac5 (23) programs. A Ramachandran plot calculated 3.9% and 3.0% of residues in the allowed region and 0.4% and 0.2% as outliers, for AsH57 and ApH87 TbpB, respectively. The numbering of residues initiates with the first mature amino acid (Cys-1) remaining after signal peptide cleavage.
Surface Plasmon Resonance
Surface Plasmon resonance experiments were performed using BiacoreX (GE Healthcare) at 25 °C. Porcine transferrin was coupled to the sensor chip (CM5 research grade) via standard N-hydroxysuccinimde and N-ethyl-N-(dimethylaminopropyl) carbodiimide activation. Wild-type TbpB and mutant variants were diluted at various concentrations with the mobile phase buffer (10 mm Hepes pH 7.5, 150 mm NaCl, and 0.005% surfactant P20). Samples were injected as analytes at a flow rate of 20 μl/min, and bound analytes were subsequently removed by washing with the mobile phase at 90 or 240 s after the injection. Regeneration buffer (10 mm Hepes, pH 7.5, 2 m NaCl) was injected prior to each analyte injection. Kinetic constants were calculated from the sensorgrams using the simulated BiacoreX evaluation software, version 4.0.1 (Biacore). The AsH57, ApH87, and ApH49 TbpBs data were fitted to a “heterogeneous ligand” binding model. The ApH87 TbpB R179E mutant was analyzed using the saturation method and fitted with a quadratic equation with BiacoreX evaluation software.
Docking Simulation
As the structure of TbpB bound to pTf has not yet been elucidated, the interaction was modeled using RosettaDock (24) incorporating the information generated by SPR analysis. RosettaDock was run in full atom mode, allowing spin around the axis connecting the two proteins, and using standard Monte Carlo movements. A custom program was written to allow experimentally determined important residues to be taken into account during a docking run. The general algorithm of the custom program was to first run RosettaDock, generating 3,000 high-resolution decoys. Each decoy was then parsed, checking whether each TbpB residue designated as important was within 10 Å of pTf, and deleting those decoys that failed on any residue. RosettaDock was then run again to generate as many new decoys as necessary to bring the total back up to 3,000, and the cycle repeated until all decoys met each of the designated distance constraints.
This program was used to generate models for both of the new structures ApH87 and AsH57 TbpB, as well as the previously solved structure from ApH49 (16). For ApH87, the residues deemed important were Tyr-95, Tyr-121, Tyr-174, and Arg-179, for AsH57 the important residues were Phe-63 and Phe-152, and for ApH49 they were Glu-112 and Phe-171. Initial perturbations were set to randomize both partners, move 3° and 8 Å, with a spin along the protein-protein axis. Monte Carlo minimization was used, and the decoys were generated in full-atom mode using multiple threads in parallel. For both the N-lobe and the full-length ApH87 TbpB with pTF 1500 decoys were generated, running on 6 threads at once. The 10 decoys were chosen by their total score, which is determined by the scoring function.
Hydrogen/Deuterium Exchange (H/DX-MS) Experiments
Each H/DX-MS experiment consisted of two samples: free TbpB (AsH57, ApH49, or ApH87) and the TbpB-pTf complex (AsH57, ApH49, or ApH87). Each experiment was completed in quadruplicate, alternating between the receptors and complexes, therefore making future receptor cross-comparison possible. Ten microliters of TbpB was mixed with 5 μl HEPES buffer (10 mm HEPES, pH 7.0). Five microliters of heavy water (deuterated water, D20) was then added to this solution, mixed, and incubated immediately in a water bath at 20 °C, for 2 min. The labeled sample was then removed from the water bath, and 10 μl of cold quench solution (100 mm glycine hydrochloride, pH 2.3) was added. This sample was immediately injected onto a 10-μl loop, connected to an online immobilized pepsin reactor (2.5 cm by 200 μm internal diameter, assembled in-house) (Applied Biosystems, Concord, ON, Canada). An isocratic gradient of 3% ACN, 0.03% TFA, and 0.02% FA, at 4 μl/min was used to flow the intact labeled protein through the digestor column, and onto a C18 reversed-phase chromatography column (assembled in-house) (Applied Biosystems, Concord, ON, Canada). The peptides were separated with a step gradient of 5 to 90% ACN in 0.03% TFA and 0.02% FA over 14 min, and directly infused into a QSTAR Pulsar i quadrupole time-of-flight (QqTOF) mass spectrometer. All chromatography was carried out under ice, at 0–4 °C. Data were collected using Analyst QSTM version 1.1 (Applied Biosystems).
To determine the TbpB peptides arising from the online pepsin digests, unlabeled peptides were prepared under the same conditions as mentioned above. Separated peptides were subjected to tandem mass spectrometry (MS/MS). The MS/MS spectral data were searched against the TbpB sequence (either ApH49, AsH57, or ApH87), using an in-house licensed version of MASCOT 2.1, with conventional identification criteria (±0.5amu mass tolerance in MS and 0.8amu mass tolerance in MS/MS). The MASCOT search identified 58 usable peptides for AsH57 and peptides were then manually verified. Mass spectral data from each replicate H/DX-MS run was analyzed using Hydra software (25), using the method described by Ling et al. (26).
RESULTS
Phylogenetic Analysis
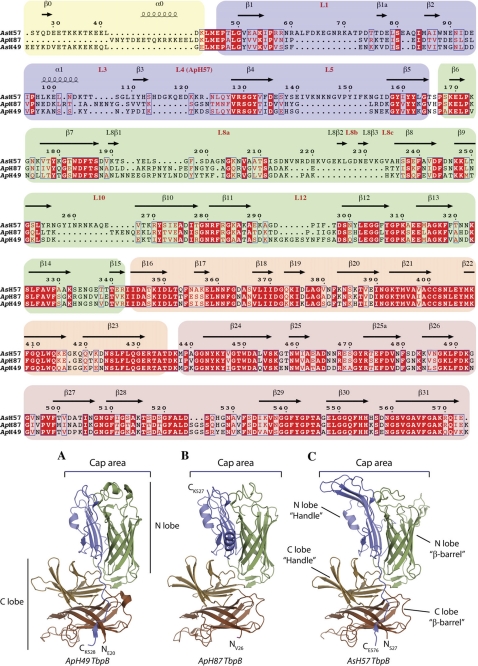
Through sequencing tbpB genes from our strain collection and combining them with sequences available from public databases, we compared TbpBs from A. pleuropneumoniae and A. suis to gauge the degree of sequence diversity (supplemental Fig. S1). Three TbpBs divergent from the one previously structurally characterized (16) were produced for structural studies and the structures of two of these TbpBs were obtained. Together with the published structure, these provide a reasonable representation of the sequence diversity (red arrows, supplemental Fig. S1). A sequence alignment between the three TbpBs for which structural information is available is shown in Fig. 1. The results illustrate that there is significant sequence variation in the handle (blue color) and barrel (green color) domains of the N-lobe of TbpB (Fig. 1, upper panel) that primarily resides in loops on cap area of the N-lobe (lower panel).
FIGURE 1.
Structural alignment of three TbpBs from the swine pathogenic strains ApH49, AsH57, and ApH87. Background colors yellow, blue, green, orange, and peach denote the domain architecture of the N-terminal anchoring peptide, then the handle and β-barrel domain from the N- and C-lobe, respectively. The secondary structure elements and loops from the cap area are shown above the sequences according to the TbpB labeling nomenclature is described in the supplementary Fig. S1. Identical residues and conserved residues are highlighted and boxed red. A, B, and C, respectively are cartoon representations of ApH49 TbpB from residues 24–528 (PDB code: 3HOL Moraes et al., 16) (A), ApH87 TbpB from residues 26–528 (PDB code: 3PQS determined in this study) (B), and AsH57 TbpB from residues 27–577(PDB code: 3PQU determined in this study). The N-lobe β-barrel and handle domain are, respectively, colored in green and blue; the C-lobe β-barrel and handle domain are, respectively, colored in brown and sand. The cap area is labeled.
Overall Structural Features
The structures of TbpBs from A. suis strain H57 (AsH57) and A. pleuropneumoniae strain H87 (ApH87) (Table 1) have a similar fold to the previously characterized TbpB from A. pleuropneumonae strain H49 (ApH49), which consists of two globular domains, the N and C lobes (N-lobe: AsH57 TbpB residues 27–342, ApH87 TbpB residues 26–294; C-lobe: AsH57 TbpB residues 343–577, ApH87 TbpB residues 295–528). Each lobe is comprised of a β-barrel element flanked by an adjacent handle domain (Fig. 1 and supplemental Fig. S2). The β-barrel configuration of N- and C-lobes consist of an eight-stranded β-barrel with, in case of AsH57 TbpB, two additional surface-exposed anti-parallel β-strands in the large N-lobe loop region between the β-strand L8β1 and β8 (Fig. 1). In both AsH57 and ApH87 TbpBs, the handle domain from N-lobe (AsH57 TbpB: residues 49–172; ApH87 TbpB: residues 49–141) consists of a four stranded anti-parallel β-sheet supported by an short α-helix, which packs together with the adjacent β-barrel forming a globular core, N-lobe, that is linked to the C-lobe handle domain (Fig. 1).
The C-lobe handle domains (AsH57 TbpB residues 343–431, ApH87 TbpB residues 295–382), as in ApH49 TbpB, are composed of six-stranded anti-parallel β-sheets stacked between the C-lobe β-barrel core (AsH57 TbpB residues 432–577, ApH87 TbpB residues 383–528) and two surface-exposed anti-parallel β-strands (Fig. 1). The N- and C-lobe of both TbpB structures form two independent structural domains that are maintained in an orthogonal orientation by an evolutionary conserved hydrophobic core flanked by two salt bridges.
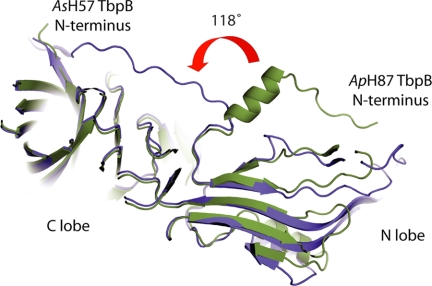
The N-terminal Anchoring Peptides Have Different Conformations
The conformation of the N-terminal anchor peptide (the residues preceding the leucine 50 residue of the first β-strand) of the ApH57 TbpB differs from the two other TbpBs. The ApH87 TbpB N terminus rotates 118 degrees at Lys-43 and extends into a long α-helix stacked on the N-lobe (Fig. 2). In the crystal, the N-terminal peptide of ApH49 and AsH87 TbpB (respectively residues 1–41, and 1–43) is oriented toward the C-lobe, interacting the C terminus by intra-chain contact. Crystallized in a different space group C2221, the ApH57 TbpB N terminus (residues 1–43) extends along the N-lobe and also interacts the C-lobe terminal residues but from the adjacent monomer in the unit cell. These two alternative conformations support the argument that TbpB uses its N-terminal region as a flexible anchoring peptide to extend above the capsular layer to bind Tf from the extracellular millieu and recruit it toward the outer membrane surface.
FIGURE 2.
The flexible N terminus-anchoring region of TbpB. Clipped schematic representation of superimposed AsH57 TbpB (colored in blue) and ApH87 TbpB (colored in green). The N- and C-lobe are indicated, and the N terminus of both TbpBs is labeled. The red curved arrow illustrates the 118° rotation between N-terminal extremities of AsH57 and ApH87 TbpBs. This angle is defined by the position of Thr-33 with respect to Lys-43 in the ApH87 and AsH57 TbpBs.
N-lobe of TbpB Varies in Sequence and Structure between Porcine Pathogens
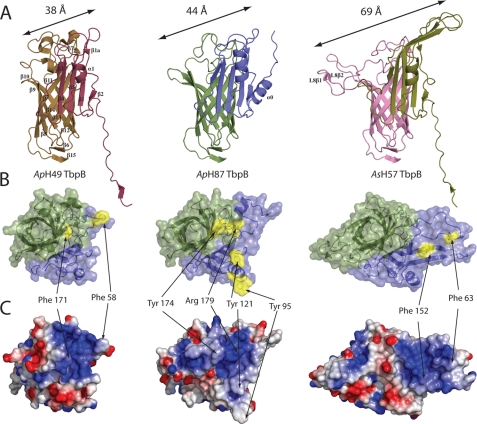
The “cap” area is composed of the loops protruding from the N-lobe β-barrel and handle domain (loop 1, 3, 5, 8, 10, 12 in addition to the long insertion within the loop 4 that points toward the Cap area in ApH57 TbpB) (Fig. 1 and supplemental Fig. S2). This cap region represents the most variable region between the three known TbpB structures, and between all TbpBs in general. Structural alignments of N-lobe domain of AsH57 and ApH87 TbpBs yield an RMSD of 1.7 Å between these two domains, and 3.4 Å and 0.78 Å when respectively compared with ApH49 TbpB.
Focusing on the putative Tf-binding site previously proposed on the cap area (16), a comparison of the cap surfaces reveals many differences between AsH57 TbpB and the two other TbpB receptors. Long insertions in the outward facing loop region within the N-lobe β-barrel and handle domain of AsH57 TbpB results in a larger surface exposed area that is 69 Å in length compared with that of ApH49 or ApH87 TbpB, whose cap areas extend 38 and 44 Å, respectively (Fig. 3A).
FIGURE 3.
A comparison of TbpB N-lobes. A, cartoon representation of TbpB N-lobe from ApH49, ApH87, and AsH57 TbpBs. The common secondary structure elements are labeled on ApH49 TbpB, and additional secondary structural elements found in ApH87 or AsH57 are labeled. B, Cap Tf-binding region of the TbpB N-lobe. Structures are all in the same orientation with a focus on the “cap” region and loops of N-lobe viewed from the top. The β-barrel domain and handle domain are, respectively, colored in green and blue. The aromatic surface exposed amino acids from the cap area are highlighted in yellow and labeled. C, cartoon and electrostatic surfaces are shown for each of ApH49, ApH87, and AsH57 TbpBs with blue and red denoting positive and negative potential.
ApH49 TbpB-pTf Sites of Interaction
Based on our previous model of the Tf-ApH49 TbpB recognition, we mapped the key binding elements involved in the pTf-binding site onto the two new TbpB structures. The first common characteristic shared between the three putative binding pockets is the large positive surface spread on the cap region, which was previously proposed to interact with the Tf C-lobe. This positively charged interface involved loops from both the β-barrel and the handle domain but, in the case of AsH57 TbpB, the site is exclusively localized on its large loop and β-strand (β4 and β5) insertions extended from the handle domain (Fig. 3B).
Characterization of ApH49 TbpB demonstrated Phe-171 as a crucial residue for Tf-binding function. Based on structure overlapping, we detected an equivalent aromatic residue, Tyr-174 in ApH87 at the similar position to ApH49 TbpB Phe171 (Fig. 3B), which is not apparent from sequence alignments (Fig. 1, upper panel). The cap region of AsH57 TbpB is more divergent than the two other crystallized TbpBs and there were no obvious residues in the binding interface that corresponded to Phe-171 or other critical residues from ApH49 TbpB. These variations are essentially due to major structural differences in loop organization of the cap region (Fig. 3B).
Although the cap interface of AsH57 TbpB does not appear to be conserved with ApH49 and ApH87, it still has a conserved feature of two surface exposed hydrophobic residues (Phe-63 and Phe-152) located within the positively charged cap area (Fig. 3B). However, these phenylalanines are localized on the handle domain loop instead of β-barrel as in the ApH87 and ApH49 TbpB structures.
Surface Plasmon Resonance Reveals the Binding Affinities of TbpBs with pTf
Surface plasmon resonance was used to quantify binding affinity between different TbpB preparations and covalently immobilized pTf on a BiacoreX (Table 2 and the supplemental Fig. S3). ApH49 and ApH87 TbpB share closely related cap areas, and an equally similar pTf affinity of 44 and 60 nm, respectively. The AsH57 TbpB has a lower binding affinity of 120 nm. Our SPR data are in agreement with a previous characterization of ApH49 TbpB by isothermal titration calorimetry that revealed a dissociation constant of 55 nm (16).
TABLE 2.
Rate and affinity constants for binding of ApH49, AsH57, and ApH87 TbpB wild types and mutants to pTf
The rate and affinity constants were determined from sensorgrams recorded for the binding of wild type and mutant TbpBs to pTf according to the “heterogeneous ligand” binding model.
| Protein | kon1 | koff1 | Kd |
|---|---|---|---|
| m−1s−1 | s−1 | ||
| ApH49 TbpB | 1.75 105 | 7.7 10−3 | 44 ± 1 nm |
| ApH87 TbpB | 2.12 105 | 1.28 10−2 | 60 ± 2 nm |
| ApH87 TbpB Y121A | 1.50 105 | 3.06 10−2 | 203 ± 8 nm |
| ApH87 TbpB Y95A | 2.94 104 | 1.17 10−2 | 585 ± 11 nm |
| ApH87 TbpB Y174A | 2.61 102 | 2.33 10−3 | 8.9 ± 0.4 μm |
| ApH87 TbpB R179E | nda | nda | 6.1 ± 0.6 μm |
| AsH57 TbpB | 2.52 105 | 3.06 10−2 | 120 ± 4 nm |
| AsH57 TbpB F152A | 6.10 104 | 2.43 10−2 | 405 ± 8 nm |
| AsH57 TbpB F63A | 3.29 105 | 1.07 10−1 | 326 ± 11 nm |
a nd, not determined.
SPR data from the TbpB receptors did not match to a 1:1 Langmuir binding model. Difficulty in fitting our kinetic data to a 1:1 model could be a consequence of ligand/analyte heterogeneity, or two-step binding kinetics. To discriminate between these models we monitored the effect of several saturation times on the dissociation events of TbpBs (supplemental Fig. S3 and Table S1): in case of heterogeneous analyte or two-step binding kinetics, the dissociation will slow as the duration of the injection is increased. Comparing the dissociation phase that occurs after variation in the saturation time, we show that the dissociation curves are not affected by the contact time (supplemental Fig. S3) and that the “heterogeneous ligand” binding model best fits the data.
Mutational Analysis of AsH57 and ApH87 TbpB
The pTf-binding site determined for the previously characterized ApH49 TbpB has common features with the two homologous TbpB structures presented herein. They all share the feature of a positively charged region in the N-lobe cap surface and all contain surface exposed aromatic residues within this region. In ApH49 TbpB this surface exposed aromatic was shown to be essential for binding pTf. Thus all exposed aromatic amino acids from the putative binding sites of AsH57 and ApH87 TbpBs were mutated to alanine and the resulting mutant proteins tested for binding by SPR. Table 2 displays the affinity constants from the wild type and mutant TbpBs. The SPR experiments indicate that Tyr-174, Tyr-121, and Tyr-95 from ApH87 TbpB are involved in pTf recognition, with respective pTf affinities of 8.9 μm, 203 nm, and 585 nm compared with 60 nm for the wild type ApH87 TbpB. Tyr-174 from ApH87 has the strongest impact on the pTf interaction and is localized near to the essential Phe-171 from ApH49 TbpB (Fig. 4B) (16). In the case of AsH57 TbpB, direct binding experiments identify two phenylalanines involved in pTf recognition; Phe-63 and Phe-152 are located in the large protruding loop L1 and the extended β5 strand from the N-lobe handle domain with their side-chains solvent exposed (Fig. 4C).
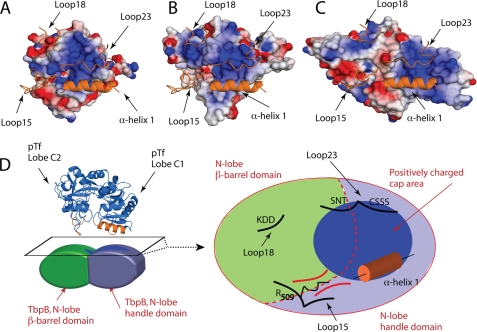
FIGURE 4.
Rosetta-docked models of TbpBs bound to pTf, supported by H/DX-MS. A–C, representation of H/DX-MS experiments (right panel) and Rosetta-docked models (left panel) of ApH49 TbpB (A), ApH87 TbpB (B), and AsH57 TbpB (C) bound to pTf. On the right is the schematic representation of the TbpBs: colored in red are the regions protected in presence of transferrin, in orange are indicated the unprotected sequence, and in light green are regions where there is no information by mass spectrometry (sequence coverage that are missing). At the left are shown the docking models with TbpB electrostatic potential surface and carton-illustrated pTf C-lobe. The pTf C-lobe is colored in blue and binding interface with TbpBs is in orange. All TbpB structures from H/DX-MS and Rosetta-docked models are shown in the same orientation. D, TbpB binding area on pTf is proposed on ApH49, ApH87, and AsH57 TbpB the Rosetta-docked models. A top view of the electrostatic potential surface of the putative TbpB-binding site on pTf is illustrated with structural elements in the binding site (α-helix H1 and the loop elements L15, L18, and L23) labeled.
The loops protruding from the N-lobe β-barrel were also proposed to mediate electrostatic interactions between pTf and TbpB (16). To probe the role of the electropositive surface of the TbpB N-lobe Cap region on pTf affinity, we mutated the conserved, solvent-exposed Arg-179 to Glu within AsH87 TbpB, (equivalent to Arg-175 in ApH49 TbpB). The resultant mutant protein was purified and examined using direct binding assays by SPR. The pTf-binding experiments indicate a drastic lost in binding efficiency of the R179E mutant compared with wild type (60 nm to 6.1 μm binding affinities) (Table 2). The substitution of a single positively charged residue in the cap area to glutamic acid is sufficient to abrogate the pTf binding activity of the ApH87 TbpB.
Models of TbpB AsH57 and ApH87 Bound to pTf
Rosetta docking was performed using the C-lobe of pTf (iron loaded) and the structures of the N-lobes of ApH87 or AsH57 TbpBs. In these docking experiments the mutational data were used to add constraints to the docking algorithm. For both TbpBs, the top 1% of the highest scoring docked models were examined manually, and one heavily favored orientation was found for each (supplemental Fig. S4). These predominant models were homologous within 20 of the top 30 decoys for their respective docking runs. The second most favored orientation had only 3 and 4 homologues each. Though ApH87 and AsH57 TbpB have many major differing structural features, especially around the proposed pTf binding site, the predicted docking orientations of both structures were nearly identical (Fig. 5). Because of this similarity, a new docked model was generated for the previously reported ApH49 TbpB structure, incorporating important residue data as constraints (described under “Experimental Procedures”). The majority of the models (17 of the top 30) resulted in a nearly identical orientation to those of AsH57 and ApH87 models.
FIGURE 5.
The conserved elements of the TbpB-pTf Binding Interface. A–C, N-lobe Cap areas of ApH49, ApH87, and the AsH57 TbpBs are represented in A, B, and C, respectively. An orange cartoon representation is the pTf predicted TbpB-binding elements from the Rosetta-docked TbpB-pTf models. In each model, the arginine 509 from the loop 15 of pTf is shown in stick representation. D, top surface of the TbpB N-lobe demarking the barrel and handle domain in red-dotted line. The negatively charged cap area is represented in dark blue, the β-barrel in green, and the handle domain in blue. Structural elements from the pTf C-lobe localized at the interface with TbpBs are drawn in black (loop 15, 18, 23, and α-helix1). The loop 23 and the helix H1 (orange) dock on the TbpB positively charged cap area, driven by electrostatic interaction. pTf loop 15 makes contact with TbpB at the between the β-barrel and handle domain, where the Arg-509 seats deeply in the canyon formed by the two domains of the N-lobe.
In our three independently generated models, the TbpB-binding interface on pTf is conserved. The C1 and C2 domains of the transferrin C-lobe bind to the handle and β-barrel domain of TbpBs, respectively (Fig. 5). Porcine transferrin docks on TbpB with the α-helix H1 (residues 352–366) and the loop elements L15 (residues 507–513 and 519–521), L18 (residues 555–559), and L23 (residues 624–627 and 633–636). A conserved binding pattern emerges from the Rosetta docked models, which suggests the helix and loop elements H1, L15, L18, and L23 of pTf form the TbpB binding interface on pTf (Fig. 5). The negatively charged α-helix H1 and loop23 (consisting of the motif CSSS, residues 624–627) of pTf binds the positively charged cap area of the TbpBs. Loop15 of pTf makes several contacts with TbpBs, but one key residue contact that is central to each model is the positively charged Arg-509, which is oriented toward a negative pocket in the canyon formed between the N-lobe β-barrel and handle domains.4 Loop18 (non uniformly charged) of pTf makes contact on the β-barrel domain but we note that the electrochemical environment on TbpB is not maintained between the three-studied TbpBs.
Porcine Transferrin Binds TbpB on the Cap Area
To localize and confirm the Tf-binding site location, we performed H/DX-MS. Results from experiments with TbpB from three strains; A. pleuropneumoniae H49 and H87, and A. suis H57 alone, and upon complex formation with pTf, are presented in Fig. 5. For the ApH49, AsH57, and ApH87 TbpBs, a total of 57, 58, and 54 pepsin-digested peptides, respectively, could be utilized, covering 46, 45, and 43% of the protein sequences and 69, 60, 61% of the respective cap region (supplemental Fig. S5). The isotopic mass shift due to deuterium incorporation in TbpB fragments were detected using a QStar pulsar mass spectrometer, and analyzed via Hydra software, as described under “Experimental Procedures.” Despite incomplete sequence coverage of the receptors with a pepsin digestion, the protected sequences of the three-studied TbpBs are all located on the N-lobe domain and more precisely localized along the cap area of each of the porcine TbpBs studied (Fig. 4).
DISCUSSION
Structural Features of TbpBs
The two new structures reported in this study demonstrate that the overall structural features of TbpBs are conserved despite considerable sequence variation and size differences. Each lobe is comprised of an 8-stranded β-barrel with a 5-stranded handle domain in the N-lobe and a 6-stranded handle domain in the C-lobe (supplemental Fig. S2). The size and sequence differences are largely manifested by variation in the loops that constitute the Cap region of the N-lobe. There is a conserved interaction between the N-terminal lobe that contains the Tf binding interface and the relatively conserved C-lobe, in an orthogonal orientation. Although the C-lobe is not directly involved in Tf binding, its presence and conserved interaction with the N-lobe could be important for the receptor function in vivo. The lack of sequence variation in the C-lobe compared with the N-lobe may infer a conserved interaction with other surface components.
In the ApH49 and AsH57 proteins the extended N-terminal anchoring peptide interacts through hydrogen bonds with the C-lobe. In the ApH87 structure the anchoring peptide assumes a different conformation that appears to be stabilized by interchain contacts with the adjacent TbpB in the crystal lattice. The complete N-terminal anchoring peptides consist of more than 43 residues that could equate to ∼100 Å in length if in an extended conformation in solution. Thus the two observed conformations (Fig. 2) suggest that the N-terminal membrane-anchored peptide may allow the TbpB receptor to adopt an extended orientation to more effectively capture host Tf in the presence of the polysaccharide capsular layer.
TbpB Conservation of the pTf-binding Sites
Based on our pTf-TbpB model, there are substantial variations in the TbpB N-lobe Cap regions, however all strains studied permit high affinity specific binding interactions with Tf. The cap region involved in pTf recognition is directly exposed to the host environment, including the immune defense elements. The sequence and structural variation of this cap region likely is a mechanism for immune evasion, providing decoy epitopes that vary between strains so that a primarily strain-specific immune response is induced (27). Because the different receptor proteins need to retain the ability to bind to transferrin in a similar fashion, there should be conserved features that could otherwise induce a cross-reactive immune response. Thus there would be selective pressure for directing the immune response to variable regions or to accomplish a somewhat conserved Tf interaction by variable means.
Based on the three solved structures of TbpB we can classify the pTf-binding sites into at least two groups: ApH49 and ApH87 TbpBs that share similarities in the loop's cap area, and AsH57 TbpB that has a large extended loop containing some new secondary structural elements. The results from this study suggest that AsH57 TbpB adopts a novel interaction pattern with pTf, as the binding interface appears highly modified compared with ApH87 and ApH49 (Fig. 3A). The cap area of AsH57 TbpB is also identified by H/DX-MS experiments as an interfacial region with pTf. The two protruding loop regions of AsH57 TbpB create a larger surface area buried in the model (∼4400 Å2) with pTf (Fig. 4C). The strikingly different loop structure while maintaining interactions with the same regions of pTf has important implications on how this diversity is created and on potential barriers to horizontal exchange of antigenic diversity.
Using a structure-based approach, TbpB mutants were selectively designed to probe related effects on function with the goal to more precisely define the binding activity of these prokaryote pTf receptors. Based on previous studies of ApH49 TbpB (16), we first focused on the solvent exposed aromatic residues accessible from the cap area of AsH57 and ApH87 TbpBs. SPR binding experiments with all the alanine substitutions of phenylalanine and tyrosine residues within the putative binding sites, revealed that similar amino acids to the ApH49 TbpB Phe-171 are also required for the pTf binding of AsH57 and ApH87 TbpBs. These binding experiments point to the importance of the Tyr-95 and Tyr-121 and the structurally conserved Tyr-174 on ApH87 TbpB (Fig. 4B), and the non-conserved Phe-63 and Phe-152 on AsH57 TbpB (Figs. 4C). Interestingly, the phenylalanines 63 and 152 are localized on the protruding loop L1 and the extended β5-strand from the handle domain instead of the β-barrel as in the case of the essential aromatic residues from ApH49 and ApH87 TbpBs (Fig. 4).
Despite the loop variation between the pTf-binding sites, the swine pathogen TbpBs share similar characteristics for capturing porcine Tf. In addition to the essential Phe and Tyr hydrophobic residues on the cap area, the pTf-TbpB recognition seems to depend on a positively charged interface, which is predicted at the pTf interface by computational Rosetta docking and by experimental HDX-MS data. Indeed the putative pTf-binding sites from the three TbpB structures all maintain a positively charged interface on the cap area, which matches to the neighborhood of the essential Phe and Tyr (Fig. 3B). To test the pTf binding dependence toward this positively charged area, we prepared an ApH87 TbpB mutant in which the Arg-179 has been substituted by a glutamic acid. This mutant allowed us to measure the deficiency in pTf-binding caused by a partial charge disruption in the putative binding site. The mutation was sufficient to disrupt the pTf binding function by insertion of a single negatively charged amino acid.
All three TbpB structures present similar characteristics involving aromatic and positively charged residues, but they show variability in localization. The binding contributors from both ApH49 TbpB and ApH87 have partial similarity in arrangement (Fig. 3B), but differ more drastically with the structurally non-conserved AsH57 TbpB binding site (Fig. 3B). The independent Rosetta docked models from ApH49, ApH87, and AsH57 predict the N-lobe Cap area as the pTf-binding pocket of the swine-related TbpBs. The H/DX-MS experimental data validates our three Rosetta docked models, confirming the Cap area as the TbpB/pTf interface. Although H/DX-MS overall TbpB sequence coverage of the three TbpB strains was low, coverage of the cap regions was quite high, with most of the missing coverage being attributed to the rigid β-barrels in each of the TbpB lobes. In agreement with our models, most of the protruding loops from the cap area are protected in presence of pTf (Fig. 4).
Despite the divergence in the TbpB interface, the conserved physicochemical properties of the binding sites support idea that these TbpB could target a common binding-site on pTf. Based on our Rosetta docking experiments, TbpB receptors bind to the same surface on pTf (Figs. 4 and 5). Our three independently generated TbpB/pTf binding models suggest the loop elements L15, L18, L23, and the α-helix H1 as the TbpB-binding site on pTf.
Despite a high sequence homology between Tfs from different species (70% between pTf and hTf C-lobes), the TbpB binding function for Tf is host specific. Interestingly, amino-acids from pTf that are predicted to contact the porcine pathogen TbpBs are not conserved when compared with hTf (supplemental Fig. S6), suggesting that pathogens have adapted to discriminate specific host Tf with higher affinity; or a host evolution as defense mechanism to avoid infection from Tf-based pathogens.
Implication in Vaccine Design
TbpBs have been targeted for development of vaccines for a number of human and veterinary pathogens due to the critical role they mediate and their resulting invariant presence in clinical disease isolates (14, 27–29). Although they are capable of inducing a protective immune response, there is an insufficient cross-protective response induced by individual intact receptor proteins to be considered as suitable vaccine antigens. The fact that these proteins mediate a conserved interaction with the host Tf provided some optimism of ultimately targeting conserved regions of the proteins for developing a more cross-protective antigen. Furthermore, if common Tf-binding regions could be targeted by the immune response, the bacteria would be unable to evade the immune response by ongoing antigenic variation as this could also compromise the critical function mediated by TbpB.
The results from this study demonstrate that there is no obvious conserved Tf-binding region in the N-terminal lobe common to all the TbpBs from porcine pathogens that could simply be used as vaccine antigen capable of inducing a cross-protective immune response. It seems evident that the divergent TbpBs accomplish binding of porcine Tf with common, but not identical, features and that further study will be needed to fully appreciate the molecular details of this interaction. Clearly the most informative approach would be determining structures of pTf-TbpB complexes but these have been evasive to date, and a combination of alternate approaches may need to be considered. A detailed appreciation of the pTf-TbpB interaction may ultimately provide insights on how to develop one or more receptor derivatives or mimics capable of inducing a cross-protective response against all porcine pathogens. Expanding this approach to developing vaccines against human pathogens will require gathering structural information on a large set of TbpB homologues to develop broad-spectrum protection again multiple pathogenic species.
Acknowledgments
We thank members of Canadian Light Source (CLS) beamline staff at CMCF-08ID-1 for assistance with data collection and members of the Moraes and Schryvers laboratories for valuable discussion.
This research was funded with operating and infrastructure support provided by Canadian Foundation for Innovation (CFI), Alberta Heritage Foundation for Medical Research (AHFMR), and Canadian Institutes of Health Research (CIHR).
The atomic coordinates and structure factors (codes 3PQS and 3PQU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
L. P. Silva, R. Yu, C. Calmettes, X. Yang, T. F. Moraes, A. B. Schryvers, and D. C. Schriemer, in preparation.
- Tf
- transferrin
- pTf
- porcine transferrin
- hTf
- human transferrin
- TbpB
- Tf-binding protein B
- TEV
- Tobacco Etch Virus.
REFERENCES
- 1. Ratledge C., Dover L. G. (2000) Annu. Rev. Microbiol. 54, 881–941 [DOI] [PubMed] [Google Scholar]
- 2. Gray-Owen S. D., Schryvers A. B. (1996) Trends Microbiol. 4, 185–191 [DOI] [PubMed] [Google Scholar]
- 3. Cornelissen C. N., Kelley M., Hobbs M. M., Anderson J. E., Cannon J. G., Cohen M. S., Sparling P. F. (1998) Mol. Microbiol. 27, 611–616 [DOI] [PubMed] [Google Scholar]
- 4. Yu R. H., Schryvers A. B. (1993) Microb. Pathog. 15, 433–445 [DOI] [PubMed] [Google Scholar]
- 5. Cornelissen C. N., Biswas G. D., Tsai J., Paruchuri D. K., Thompson S. A., Sparling P. F. (1992) J. Bacteriol. 174, 5788–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Retzer M. D., Yu R., Zhang Y., Gonzalez G. C., Schryvers A. B. (1998) Microb. Pathog. 25, 175–180 [DOI] [PubMed] [Google Scholar]
- 7. Zak O., Aisen P. (1986) Blood 68, 157–161 [PubMed] [Google Scholar]
- 8. Baltes N., Hennig-Pauka I., Gerlach G. F. (2002) FEMS Microbiol. Lett. 209, 283–287 [DOI] [PubMed] [Google Scholar]
- 9. Schryvers A. B., Gonzalez G. C. (1990) Can J. Microbiol. 36, 145–147 [DOI] [PubMed] [Google Scholar]
- 10. Cheng Y., Zak O., Aisen P., Harrison S. C., Walz T. (2004) Cell 116, 565–576 [DOI] [PubMed] [Google Scholar]
- 11. Kaplan J. (2002) Cell 111, 603–606 [DOI] [PubMed] [Google Scholar]
- 12. Pizza M., Scarlato V., Masignani V., Giuliani M. M., Aricò B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., Galeotti C. L., Luzzi E., Manetti R., Marchetti E., Mora M., Nuti S., Ratti G., Santini L., Savino S., Scarselli M., Storni E., Zuo P., Broeker M., Hundt E., Knapp B., Blair E., Mason T., Tettelin H., Hood D. W., Jeffries A. C., Saunders N. J., Granoff D. M., Venter J. C., Moxon E. R., Grandi G., Rappuoli R. (2000) Science 287, 1816–1820 [DOI] [PubMed] [Google Scholar]
- 13. Lissolo L., Maitre-Wilmotte G., Dumas P., Mignon M., Danve B., Quentin-Millet M. J. (1995) Infect. Immun. 63, 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Danve B., Lissolo L., Mignon M., Dumas P., Colombani S., Schryvers A. B., Quentin-Millet M. J. (1993) Vaccine 11, 1214–1220 [DOI] [PubMed] [Google Scholar]
- 15. Gray-Owen S. D., Schryvers A. B. (1993) Microb Pathog 14, 389–398 [DOI] [PubMed] [Google Scholar]
- 16. Moraes T. F., Yu R. H., Strynadka N. C., Schryvers A. B. (2009) Mol. Cell 35, 523–533 [DOI] [PubMed] [Google Scholar]
- 17. del Río M. L., Gutiérrez-Martin C. B., Rodríguez-Barbosa J. I., Navas J., Rodríguez-Ferri E. F. (2005) FEMS Immunol. Med. Microbiol. 45, 75–86 [DOI] [PubMed] [Google Scholar]
- 18. Bahrami F., Ekins A., Niven D. F. (2003) Vet. Microbiol. 94, 79–92 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalez G. C., Caamano D. L., Schryvers A. B. (1990) Mol. Microbiol. 4, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 25. Slysz G. W., Baker C. A., Bozsa B. M., Dang A., Percy A. J., Bennett M., Schriemer D. C. (2009) BMC Bioinformatics 10, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ling J. M., Shima C. H., Schriemer D. C., Schryvers A. B. (2010) Mol. Microbiol. 77, 1301–1314 [DOI] [PubMed] [Google Scholar]
- 27. Rossi-Campos A., Anderson C., Gerlach G. F., Klashinsky S., Potter A. A., Willson P. J. (1992) Vaccine 10, 512–518 [DOI] [PubMed] [Google Scholar]
- 28. Myers L. E., Yang Y. P., Du R. P., Wang Q., Harkness R. E., Schryvers A. B., Klein M. H., Loosmore S. M. (1998) Infect. Immun. 66, 4183–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potter A. A., Schryvers A. B., Ogunnariwo J. A., Hutchins W. A., Lo R. Y., Watts T. (1999) Microb. Pathog. 27, 197–206 [DOI] [PubMed] [Google Scholar]