Abstract
Inositol-requiring enzyme 1 (IRE1) is the most highly conserved signaling node of the unfolded protein response (UPR) and represents a potential therapeutic target for a number of diseases associated with endoplasmic reticulum stress. IRE1 activates the XBP-1 transcription factor by site-specific cleavage of two hairpin loops within its mRNA to facilitate its nonconventional splicing and alternative translation. We screened for inhibitors using a construct containing the unique cytosolic kinase and endoribonuclease domains of human IRE1α (hIRE1α-cyto) and a mini-XBP-1 stem-loop RNA as the substrate. One class compounds was salicylaldehyde analogs from the hydrolyzed product of salicylaldimines in the library. Salicylaldehyde analogs were active in inhibiting the site-specific cleavage of several mini-XBP-1 stem-loop RNAs in a dose-dependent manner. Salicyaldehyde analogs were also active in inhibiting yeast Ire1 but had little activity inhibiting RNase L or the unrelated RNases A and T1. Kinetic analysis revealed that one potent salicylaldehyde analog, 3-ethoxy-5,6-dibromosalicylaldehyde, is a non-competitive inhibitor with respect to the XBP-1 RNA substrate. Surface plasmon resonance studies confirmed this compound bound to IRE1 in a specific, reversible and dose-dependent manner. Salicylaldehydes inhibited XBP-1 splicing induced pharmacologically in human cells. These compounds also blocked transcriptional up-regulation of known XBP-1 targets as well as mRNAs targeted for degradation by IRE1. Finally, the salicylaldehyde analog 3-methoxy-6-bromosalicylaldehyde strongly inhibited XBP-1 splicing in an in vivo model of acute endoplasmic reticulum stress. To our knowledge, salicylaldehyde analogs are the first reported specific IRE1 endoribonuclease inhibitors.
Keywords: Enzyme Inhibitors, ER Stress, Protein Folding, RNA Processing, Transcription Factors, IRE1, RNase, XBP-1
Introduction
Protein folding perturbations resulting in endoplasmic reticulum (ER)2 stress are thought to play a role in the pathogenesis of diseases as diverse as neurodegeneration, diabetes, and cancer. The unfolded protein response (UPR) coordinates the ability of a cell to respond to ER stress by altering protein translation, folding, and post-translational modification of all secreted and membrane proteins. Terminally unfolded proteins are retrotransported to the cytosol by the ER-associated degradation machinery for proteolysis by the proteasome. The ER is also the site of lipid biosynthesis and membrane expansion. These activities are linked physiologically to specialized secretory cells; however, depending on stress levels, the UPR can control cellular survival or death via autophagy and apoptosis (1). Tumors may utilize the UPR as a survival mechanism against nutrient-related stress as a result of poor vascularization and rapid growth (2). Secretion and membrane composition are balanced by the high caloric demand of these activities against the energy homeostasis of the cell (1) and perturbations can result in activation of innate immunity and inflammation (3, 4).
Inositol-requiring enzyme 1 (IRE1) is the most highly conserved signaling node of the unfolded protein response (5, 6). A unique ER-resident transmembrane kinase with a novel C-terminal endoribonuclease domain, IRE1 is activated in part by the disassociation of BiP/GRP78 in the presence of unfolded protein in the ER lumen (1). The signal is transduced to the cytosol by the sequential dimerization/multimerization, trans-autophosphorylation, and activation of its endoribonuclease (7). The specific activity of the endoribonuclease is responsible for the unconventional cytosolic splicing of HAC1 in yeast (7) and excision of the 26-nucleotide intron of the X-box-binding protein 1 (XBP-1) transcription factor in metazoan organisms (8, 9). In mammalian cells IRE1 acts in concert with companion UPR signaling molecules: PKR-like ER resident kinase and ATF6 (1).
In response to ER stress, the PKR-like ER resident kinase dimerizes in a manner similar to IRE1 to promote its autophosphorylation activity. Once activated, PKR-like ER resident kinase phosphorylates the eukaryotic translation factor eIF2α to reduce protein translation and translocation into the ER (10). This mechanism quickly relieves the load on the folding machinery in the ER allowing time to readjust and coordinate upstream activities by transcriptional induction. Highly specific translation of the ATF4 transcription factor during the eIF2α enforced state leads to up-regulation of ER resident chaperones, redox enzymes, and proapoptotic and autophagic factors such as CHOP and MAPKC3B, respectively (11).
After experiencing ER stress, ATF6, the third UPR signaling regulator, is translocated to the Golgi and cleaved by the dual sterol regulatory element-binding protein proteases called site-1 protease (S1P) and site-2 protease (S2P) (1). Release of the cytosolic portion produces the active ATF6 transcription factor, which targets transcriptional up-regulation of an overlapping and distinct set of ER resident chaperones such as GRP78/BiP and GRP94 (12, 13).
XBP-1 mRNA, a major substrate of the IRE1 endoribonuclease, is cleaved specifically at two conserved stem-loop sites. Each site is located 3′ to a mirrored guanosine residue in the 7-base loop (8, 9). The resulting internal fragment, a 26-nucleotide intron, is removed and the two exon ends are ligated by an unknown mechanism in mammalian cells (14) and tRNA ligase in yeast (15). The rejoined mRNA shifts the open reading frame (ORF) and extends the C-terminal domain of XBP-1 from amino acid 164 with an alternative 212-amino acid reading frame producing the active “spliced” transcription factor, XBP-1s, which regulate a broad range of ER resident chaperones, ER translocon channels, ER-associated degradation components (16), and lipid metabolic enzymes (17, 18). XPB-1s acts alone as a homodimer but can also heterodimerize with ATF6 (12). The unspliced form of XBP-1 (XBP-1u) is short lived and rapidly degraded by the proteasome (19). It is thought to negatively regulate XBP-1s and ATF6 (19, 20). In this manner the activity of IRE1 can rapidly control downstream activities of the UPR. An emerging and important activity of IRE1 is the degradation of mRNAs encoding ER-targeted membrane and secreted proteins during stress analogous to the less discriminate activities of evolutionary homologue of the IRE1, RNase L (21). This activity in combination with expression of the XBP-1 has the potential to alter the surface composition of stressed cells and the extracellular proteome.
The cytosolic portion of IRE1 contains an active kinase domain and a novel endoribonuclease (7): the kinase-extension endoribonuclease (KEN) domain (22). The cytosolic kinase and endoribonuclease crystal structure was solved for the yeast Ire1 (22, 23) and the structures of the luminal domains of both yeast (24) and human (25) were determined previously; therefore, a near complete structure of IRE1 can be assembled. The activation of IRE1 endoribonuclease is controlled by dimerization and trans-autophosphorylation of the kinase. In yeast, ADP serves as a cofactor after activation, where binding to the kinase cleft promotes a closed conformation, which in turn stabilizes the dimer of the endoribonuclease KEN domain and promotes endoribonuclease activity (22). Using a mutant yeast Ire1 Papa et al. (26) showed that an ATP competitive drug could activate the endoribonuclease in an analogous manner. Additional studies confirmed that ATP competitive kinase inhibitors can act as yeast Ire1 endoribonuclease activators (23), a potential therapeutic modality to induce the cytoprotective activities of XBP-1s. Recent studies have demonstrated that small molecules such as quercetin can act as agonists by binding to sites remote from the ATP binding site of the kinase domain but still act by promoting dimerization (27).
In an attempt to discover inhibitors of XBP-1 mRNA splicing, we produced the soluble cytosolic fragment of human IRE1 (hIRE1α-cyto) as a GST fusion protein in insect cells. The purified and GST-free hIRE1α-cyto protein was active and cleaved XBP-1 substrates in a sequence-specific manner. We screened 220,000 compounds using a fluorescently labeled mini-XBP-1 stem-loop RNA substrate. One class of inhibitor found was salicylaldimine analogs. We found that the active component of these library compounds was the salicylaldehyde form of the salicylaldimine. These salicylaldehyde compounds were specific for inhibiting the IRE1 endoribonuclease activity, and were active in cells to inhibit XBP-1 splicing as well as in ER stress models in vivo.
EXPERIMENTAL PROCEDURES
Baculovirus Expression and Purification of hIREα-cyto
Expression of GST-hIRE1α-cyto was performed using the Bac-N-Blue Baculovirus system and SF9 insect cells essentially as described by the Bac-N-Blue transfection manual (Invitrogen). GST was fused to the N-terminal end of the cytosolic human IRE1α (amino acids 462–977) in the pBacPac4.5 transfer vector and included a PreScission Protease cleavage site in the linker. Recombinant baculoviruses were obtained by picking clear plaques after co-transfection of bacmid DNA and amplified 3 times to obtain high titer P3 stocks. Insect HiFive cells were infected at multiplicity of infections in the 2 to 5 range and were grown from a 500-ml shaking insect cell culture for 48 h at 28 °C. The insect cells were lysed by suspending the cells in Buffer A (25 mm Tris-HCl, pH 7.5, 50 mm KCl, 5 mm MgCl2, 1 mm EDTA, 2.5 mm DTT, 0.1 mm ATP, 10% sterile glycerol, 0.005% Nonidet P-40, 1 μg/ml of leupeptin, 100 mm NaF, 100 mm NaVO4, 100 mm PMSF; 30 ml/500-ml culture), transferring the suspension to a high speed centrifuge tube, and sonicating the suspension on ice. The sonicated preparation was spun at 13,000 × g for 30 min at 4 °C. The supernatant was combined with glutathione-Sepharose beads in a tube and gently mixed on a rotator for 1–2 h at 4 °C. After binding, the bead mixture was transferred to a PD-10 column from Amersham Biosciences. The column was washed five times with Buffer A followed by two washes with Buffer B (25 mm Tris-HCl, pH 7.5, 50 mm KCl, 2.5 mm MgCl2, 1 mm EDTA, 2.5 mm DTT, 10% sterile glycerol, 0.0025% Nonidet P-40). The GST tag was removed using Prescission protease (GE Healthcare) cleavage. Cleavage buffer (825 μl of Buffer B, 350 μl of sterile glycerol, and 35 μl of PreScission protease/ml of beads) was added to the column and incubated for 4 h at 4 °C with tumbling. The final product was collected in the final eluate. hIREα-cyto preps were dialyzed in storage buffer (17.0 mm Tris-HCl, pH 7.5, 34.0 mm KCl, 1.7 mm MgCl2, 2.0 mm DTT, 0.0017% Nonidet P-40, and 20% glycerol). Typically, 500-ml insect cell cultures produced roughly 0.5 mg of purified hIRE1α-cyto, which was concentrated, titrated for activity, pooled, re-aliquoted, and stored at −80 °C.
Bacterial Expression and Purification of RNase L Catalytic Fragment
Residues 333–651 of mouse RNase L was expressed as a polyhistidine-tagged fusion in Escherichia coli using the pPROX-HTA vector system (Invitrogen) and purified as described previously for the expression of yeast Ire1cyto for structural studies (22).
In Vitro Endoribonuclease Assays
Endoribonuclease assays were performed as previously described for yeast (27) and human IRE1 (29). Briefly, reactions were run in 10- or 20-μl volumes using IRE reaction buffer (20 mm HEPES, pH 7.5, 50 mm KOAc, 0.5 mm MgCl2, 3 mm DTT, and 0.4% polyethylene glycol) and various amounts of hIRE1α-cyto (typically 0.01 to 1 μm) and substrate concentrations ranging from 0.1 to 10 μm at 30 °C. Fluorescently tagged RNA oligos were read using an Acquest 384 plate reader (LJL Biosystems). In addition, reaction products were visualized by denaturing 15% TBE urea in 12-well gels (Invitrogen) using a Bio-Rad molecular Imager FX. Unlabeled oligos were stained with SYBR Gold (Invitrogen). RNA oligos were purchased from IDT DNA Technologies. RNase A and T1 were purchased from Sigma.
High-throughput Screening
The MannKind chemical library of 220,000 individual compounds was screened in 384-well Greiner Bio-one polypropylene plates (Greiner). Columns 1 and 2 of each plate served as positive controls (no compound) and rows 23 and 24 as negative controls (no compound, no hIREα-cyto). First, the reaction buffer was loaded in plates using a Beckman-Coulter Biomek FX robot. Next, 25 nl of each compound from a 10 mm DMSO compound stock was pinned (using a V&P scientific pinhead on the Biomek FX pin tool) into the reaction mixture (final concentration = 20 μm) singly from the library stock plates stored at −20 °C. Next, 1 μl of hIREα-cyto was added to each reaction well using fresh tips. Plates were incubated at room temperature for 5 min before adding 1 μl of Cy5-labeled (Fig. 1C) mini-XBP-1 stem-loop RNA. The final volume for each well was 10 μl. Buffer concentrations were the same as for the endoribonuclease assay except for the presence of 5% DMSO; the hIRE1α-cyto concentration was ∼10 nm and the RNA substrate was 100 nm. Plates were then placed in an incubator at 30 °C for 1 h. After incubation, plates were immediately transferred to an Acquest 384-well plate reader with settings and filters for the Cy5 fluorophore. Data were acquired, transferred to an ActivityBase database, and analyzed using ActivityBase discovery software (IDBS). Activity was expressed as the percentage of the compound treated well minus the negative control divided by the positive control minus the negative control ((test − negative)/(positive − negative) × 100). The cut-off for hits was considered at 60% of positive control. Hits were confirmed in triplicate using the above procedure. If compounds remained active in two of three wells they were considered bona fide hits. The integrity of the hits was analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Shimadzu LCMS-2010.
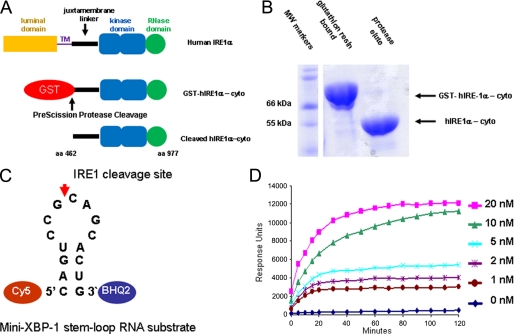
FIGURE 1.
Recombinant expression and activity of the purified cytosolic human IRE1α kinase endoribonuclease. A, expression strategy for a cytosolic fragment (amino acids 462–977) of human IRE1α (hIRE1α-cyto) using a baculovirus/insect cell system. hIRE1α-cyto was liberated from GST using PreScission protease. B, SDS-polyacrylamide gel electrophoresis analysis of GST-fused and PreScission-liberated hIRE1α-cyto. C, schematic of the mini-XBP-1 stem-loop used as the hIRE1α-cyto substrate for HTS and IC50 analysis; the Cy5 fluorophore was linked to the 5′ end and black hole quencher 2 (BHQ2) was linked to the 3′ end. D, time course analysis of mini-XBP-1 substrate cleavage (100 nm starting concentration) by the indicated concentrations of hIRE1α-cyto.
IC50 Analysis
Several salicylaldimine hit compounds were re-purchased from the library supplier (Maybridge, UK). Salicylaldehyde compounds described in this study were purchased from Sigma or Princeton Biomolecular Research. Compounds were resuspended to 10 mm in DMSO and stored at −20 °C as a stock solution. Compounds were diluted 1:3 in IRE1 reaction buffer 9 times, starting at 20 μm and down to 1 nm. Assays were run on triplicate plates and analyzed as described for HTS. IC50 values were calculated as the mean of 3 reactions that inhibited 50% of the positive control.
Kinetics Assays
In black clear bottom 384-well plates, 10 nm hIRE1α-cyto was combined with a range of compound concentrations (0, 12.5, 25, 50, 100, and 200 nm) in IRE1 reaction buffer (20 mm HEPES, pH 7.4, 50 mm KOAc, 0.5 mm MgCl2, 0.4% polyethylene glycol, 3 mm DTT), with a total volume of 10 μl. The plate was incubated at RT for 10 min, and the fluorescent substrate was added (0.5, 1, 2, 3, 5, and 7 μm). The plate was immediately read at 45-s kinetic reads for 1 h at 30 °C, excitation 620 nm and emission 680 nm on a FLUOstar OPTIMA plate reader (BMG LABTECH). Data were analyzed using Visual Enzymics software (SoftZymics Inc.).
Surface Plasmon Resonance
Compound Binding with a Protein-immobilized Chip
Protein (hIRE1α-cyto or RNase A) was immobilized with the sample chamber set at 4 °C and analysis chamber at 30 °C. A CM5 sensor chip (GE Healthcare) was primed once with pure water and twice with IRE1 reaction buffer. Immobilization was performed using the amine coupling kit (GE Healthcare), following the manufacturer's instructions. All injections were at 10 μl/min. RNase A, used as a control, was injected to 3000 response units; hIRE1α-cyto to 12,000–15,000 response units. Immediately after the protein was bound, running buffer was switched to IRE1 reaction buffer plus 5% DMSO, the sample compartment was set at 25 °C and the analysis compartment at 30 °C, and the instrument was primed twice. The injection series was as follows: 6–8 startup cycles with buffer, followed by a solvent correction cycle and 2 DMSO only samples to stabilize baseline. Compounds were prepared so the final solution has 5% DMSO in IRE1 reaction buffer. Compounds were injected over the surface of the chip at 30 μl/min for 3 min. Between injections, compound bound to the chip was removed with a 50% DMSO regeneration solution.
Biacore Substrate Binding with Protein-immobilized Chip
This procedures was performed the same as above except DMSO was not used; the running buffer was IRE1 reaction buffer. Two concentrations of substrate (0.8 and 4 μm) were diluted in IRE1 buffer and run on the chip. No solvent correction curve was run.
Biacore hIRE1α-cyto Binding Experiments with Substrate-bound Chip
The biotinylated RNA substrate was prepared as follows: RNA was diluted in RNA running buffer (10 mm Tri-HCl, pH 8.0, 150 mm NaCl) to 1 μm and heated at 80 °C for 10 min, cooled to RT, then diluted 500 times in immobilization buffer (RNA RB plus 0.005% P-20, 62.5 μg/ml of BSA, 125 μg/ml of tRNA, 1 mm DTT, and 5% glycerol). RNA was then bound to the streptavidin sensor chip at about 200 response units (GE Healthcare), according to the manufacturer's instructions. hIRE1α-cyto was diluted to 100 nm in IRE1 reaction buffer, and run over the surface of the chip at 100 μl/min for 2 min. Data analysis was performed using Biaevaluation software (GE Healthcare) and Excel.
Biacore Compound Binding Experiments with Substrate-bound Chip
The procedure for substrate immobilization was the same as above. Compounds were injected over the surface of the chip at 30 μl/min for 3 min. Experiments were performed on a Biacore T100 (GE Healthcare); data analysis was performed using Biaevaluation software (GE Healthcare) and Excel for all of the above experiments.
Cell Culture
HEK293, MM1.s, and U266 cells were grown in monolayer culture using Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), unless specified otherwise, at 37 °C and 5% CO2. Salicylaldehyde compounds were kept as 10 mm stocks in DMSO at −20 °C and diluted in medium as indicated. Tunicamycin (Tm) and thapsigargin (Tg) were resuspended in DMSO and diluted in medium. DTT was made fresh in sterile deionized H2O as a 1 m stock.
RT-PCR
Procedures for measuring XBP-1s and XBP-1u mRNAs have been described previously (19). Briefly, total RNA was harvested from cells or tissue using TRIzol according to the manufacturer's procedures. After ethanol precipitation and resuspension of the RNA, RiboGreen (Invitrogen) was used to quantify the yield and normalize the RNA concentration in the source tube containing isolated RNA. RT-PCR was performed by Oligo(dT) priming, and SuperScript II (Invitrogen) transcription using the Amplitaq Gold Kit (Applied Biosystems) according to the manufacturer's protocols. Primers for human XBP-1 were 5′-CCTGGTTGCTGAAGAGGAGG-3′ (forward) and 5′-CCATGGGGAGATGTTCTGGAG-3′ (reverse), and for mouse were 5′-ACACGCTTGGGAATGGACAC-3′ (forward) and 5′-CCATGGGAAGATGTTCTGGG-3′. All DNA oligos were purchased from IDT DNA Technologies. PCR were run on a Bio-Rad PTC-100 96-well thermocycler with heating at 94 °C for 30 s, annealing at 58 °C for 30 s, and polymerizing at 72 °C for 30 s for 35 cycles. Reactions were run on 4% precast NuSieve gels from Cambrex and visualized by ethidium bromide staining and UV excitation.
Western Blot Analysis
Cells were lysed in RIPA buffer containing 150 mm NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0, 1 mm PMSF, 1× phosphatase inhibitor mixture (Sigma) for 20 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis after solubilizing the samples by SDS sample buffer (Invitrogen) and heating at 95 °C for 5 min. A NuPAGE 12% Tris gel (Invitrogen) was used to separate the proteins. Proteins was transferred to a PVDF membrane and probed with rabbit anti-hIRE1α-cyto polyclonal antibody (18) or anti-hIRE1α phospho-specific (Ser-724) antibody (Novus Biologicals). Blots were incubated with anti-rabbit IgG HRP-conjugated secondary antibody and proteins were visualized using enhanced chemiluminescent substrate (Thermo Scientific). Anti-α-tubulin and anti-β-actin antibodies (Sigma) were used to normalize the protein amount loaded on the gel.
Animal Studies
Pathogen-free 8–10-week-old female SCID CB17 mice (Taconic Farms, Oxnard, CA) were housed at MannKind Corporation according to IACUC guidelines. All animal studies were reviewed and approved by an IACUC institutional review committee. Mice were housed in HEPA-filtered cage racks, fed ad libitum, and euthanized by CO2 asphyxiation at the time of organ harvest at the end of the experimental treatment. Tm (Sigma) was made fresh before the experiment by dissolving in DMSO.
RESULTS
High-throughput Screen
Studies with yeast Ire1 have demonstrated that kinase inhibitors can act as allosteric agonists for the endoribonuclease (23, 26, 27). In addition, quercetin, a flavonol with known kinase inhibitory activity, binds to a novel and distinct allosteric site on the dimer interface of the KEN domain stabilizing the dimer to promote endoribonuclease activity (27). Yeast Ire1 endoribonuclease shares 39% amino acid identity with human IRE1 (28) and they are functionally interchangeable as yeast Ire1 will cleave a human mini-XBP-1 stem-loop RNA in vitro (22) and human IRE1α will cleave the yeast XBP-1 ortholog HAC1 RNA in cells (28) and in vitro (29). To screen for potential IRE1 endoribonuclease inhibitors, we produced over 50 mg of the purified human IRE1α-cytosolic domain (hIRE1α-cyto), amino acids 462–977 (Fig. 1A), using baculovirus expression in insect cells. hIRE1α-cyto was expressed with an N-terminal GST tag, purified using glutathione resin, and cleaved off the resin with PreScission protease (Fig. 1, A and B). The protein was phosphorylated on serine 724 as measured by treatment with λ-phosphatase (λ-PPase) and Western blotting with an anti-hIRE1α phosphoserine 724-specific antibody (supplemental Fig. 1A). We determined that our IRE1 protein preparations contained specific activity of IRE1 by using a series of wild type and mutant mini-XBP-1 stem-loop RNA substrates (supplemental Fig. S1, B and C) in both fluorescence quenching (FQ)-based and urea polyacrylamide gel electrophoresis-based cleavage assays (see “Experimental Procedures” for details). The absence of significant cleavage of the mutant stem-loop RNA substrates versus wild type stem-loop substrates further proved our protein preparations contained no contaminating RNases (supplemental Fig. S1C). Interestingly, in contrast to yeast Ire1, ADP had no effect on activity levels of human IRE1 even at millimolar concentrations, nor did ATP. Therefore, ATP was removed from the reaction mixture and no ADP was added; however, small amounts may remain from the IRE1 purification buffer. Using the FQ assay and a variant substrate with a Cy5 fluorescent probe more suitable for high throughput screening (Fig. 1C) we observed enzyme concentration-dependent cleavage of the labeled substrate; at 20 nm concentration, purified hIRE1α-cyto protein cleaved >90% of the mini-XBP-1 RNA substrate after a 1-h incubation giving rise to a greater than 5-fold increase in signal over background (Fig. 1D). We automated the FQ cleavage assay using liquid handlers and robotics and optimized the procedure to screen our library. Each well in the 384-well plate contained ∼10 nm hIRE1α-cyto protein and 100 nm substrate in a 10-μl total volume. Plates were incubated at 30 °C for 1 h until the reaction was stopped. Each plate had a negative control row (no enzyme) and a positive control row with no compound. Z values were calculated to be 0.8 or greater. Compounds were screened singularly at 20 μm. Hits were scored by inhibition activities of 60% or more of positive control (no compound) (supplemental Fig. S1D). One cluster of hits was salicylaldimines (Fig. 2Ai) based on our internal compound registry. Hit confirmation in triplicate showed that these compounds were active (data not shown). Salicylaldimine hit compounds were typically dihalogenated with bromine, chlorine, or iodine, at positions 3 and 5 of the aromatic ring (Fig. 2A, i) denoted as X. Furthermore, the salicylaldimine compounds were active in inhibiting the endoribonuclease activity of hIRE1α in a dose-dependent fashion as confirmed by IC50 analysis (see Fig. 2A, ii, for a representative IC50 profile). Upon analytical analysis by liquid chromatography-mass spectrometry (LC-MS), it was noted these library compounds had hydrolyzed into their respective salicylaldehydes in the presence of water (Fig. 2B). When diluted into an aqueous solution from their 10 mm DMSO stock solution, the aldimine group of the re-synthesized salicylaldimine compounds were also hydrolyzed to their respective aldehydes. When the salicylaldehyde analogs of the hit compounds (Fig. 2C, i to iii) where tested alone, all were active. To ensure that the salicylaldehyde analogs were not interfering with the FQ-based RNA cleavage assay by an unexpected mechanism, we confirmed the inhibitory activity for hIRE1α-cyto using several different substrates; one was a similar mini-XBP-1 stem-loop RNA with fluorescein isothiocyanate (FITC) as the fluorophore and black hole quencher 1 (BHQ1)(supplemental Fig. S1B) as the quencher, and IC50 values were determined using a shorter excitatory wavelength for the FITC probe (data not shown). The other substrate had a longer stem without a tag (supplemental Fig. S1B). Inhibition of cleavage of the longer stem substrate was visualized by urea PAGE and SYBR Gold staining (data not shown).
FIGURE 2.
Active salicylaldimine hit compounds, IC50 curves of their respective salicylaldehydes, and additional analogs used in these studies. Salicylaldimine hit compounds were of the characteristics shown in A, i, where substitutions of X at positions 3 and 5 were di-iodine, di-bromine, or di-chlorine and R was various constituents linked by an imine. ii shows the structure and 50% IC50 profile for a reproduced hit. Performed in triplicate, each compound concentration on the curve is expressed as the mean ± S.D. We determined by LC-MS that the salicylaldimine analogs were hydrolyzing to salicylaldehydes (B) and tested the purified forms without the R groups (C, i–iii). Representative IC50 analysis of salicylaldehyde (C, iv) and a number of analogs (C, v–x) with substituents at positions 3, 5, and 6. PLP (C, xi) and pyridoxal (C, xii), compounds related to salicylaldehydes, were not active.
Structure-Activity Relationship
To determine what moieties of the salicylaldehyde compounds were required for hIRE1α-cyto inhibitory activity, we purchased a select set of salicylaldehyde analogs (Fig. 2C). Initially we tested salicylaldehyde itself (Fig. 2C, iv), which was not active at 20 μm. The addition of a methoxy group at position 3 (ortho-vanillin) enhanced activity (Fig. 2C, v) compared with salicylaldehyde (Fig. 2C, iv) alone but this compound was not as potent as those bearing dihalogens at positions 3 and 5 (Fig. 2C, i to iii). However, bromine addition to position 5 on the methoxy analog significantly enhanced activity (Fig. 2C, vi versus v). A similar potency increase was observed upon bromine addition at position 6 (Fig. 2C, vii). These effects were additive as demonstrated by the most potent compound, 3-ethoxy-5,6-dibromosalicylaldehyde with an IC50 of ∼0.12 μm (Fig. 2C, viii). The methoxy version (position 3) of this compound (Fig. 2C, viii), difficult to obtain and limited in quantity, was also highly potent and comparable in activity by IC50 analysis (data not shown). When the formyl group was removed, all activity was lost (Fig. 2C, ix versus vi). Removal of the hydroxyl group also led to a complete loss in activity (Fig. 2C, x versus vii). If the hydroxyl group at position 2 was moved to position 4, such as in the compound vanillin, compared with ortho-vanillin (Fig. 2C, v), the activity was abolished (data not shown). Pyridoxal phosphate (PLP) (Fig. 2C, xi) and pyridoxal (Fig. 2C, xii), corresponding to vitamin B6 and its precursor, respectively, each possessing a hydroxyl group at position 2 and a formyl group at position 1, para to a nitrogen in the pyridine ring, were found to be completely inactive at 20 μm. In addition to the hydroxyl and adjacent formyl group, these data suggest additional lipophilic groups around the ring at positions 3, 5, and 6 are required for activity with all three in combination being optimal.
Selectivity
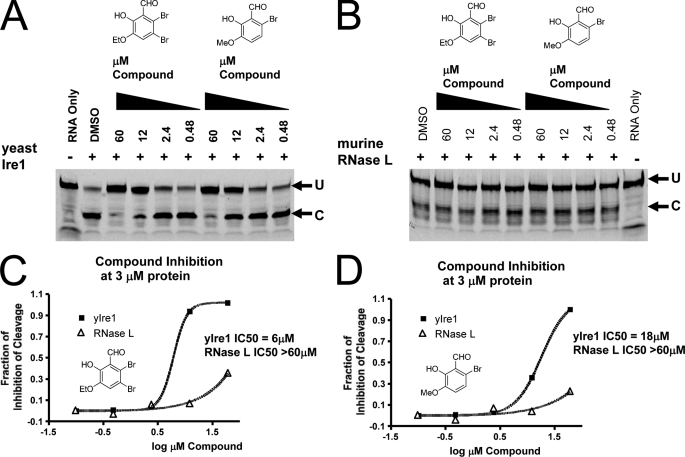
Eukaryotic IRE1s and the vertebrate RNase Ls encode unique endoribonucleases across all species (30). The KEN domains of such enzymes are C-terminal to the kinase domain (22). Although, the endoribonuclease function and catalytic residues are conserved between IRE1 and RNase L (22), IRE1 is competent for phosphotransfer function and RNase L is not (31). Yeast and human IRE1 are interchangeable with respect to the XBP-1 and HAC1 RNA substrates in cells (28, 29) and yeast Ire1 can cleave human mini-XBP-1 substrates as a purified enzyme (22); however, RNase L will not cleave XBP-1 RNA and has its own RNA selectivity requirements (30). To determine the selectivity of the salicylaldehydes for IRE1, we tested two potent compounds (Fig. 2C, vii and viii) on both yeast Ire1 and murine RNase L using in vitro enzyme assays. As shown in Fig. 3A, both compounds were active at inhibiting yeast Ire1 when using a fluorescently tagged mini-XBP-1 substrate. The 50- and 44-fold weaker potency of 3-ethoxy-5,6-dibromosalicylaldehyde (Fig. 3C) and 3-methoxy-6-bromosalicylaldehyde (Fig. 3D), respectively, against the yeast relative to the human IRE1 enzyme likely reflects the considerable divergence in primary structure (39% amino acid identity) of the endoribonuclease. We then assayed the compounds directly with murine RNase L (Fig. 3B) under similar reaction conditions using an RNase L-specific RNA substrate (32). These two compounds had little if any RNase L inhibitory activity, even at 60 μm, the highest concentration tested (Fig. 3, B–D). In addition, we tested the unrelated RNases, A and T1, using the FQ assay substrate (Fig. 1C). Bovine RNase A cleaves preferentially 3′ to cytidine and uridine in single-stranded RNA and fungal RNase T1 cleaves 3′ to guanosine in single-stranded RNA, which would include the same site that IRE1 cleaves in the mini-XBP-1 stem-loop RNA substrate; both enzymes were highly active and were titrated down to similar activity levels as hIRE1α-cyto in the FQ IC50 assay. Even the most potent compounds had no activity inhibiting these two enzymes at 20 μm, the highest concentrations tested (supplemental Fig. S2). These results further suggested the salicylaldehydes were not binding to or modifying the RNA substrate but were indeed inhibiting the catalytic activity of IRE1 endoribonuclease specifically.
FIGURE 3.
Cross-reactivity analysis of salicylaldehyde analogs against yeast Ire1 and murine RNase L. Both 3-ethoxy-5,6-dibromosalicylaldehyde and 3-methoxy-6-bromosalicylaldehyde displayed cross-reactivity against yeast Ire1 (A). Yeast Ire1-cyto was preincubated with the indicated concentrations of compound for 1 h at room temperature. Then the 5′ FITC-labeled single hairpin RNA substrate (5′-CAUGUCCGCAGCGCAUG-3′) was added and the reaction incubated for 90 min at room temperature. Reaction mixtures were then resolved by PAGE and fluorescence was visualized by a Typhoon imager. Neither 3-ethoxy-5,6-dibromosalicylaldehyde nor 3-methoxy-6-bromosalicylaldehyde show cross-reactivity against murine RNase L (B). A catalytic fragment (residues 333–651) of murine RNase L expressed and purified from bacteria was preincubated with the indicated concentrations of compound for 1 h at room temperature. Then 5′ FITC-labeled RNase L RNA substrate (5′-C11U2C7-3′) was added and the reaction was incubated for 90 min at room temperature. Reaction mixtures were then resolved by PAGE and fluorescence was visualized by a Typhoon imager. IC50 profiles for 3-ethoxy-5,6-dibromosalicylaldehyde (C) and 3-methoxy-6-bromosalicylaldehyde (D) against yeast Ire1 and RNase L are indicated. Quantification of cleavage was performed by phosphorimager analysis and graphed using GraphPad.
Mechanism of Action
To understand the mechanism of binding, a series of kinetic experiments were performed as described under “Experimental Procedures” (33). The Km of hIRE1α-cyto for the mini-XBP-1 stem-loop RNA substrate (Fig. 4A) was calculated to be 0.8 μm using Visual Enzymics software (Fig. 4A, i and ii) with a Kcat of 200 s−1. Using this information we conducted enzymatic analysis of 3-ethoxy-5,6 dibromosalicylaldehyde, the most potent compound by IC50 analysis. Fitting the resulting data indicated a non-competitive mechanism with respect to the mini-XBP-1 stem-loop RNA substrate. Both the fitted data (Fig. 4B, i) and a Lineweaver-Burk plot (Fig. 4B, ii) are shown. In a non-competitive type inhibition, the compound binds both the enzyme (as described by Kis) and the enzyme-substrate complex (as described by Kii), as shown under supplemental Fig. S3. The Kis for 3-ethoxy-5,6-dibromosalicylaldehyde was calculated to be 88 nm and the Kii was 71 nm, demonstrating the compound bound both hIRE1α-cyto and hIRE1α-cyto-RNA complex with approximately equal affinity.
FIGURE 4.
Enzyme kinetics of hIRE1α-cyto, mini-XBP-1 RNA stem-loop, and the inhibitor compound 3-ethoxy-5,6-dibromosalicylaldehyde. Titration of the substrate Cy5-labeled mini-XBP-1 RNA stem-loop as shown in Fig. 1, with constant enzyme (hIRE1α-cyto) concentration and measuring the slope of the initial reaction rates (A, i) determined the Michaelis-Menten constant (Km). Using a Lineweaver-Burk plot (A, ii), the x-axis intercept therefore specified the Km of 0.8 μm for a single mini-XBP-1 RNA stem-loop substrate (Fig. 1C). Titration of both substrate and 3-ethoxy-5,6-dibromosalicylaldehyde demonstrated a non-competitive mode of inhibition relative to the substrate (B). Data in B, i, were plotted as 1/velocity against 1/substrate concentration as a Lineweaver-Burk plot (B, ii). The binding constants Kii and Kis were 71 and 88 nm, respectively. Data are shown from a representative of three separate experiments. Reaction kinetics data were fit using Visual Enzymics software.
To confirm and complement the kinetics data, a series of studies using Biacore surface plasmon resonance technology was then performed. An XBP-1 stem-loop with an artificially extended stem (Fig. 5A) and 5′ biotin label was used for binding studies. When immobilized to a streptavidin-coated sensor chip, the longer stem provided a greater separation between the sensor chip and the loop sequence recognized by IRE1. In addition, the longer stem provided greater mass to increase the binding signal when used as the soluble binding partner. As shown in Fig. 5B, the hIRE1α-cyto protein when linked to the chip was able to bind the extended stem-loop RNA in a dose-dependent manner indicating that hIRE1α-cyto was immobilized on the sensor chip in a functional form. Using this system, we showed that 3-ethoxy-5,6-dibromosalicylaldehyde binds reversibly to hIRE1α-cyto alone without the presence of the mini-XBP-1 stem-loop substrate with a calculated Kd of ≈100 nm (Fig. 5C), similar to what was observed in our kinetic analysis. Furthermore, when the extended stem-loop was immobilized on the chip, 100 nm hIRE1α-cyto demonstrated a robust binding signal (Fig. 5D), whereas 3-ethoxy-5,6-dibromosalicylaldehyde did not (Fig. 5E). When RNase A was amine coupled to the sensor chip, 3-ethoxy-5,6-dibromosalicylaldehyde gave no detectable binding signal (Fig. 5F). Together these results strengthened the notion that the salicylaldehyde analogs bind directly and selectively to IRE1 to inhibit the cleavage of RNA substrates.
FIGURE 5.
Surface plasmon resonance of hIRE1α-cyto, substrate, and compound binding characteristics. Schematic of the XBP-1 RNA stem-loop with an extended stem used for surface plasmon resonance binding experiments is shown (A). Surface plasmon resonance binding profiles with the immobilized reagent on the solid chip surface are indicated at the bottom and the soluble binding partner indicated at the top (A–F). Active hIRE1α-cyto was linked to the Biacore chip by amine coupling and demonstrated specific and dose-dependent binding to the substrate confirming that hIRE1α-cyto was active on the chip (B). When compound 3-ethoxy-5,6-dibromosalicylaldehyde was passed over hIRE1α-cyto on the chip, a specific dose-dependent binding was observed with fast on-fast off kinetics (C). The disassociation constant (Kd) was calculated to be ≈100 nm. Biotinylated stem-loop RNA was immobilized to a streptavidin-coated chip and a large mass change was observed when hIRE1α-cyto was passed over (D). Repeated exposure to soluble hIRE1α-cyto degraded the signal likely due to site-specific cleavage of the stem-loop RNA on the solid surface, therefore, only a single concentration is shown. Passage of 3-ethoxy-5,6-dibromosalicylaldehyde over a chip immobilized with the XBP-1 stem-loop RNA did not give rise to a detectable binding signal (E). When RNase A was coupled to the chip and compound 3-ethoxy-5,6-dibromosalicylaldehyde was passed over the surface, no detectable binding was observed (F).
Cellular Activity
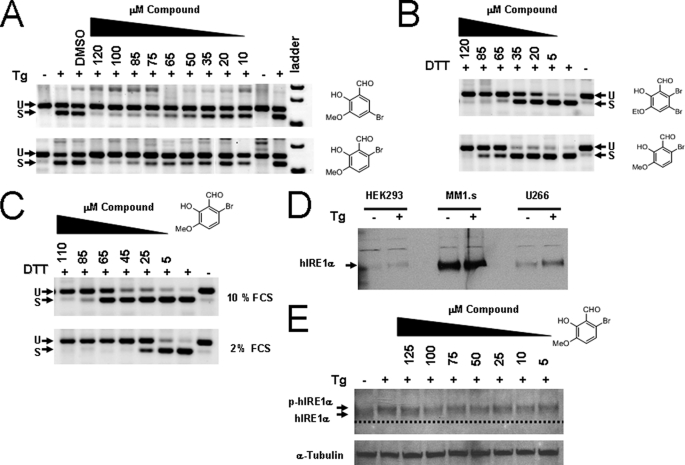
Although in vitro IRE1 activity using a single mini-XBP-1 stem-loop RNA substrate, at least in part, reproduces XBP-1 splicing activity in the cell, cleavage of the natural substrate in cells is likely more complex. IRE1 oligomerization (34), IRE1 protein complexes (35, 36), and a highly structured RNA substrate with dual hairpin loops (22, 37) may represent a significant difference between the activities in the cell and our in vitro reconstituted system. It is clear from the crystal structure of the active form of yeast Ire1 that the endoribonuclease has two mirrored catalytic sites that may operate cooperatively to cleave both sites in the dual hairpin loops, which are in close proximity, potentially in the form of a kissing hairpin (22). We therefore tested the most potent compounds in a cell-based assay using XBP-1 splicing as the direct cellular readout. We first tested compounds 3-methoxy-5-bromosalicylaldehyde and 3-methoxy-6-bromosalicylaldehyde in HEK293 cells using Tg as a stressor, as all cells contain this highly conserved ER stress pathway but need exogenous stress in culture to activate it. In unstressed cells, XBP-1 mRNA is detected predominantly if not entirely in its unspliced form using XBP-1-specific RT-PCR analysis. After addition of the compound for 2 h followed by a 6-h treatment with Tg, the formation of XBP-1s was inhibited in a dose-dependent manner (Fig. 6A). At high concentrations, a near complete inhibition of XBP-1s was observed, whereas the complete transcriptional repopulation of XBP-1u was achieved (Fig. 6A). These results are consistent with the compounds acting as IRE1 endoribonuclease inhibitors within cells. To confirm these findings we treated the myeloma cell line MM1.s with 3-ethoxy-5,6-dibromosalicylaldehyde and a different but powerful ER stressing agent, dithiothreitol (DTT), adding both simultaneously. Most cells rapidly activate XBP-1 splicing with this common thiol reducing agent at millimolar concentrations and respond with saturated splicing of XBP-1 (38). Again, when treated with 3-methoxy-6-bromosalicylaldehyde and 3-ethoxy-5,6-dibromosalicylaldehyde, dose-dependent and complete inhibition of XBP-1 splicing was achieved with the latter compound (top panel) appearing to be modestly more potent (Fig. 6B). Inhibition of XBP-1 splicing in MM1.s cells using Tm as the stressing agent was also observed indicating salicylaldehydes truly bind to and block IRE1 endoribonuclease activity in cells (data not shown). We determined that XBP-1 splicing could be inhibited by adding the compound before, during, or after treatment with ER stressing agents (data not shown). This suggests that salicylaldehyde inhibitors are likely acting on the activated state of the enzyme as we observe in the case of our in vitro analyses.
FIGURE 6.
Inhibition of XBP-1 splicing in human cells using salicylaldehyde analogs. HEK293 cells were left untreated or treated with 300 nm Tg for 3 h. Compounds (right of panel) were added 2 h before Tg at the indicated dose; total RNA was harvested and RT-PCR was performed using human-specific XBP-1 primers flanking the splice site 3 h after stress induction. PCR products were run on 4% agarose gels and stained with ethidium bromide and shown as the inverse image. Spliced (S) and unspliced (U) reaction products of XBP-1 mRNA are designated (A). Human myeloma MM1.s cells were treated with 2 mm DTT as the ER stressing agent and exposed to increasing concentrations of the indicated compound for 2 h (B): both DTT and the compounds were added at the same time. When MM1.s cells were treated in 2% FCS medium as in B, the potency of 3-methoxy-6-bromosalicylaldehyde increased, indicating compounds are partially absorbed to serum proteins (C). Western blot showing relative levels of IRE1α in human cell lines both untreated and treated with 300 nm Tg for 2 h (D). MM1.s myeloma cells had high steady state levels of IRE1α compared with HEK293 and the IgE secreting human myeloma cell line U266. When HEK293 cells were incubated with Tg and increasing concentrations of 3-methoxy-6-bromosalicylaldehyde for 3 h, no change in phosphorylation status was observed (E). The dashed line is a reference to observe the slight decrease in mobility due to phosphorylation (p-hIRE1α). The blot was reprobed with anti-α-tubulin antibody used as a loading control.
Because the cellular on-target 50% inhibition (EC50) of the ratio of XBP-1u to XBP-1s was shifted significantly higher than the in vitro enzyme IC50, we reasoned that these compounds might possess significant serum protein binding properties. Therefore, we treated MM1.s cells simultaneously with 2 mm DTT and increasing concentrations of 3-methoxy-6-bromosalicylaldehyde in high and low concentrations of fetal calf serum (FCS). The EC50 shifted from ∼65 μm with 10% FCS (Fig. 6C, upper panel) to below 25 μm with 2% FCS (Fig. 6C, lower panel), a nearly 3-fold decrease, confirming their serum protein binding ability. Using a rabbit polyclonal antibody generated against the baculovirus-expressed hIRE1α-cyto protein published previously (18) and Western blotting, IRE1 expression was quantified in several human cell lines; relatively high steady state levels of IRE1 expression was observed in MM1.s cells. ER stress agents induced a small decrease in mobility and an increase in intensity of the IRE1-specific band (Fig. 6D) indicating differential phosphorylation states of IRE1 and possibly reflecting the nature of the antibody raised against the activated-IRE1 immunogen. Treatment of HEK293 cells with 125 μm 3-methoxy-6-bromosalicylaldehyde did not impact the mobility shift when added 2 h before activation with 300 nm Tg indicating that IRE1 autophosphorylation was not altered (Fig. 6E). Low levels of IRE1 activation were observed in unstressed human RPMI 8226 myeloma cells as demonstrated by a slight serine 724 phosphorylation and XBP-1s expression (supplemental Fig. S4A). Compound treatment alone for 2 or 6 h increased the phosphorylation status (supplemental Fig. S4B) indicating that 3-methoxy-6-bromosalicylaldehyde induces ER stress either directly by inhibiting IRE1 endoribonuclease activity or indirectly because of ER stress resulting from the high concentration of organic compound used. When RPMI 8226 cells were treated with Tg, serine 724 phosphorylation was observed and the compound further increased phosphorylation even when XBP-1s was completely blocked (supplemental Fig. S4B). This result confirmed that salicylaldehyde analogs do not inhibit the intrinsic autophosphorylation activation step mediated by the kinase domain of IRE1. We then determined whether 3-methoxy-6-bromosalicylaldehyde affected downstream activities of IRE1 using RT-qPCR under the same conditions. Both XBP-1s transcriptional up-regulation of ERdj4 and p58IPK (16) as well as IRE1-mediated mRNA degradation of Blos1 (39) and CD59 (40) were significantly inhibited (supplemental Fig. S5).
In Vivo Studies
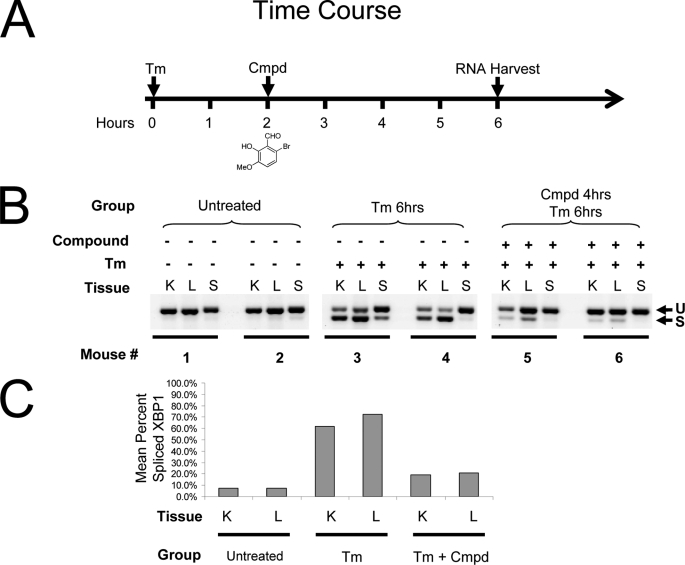
We asked the question: could salicylaldehyde analogs inhibit XBP-1 splicing in vivo using a murine model of ER stress? Although, the context of physiological ER stress and its relationship to disease are only beginning to emerge, it is clear that the IRE1/XBP-1 pathway is largely off in adult tissues with only very low levels of activation in organs or very specific activation in certain compartments such as the placenta experiencing high ER stress loads (41). Therefore, we used a chemical stressing agent, Tm, which blocks N-linked glycosylation in the ER, inducing ER stress in the liver and kidney when administered to mice (13, 42). Typically, induction of XBP-1 splicing can be observed starting at 2 h after treatment with 1 mg/kg by intraperitoneal injection, peaking at 6 h and dropping off by 24 h (13, 42). We treated mice intraperitoneally with 50 mg/kg of 3-methoxy-6-bromosalicylaldehyde 2 h after Tm administration and harvested tissues after a total of 6 h for RNA extraction (Fig. 7A). As observed in Fig. 7B, XBP-1 splicing induced by Tm was robust in the liver and kidney (and in one mouse, the spleen) and was strongly inhibited by ∼80% in both organs of the two mice treated with 3-methoxy-6-bromosalicylaldehyde (Fig. 7, B and C). Vehicle control did not induce XBP-1 splicing nor did it inhibit Tm-induced XBP-1 splicing (data not shown). These studies demonstrated that salicylaldehyde analogs inhibit XBP-1 splicing in animals.
FIGURE 7.
In vivo inhibition of XBP-1 splicing by 3-methoxy-6-bromosalicylaldehyde. Time course of in vivo treatment of mice (2 per treatment group) with Tm and 3-methoxy-6-bromosalicylaldehyde (cmpd) (A). CB17 SCID mice were treated with 1 mg/kg of Tm administered by intraperitoneal injection at time 0. Two hours later 3-methoxy-6-bromosalicylaldehyde dissolved in DMSO was delivered by the same route at 50 mg/kg and 4 h later, 6 h from time 0, the kidney (K), liver (L), and spleen (S) were harvested and snap frozen in liquid nitrogen. Tissues were homogenized and total RNA was prepared using TRIzol. RT-PCR was performed using murine-specific primers flanking the XBP-1 mRNA dual hairpin stem-loop and products were separated on a 4% agarose gel. Untreated mice had little or no evidence of XBP-1 splicing (U). Tm treatment induced robust splicing (S) at 6 h and 3-methoxy-6-bromosalicylaldehyde strongly inhibited slicing after 4 h of treatment (B). Scanning and quantification of the bands in B expressed as percent spliced XBP-1 (by dividing intensity of the spliced band by the total intensity of both) shows ∼80% inhibition for the mean of two mice per each group (C).
DISCUSSION
Regulated splicing of XBP-1 by IRE1 is believed to be a cellular response to acute and long term ER stress; distinct cellular outcomes diverge through this signaling node of the UPR depending on the magnitude and time frame of the stress (1). Because splicing of XBP-1 may benefit cellular survival and proliferation of malignant cells during stress imposed by the tumor microenvironment, the endoribonuclease activity of IRE1 may contribute to the pathogenesis and morbidity of diverse cancers (2, 43–45). We designed a direct high-throughput assay to search for inhibitors of this enzymatic activity. We discovered salicylaldehydes are potent and selective inhibitors of the IRE1 endoribonuclease directly on the enzyme, in vitro in cells and in vivo in animals.
The kinase domain of both yeast Ire1 and mammalian IRE1 is required for dimerization and activation of the endoribonuclease; however, kinase active site mutations (26) and extreme ER stress may bypass this requirement (23). Interestingly, some kinase inhibitors act as endoribonuclease agonists by promoting dimerization through mimicry of ADP binding (22), which enforces a kinase closed conformation for yeast Ire1 (23, 27). The kinase domain represents an obvious drug binding site for potentiators and possibly inhibitors of IRE1 signaling. We observed that salicylaldehyde analogs did not decrease autophosphorylation of IRE1 in cells at concentrations that completely blocked XBP-1 splicing. Therefore, it is unlikely that salicylaldehydes modulate the kinase domain directly. Our studies also suggest requirements for nucleotide co-factors may be somewhat different between yeast and human IRE1 as ADP did not potentiate activated hIRE1α-cyto endoribonuclease activity.
Although, small molecule agonists have been found for yeast Ire1 (23, 27) as well as human RNase L (46), small molecule antagonists have not been described for this family of endoribonucleases. Only substrate-like nucleoside analog inhibitors have been designed for RNase L (47). Small molecule inhibitors for the RNase A superfamily member angiogenin have been found by HTS (48), as well as transition state-like and nucleotide analog inhibitors described for RNase A (see Ref. 49 for a review) and RNase T1 (50). Our results show that salicylaldehyde analogs potently inhibit the endoribonuclease of IRE1 but do not inhibit RNases A, T1, or L. There have been no reports of salicylaldehydes as inhibitors of endoribonuclease enzymes; however, past studies showed the related PLP binds to and partially inhibits RNase A (51). PLP, the active form of vitamin B6, is a cofactor for more than 100 distinct enzymes needed for biogenesis of amino acids and neurotransmitters as well as gluconeogenesis and lipid metabolism (52). PLP provides an electrophile in the core of enzymes catalyzing reactions such as transaminations and decarboxylations. Invariably, in PLP-dependent enzymes, a central active site lysine residue forms the reactive aldimine with the formyl group of the pyridine ring. PLP binds RNase A rapidly and covalently by forming a reversible Schiff base adduct with the ϵ-NH2 of one of two key lysines residues, 41 and 7, in and around the active site, respectively (51), and to a lesser degree the α-NH2 of lysine 1 (53). Neither pyridoxal nor PLP were active as inhibitors of the IRE1 endoribonuclease. IRE1 may be evolutionarily adapted against such an inhibitory activity from pyridoxal and PLP, compounds found naturally in all organisms (52).
Salicylaldehyde analogs potent for human IRE1α were significantly less active for yeast Ire1 and inactive for murine RNase L. Combined, the data presented here suggest there is a unique and specific site on IRE1 for salicylaldehyde analog binding that differs somewhat between human IRE1α and yeast Ire1 and is not conserved on the IRE1 homologue RNase L. Our kinetic data and binding studies support the notion that salicylaldehydes are reversible non-competitive inhibitors with respect to the XBP-1 stem-loop RNA substrate. This mode of action is consistent with the difficulty of a small molecule to displace such a large protein-RNA interaction; however, it does not determine whether the binding is near the active site or a more distant allosteric site.
Aromatic aldehydes are known to interact reversibly with one or more OH-, SH-, or NH-nucleophiles of protein and/or nucleic acids. This interaction can range from a weak hydrogen bond to a stronger covalent bond. As in salicylaldehydes, the formyl group is neighbored by a hydroxyl group, their oxygen atoms can participate in varying levels of a hydrogen bond array. Alternatively, in the right arrangement of steric and stereoelectronic factors, aldehydes can form hemiacetals, hemithioacytals, or hemiaminals as a monoaddition product or, in the case of bisaddition, any uniform or mixed bisaddition product. Furthermore, aldehydes and amines can form imines called Schiff bases (54). Moreover, salicylaldehydes are known metal chelators further complicating the binding possibilities. Early kinetic results demonstrating fast on-fast off rates for the inhibitor indicate a relatively weak interaction with the enzyme. Further investigations of the exact nature of salicylaldehyde inhibitor binding are currently underway.
Inhibition of Blos1 and CD59 mRNA down-regulation at early time points with salicylaldehyde inhibitors further strengthens the idea that the RIDD activity (21, 39) is a direct cleavage mechanism of the IRE1 endoribonuclease. The findings demonstrated here will allow the use of salicylaldehyde analogs as tool compounds to dissect the divergent functions of IRE1 (55), the inflammatory signaling through JNK1 (56), and to explore the relationship between XBP-1 splicing and many disease states related to ER stress. Last, these data suggest more potent and drug-like molecules can be designed that may have therapeutic value.
Acknowledgments
We thank David Lonergan, Gary Flynn, and Peter Pallai for chemistry support. We thank Qingping Zeng and Andras Toro for assistance with chemical structures and critical reading of the manuscript.
This work was supported in part by a Biotech Investment Award from the Multiple Myeloma Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ER
- endoplasmic reticulum
- IRE1
- inositol-requiring enzyme 1
- XBP-1
- X-box-binding protein 1
- UPR
- unfolded protein response
- Tg
- thapsigargin
- Tm
- tunicamycin
- PLP
- pyridoxal phosphate
- HTS
- high-throughput screening
- DMSO
- dimethyl sulfoxide
- FQ
- fluorescence quenching.
REFERENCES
- 1. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 2. Lee A. S., Hendershot L. M. (2006) Cancer Biol. Ther. 5, 721–722 [DOI] [PubMed] [Google Scholar]
- 3. Kaufman R. J., Cao S. (2010) EMBO Mol. Med. 2, 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hotamisligil G. S. (2010) Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox J. S., Shamu C. E., Walter P. (1993) Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 6. Mori K., Ma W., Gething M. J., Sambrook J. (1993) Cell 74, 743–756 [DOI] [PubMed] [Google Scholar]
- 7. Sidrauski C., Walter P. (1997) Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 8. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 9. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 10. Harding H. P., Zhang Y., Ron D. (1999) Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 11. Ye J., Koumenis C. (2009) Curr. Mol. Med. 9, 411–416 [DOI] [PubMed] [Google Scholar]
- 12. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 13. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. (2007) Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 14. Harding H. P., Lackey J. G., Hsu H. C., Zhang Y., Deng J., Xu R. M., Damha M. J., Ron D. (2008) RNA 14, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sidrauski C., Cox J. S., Walter P. (1996) Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 16. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) Mol. Cell Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sriburi R., Bommiasamy H., Buldak G. L., Robbins G. R., Frank M., Jackowski S., Brewer J. W. (2007) J. Biol. Chem. 282, 7024–7034 [DOI] [PubMed] [Google Scholar]
- 18. Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008) Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoshida H., Uemura A., Mori K. (2009) Cell Struct. Funct. 34, 1–10 [DOI] [PubMed] [Google Scholar]
- 21. Hollien J., Weissman J. S. (2006) Science 313, 104–107 [DOI] [PubMed] [Google Scholar]
- 22. Lee K. P., Dey M., Neculai D., Cao C., Dever T. E., Sicheri F. (2008) Cell 132, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., Stroud R. M., Walter P. (2009) Nature 457, 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18773–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z., Kaufman R. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14343–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papa F. R., Zhang C., Shokat K., Walter P. (2003) Science 302, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 27. Wiseman R. L., Zhang Y., Lee K. P., Harding H. P., Haynes C. M., Price J., Sicheri F., Ron D. (2010) Mol. Cell 38, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tirasophon W., Welihinda A. A., Kaufman R. J. (1998) Genes Dev. 12, 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niwa M., Sidrauski C., Kaufman R. J., Walter P. (1999) Cell 99, 691–702 [DOI] [PubMed] [Google Scholar]
- 30. Dong B., Niwa M., Walter P., Silverman R. H. (2001) RNA 7, 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong B., Silverman R. H. (1999) Nucleic Acids Res. 27, 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carroll S. S., Chen E., Viscount T., Geib J., Sardana M. K., Gehman J., Kuo L. C. (1996) J. Biol. Chem. 271, 4988–4992 [DOI] [PubMed] [Google Scholar]
- 33. Copeland R. A. (2005) Evaluation of Enzyme Inhibitors in Drug Discovery, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 34. Li H., Korennykh A. V., Behrman S. L., Walter P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hetz C., Glimcher L. H. (2009) Mol. Cell 35, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu Y., Mao T., Zhang Y., Shao M., You J., Ding Q., Chen Y., Wu D., Xie D., Lin X., Gao X., Kaufman R. J., Li W., Liu Y. (2010) Sci. Signal. 3, ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gonzalez T. N., Sidrauski C., Dörfler S., Walter P. (1999) EMBO J. 18, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DuRose J. B., Tam A. B., Niwa M. (2006) Mol. Biol. Cell 17, 3095–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hollien J., Lin J. H., Li H., Stevens N., Walter P., Weissman J. S. (2009) J. Cell Biol. 186, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oikawa D., Tokuda M., Iwawaki T. (2007) Biochem. Biophys. Res. Commun. 360, 122–127 [DOI] [PubMed] [Google Scholar]
- 41. Iwawaki T., Akai R., Yamanaka S., Kohno K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16657–16662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hetz C., Bernasconi P., Fisher J., Lee A. H., Bassik M. C., Antonsson B., Brandt G. S., Iwakoshi N. N., Schinzel A., Glimcher L. H., Korsmeyer S. J. (2006) Science 312, 572–576 [DOI] [PubMed] [Google Scholar]
- 43. Spiotto M. T., Banh A., Papandreou I., Cao H., Galvez M. G., Gurtner G. C., Denko N. C., Le Q. T., Koong A. C. (2010) Cancer Res. 70, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carrasco D. R., Sukhdeo K., Protopopova M., Sinha R., Enos M., Carrasco D. E., Zheng M., Mani M., Henderson J., Pinkus G. S., Munshi N., Horner J., Ivanova E. V., Protopopov A., Anderson K. C., Tonon G., DePinho R. A. (2007) Cancer Cell 11, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Auf G., Jabouille A., Guérit S., Pineau R., Delugin M., Bouchecareilh M., Magnin N., Favereaux A., Maitre M., Gaiser T., von Deimling A., Czabanka M., Vajkoczy P., Chevet E., Bikfalvi A., Moenner M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15553–15558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thakur C. S., Jha B. K., Dong B., Das Gupta J., Silverman K. M., Mao H., Sawai H., Nakamura A. O., Banerjee A. K., Gudkov A., Silverman R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueno Y., Yamada Y., Nakanishi M., Kitade Y. (2003) Bioorg. Med. Chem. 11, 5069–5073 [DOI] [PubMed] [Google Scholar]
- 48. Kao R. Y., Jenkins J. L., Olson K. A., Key M. E., Fett J. W., Shapiro R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10066–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raines R. T. (1998) Chem. Rev. 98, 1045–1066 [DOI] [PubMed] [Google Scholar]
- 50. Georgalis Y., Zouni A., Hahn U., Saenger W. (1991) Biochim. Biophys. Acta 1118, 1–5 [DOI] [PubMed] [Google Scholar]
- 51. Raetz C. R., Auld D. S. (1972) Biochemistry 11, 2229–2236 [DOI] [PubMed] [Google Scholar]
- 52. Percudani R., Peracchi A. (2003) EMBO Rep. 4, 850–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riquelme P., Brown W. E., Marcus F. (1975) Int. J. Pept. Protein Res. 7, 379–387 [DOI] [PubMed] [Google Scholar]
- 54. Smith M. B., March J. (2001) March's Advanced Organic Chemistry, 5th Edition, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 55. Han D., Lerner A. G., Vande Walle L., Upton J. P., Xu W., Hagen A., Backes B. J., Oakes S. A., Papa F. R. (2009) Cell 138, 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]