Abstract
Succinate-ubiquinone oxidoreductase (SQR) and menaquinol-fumarate oxidoreductase (QFR) from Escherichia coli are members of the complex II family of enzymes. SQR and QFR catalyze similar reactions with quinones; however, SQR preferentially reacts with higher potential ubiquinones, and QFR preferentially reacts with lower potential naphthoquinones. Both enzymes have a single functional quinone-binding site proximal to a [3Fe-4S] iron-sulfur cluster. A difference between SQR and QFR is that the redox potential of the [3Fe-4S] cluster in SQR is 140 mV higher than that found in QFR. This may reflect the character of the different quinones with which the two enzymes preferentially react. To investigate how the environment around the [3Fe-4S] cluster affects its redox properties and catalysis with quinones, a conserved amino acid proximal to the cluster was mutated in both enzymes. It was found that substitution of SdhB His-207 by threonine (as found in QFR) resulted in a 70-mV lowering of the redox potential of the cluster as measured by EPR. The converse substitution in QFR raised the redox potential of the cluster. X-ray structural analysis suggests that placing a charged residue near the [3Fe-4S] cluster is a primary reason for the alteration in redox potential with the hydrogen bonding environment having a lesser effect. Steady state enzyme kinetic characterization of the mutant enzymes shows that the redox properties of the [3Fe-4S] cluster have only a minor effect on catalysis.
Keywords: Electron Transfer, Flavoproteins, Iron-Sulfur Protein, Quinones, Respiration, Site-directed Mutagenesis, X-ray Crystallography
Introduction
Succinate dehydrogenase (succinate-ubiquinone oxidoreductase (SQR),3 complex II) and Escherichia coli fumarate reductase (menaquinol-fumarate oxidoreductase (QFR)) are structurally and functionally similar enzymes that have evolved from a common evolutionary ancestor (1, 2). These membrane-bound enzyme complexes couple the interconversion of the dicarboxylates succinate and fumarate with quinone and quinol. As shown in Fig. 1, the E. coli (SQR/QFR) complexes, like their mammalian counterparts, are heterotetramers and consist of a soluble dehydrogenase fragment that is anchored to a transmembrane domain. The dicarboxylate-binding site and covalently bound FAD cofactor are found in the membrane extrinsic flavoprotein subunit (SdhA/FrdA; ∼64–66 kDa). A smaller (∼27-kDa) iron-sulfur protein subunit (SdhB/FrdB) contains three distinct iron-sulfur clusters, a [2Fe-2S]2+,1+ cluster proximal to the flavoprotein subunit, a [4Fe-4S]2+,1+ cluster near the middle of the subunit, and a distal [3Fe-4S]1+,0 cluster located near the interface with the membrane. The soluble domain is anchored to the membrane through interaction with two hydrophobic membrane-spanning subunits (SdhC/FrdC and SdhD/FrdD).
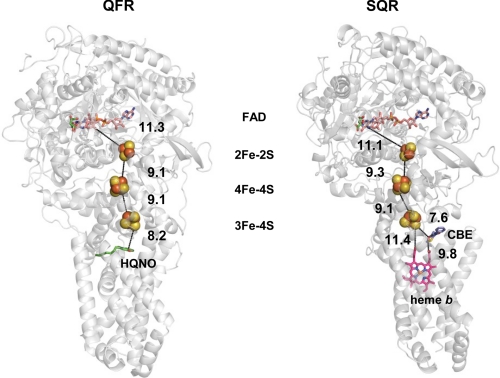
FIGURE 1.
Overall structure and spatial arrangement of redox centers in E. coli QFR and SQR. The overall structure of QFR (left side) and SQR (right side) is shown in a gray ribbon diagram. From the top of the figures, the FAD cofactor is shown as a colored stick figure with the dicarboxylate-binding site inhibitor oxaloacetate shown in green. The FeS clusters are shown as spheres with the iron atoms in red and the sulfur atoms in yellow. The edge-to-edge distances between the various redox centers are shown and are connected by dashed lines. The specific quinone site inhibitor 2-n-heptyl-4-hydroxyquinoline-N-oxide (HQNO) for QFR is shown in green, and the specific quinone site inhibitor carboxin (CBE) for SQR is shown in blue and red. The heme b found only in SQR is shown as a pink stick diagram. For preparing the diagrams, Protein Data Bank coordinates 1KF6 (QFR) and 2WDQ (SQR) were used.
The flavoprotein and iron-sulfur protein subunits are highly conserved throughout eukaryotes and prokaryotes; however, the primary sequence of the membrane-spanning subunit is more varied (1–4). In addition to the flavin and FeS redox clusters, a catalytic quinone-binding site is centrally located in both QFR and SQR at the interface of the soluble and transmembrane domain subunits. A difference in the family of complex II enzymes is that the hydrophobic peptides contain zero, one, or two b hemes (1–4). E. coli SQR, like its mammalian mitochondrial counterpart, contains one b heme sandwiched between the SdhC and SdhD subunits, whereas E. coli QFR is an example of a complex II homologue that lacks heme. Nevertheless, both enzymes can catalyze the same enzymatic reactions, demonstrating that the heme is not essential for catalysis (5–7).
In E. coli and related bacteria, different quinones are utilized during aerobic or anaerobic growth of the organism (8). EPR and redox studies have shown that a difference between QFR and SQR is that the chain of iron-sulfur clusters is of lower potential in QFR compared with SQR, consistent with the physiological role of the two enzymes in anaerobic and aerobic respiration, respectively (1–4). Consequently, for QFR, menaquinone (MQ; Em,7 = −74 mV) is the preferred physiological donor of electrons to the [3Fe-4S]1+,0 cluster, whereas in SQR, ubiquinone (UQ; Em,7 = +90 mV) is the acceptor of electrons from this cluster.
Similar to many other electron transfer proteins, a chain of redox cofactors connects the spatially separated catalytic sites where the substrates, dicarboxylates and quinones, are oxidized/reduced. Iron-sulfur clusters comprise one of the most common classes of such redox electron carriers. The midpoint potential (Em) of the redox cofactors is usually defined by the Em values of the redox transitions catalyzed. The redox potential of the [3Fe-4S] cluster in E. coli SQR is Em = +70 mV (5), near the potential of its electron acceptor UQ, whereas for QFR it is some 140 mV lower (Em = −70 mV) (9, 10), nearly isopotential with its electron donor MQ. Thus, it appears that the enzymes have evolved so that the protein environment controls the redox potential of the iron-sulfur cluster involved in accepting/donating electrons from the quinones. A number of factors have been proposed to control the Em of iron-sulfur clusters including (i) the local hydrogen bonding network, (ii) solvent accessibility, (iii) charged residues in the vicinity of the iron-sulfur cluster affecting the electrostatics of the region, and (iv) the proximity of main chain amide dipoles (11–14).
Complex II is an excellent model system to evaluate the effect of the type of quinone and potential of the iron-sulfur cluster in modulating electron transfer catalysis. Both E. coli SQR and QFR are capable of functioning with either benzo- or naphthoquinones (7, 15), and their respective protein environment has modulated the redox potential of the [3Fe-4S] cluster over a 140-mV range. In the present study, we used a combination of x-ray crystallography, EPR and visible spectroscopy, site-directed mutagenesis, and kinetic analysis to show that a side chain of an amino acid near the [3Fe-4S] cluster attenuates the Em of the cluster. We made equivalent substitutions in the SQR SdhB and QFR FrdB subunits to show that the solvent-exposed side chain of SdhB His-207/FrdB Thr-205 modulates the electronic properties of the [3Fe-4S] cluster and binding of quinones.
EXPERIMENTAL PROCEDURES
Materials
Detergents dodecylmaltoside and C12E9 ( polyoxyethylene 9-lauryl ether) were obtained from Anatrace (Maumee, OH). Carboxin (5,6-dihydro-2-methyl-1,4-oxathiin-3-carboxanilide) was obtained from Duchefa Biochemie (Haarlem, The Netherlands).
Strains, Plasmids, and Mutagenesis
E. coli strain DW35 (ΔfrdABCD, sdhC::kan) was used as the host for expression of SQR as described (16). Plasmid pH3 (frdA+B+C+D+) was used for expression of wild-type QFR (17), and plasmid pFAS (PFRDsdhC+D+A+B+) was used for expression of wild-type SQR (16). Mutations were introduced using the QuikChange (Stratagene, La Jolla, CA) mutagenesis kit using pFAS or pH3 as templates (16). Primers for mutagenesis were obtained from Qiagen (Valencia, CA).
Protein Expression, Purification, and Crystallization
The SdhB H207T substituted enzyme was expressed, purified, and crystallized following the published procedures (18). Initial crystals grown under conditions used for the wild-type enzyme were in a different space group, H3, and were perfect merohedral twins. Addition of organic additives promoted growth of untwinned crystals, in space group P212121, with cell dimensions similar to those of the crystals of wild-type enzyme.
The crystals used for data collection were from SdhB H207T enzyme mixed in a 1:10 (v/v) ratio with the inhibitor carboxin (from a 10 mm stock solution in ethanol). The reservoir solution contained 0.1 m Tris (pH 8.5), 0.1 m Li2SO4, 0.1 m NaCl, 0.009% dodecylmaltoside, 3% 1,6-hexanediol, and 10% (w/v) PEG 4000. The crystals appeared overnight and were frozen within 4 days of setup using 30% glycerol as cryoprotectant and mounted with mesh LithoLoopsTM (Molecular Dimensions Ltd., Suffolk, UK) to support the large plate crystals.
Data Collection
X-ray data were collected from frozen crystals at 100 K at the European Synchrotron Radiation Facility beamline ID23-1. The data set was collected from three different positions on the crystal, to minimize the effects of radiation damage and increase the resolution of the complete data set, using a 100 × 100-μm beam. Intensities were integrated using the program XDS (19) and merged using SCALA (20, 21). Data collection statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses refer to the highest resolution shell. r.m.s., root mean square; ESRF, European Synchrotron Radiation Facility. DPI, Diffraction Precision Index.
| Data set | SdhB H207T |
| Beamline | ESRF ID23-1 |
| Wavelength (Å) | 0.97625 |
| Space group | P212121 |
| Unit cell (Å) | a = 119.85, b = 183.80, c = 202.78 |
| Resolution (Å) | 49.63-2.70 (2.85-2.70) |
| Rmerge (%)a | 9.2 (63.5) |
| Completeness (%) | 99.9 (100.0) |
| 〈〈I〉/σ〈I〉〉 | 11.1 (2.0) |
| Observed reflections | 451,178 (65899) |
| Unique reflections | 122,882 (17824) |
| Redundancy | 3.7 (3.7) |
| Refinement statistics | |
| Resolution (Å) | 48.85-2.70 (2.77-2.70) |
| Reflections working set | 116,577 (8281) |
| Reflections test set | 6,188 (419) |
| Rcryst (%)b | 19.0 (28.6) |
| Rfree (%)b | 22.2 (32.6) |
| Total number of atoms used | 25,058 |
| B factors | |
| From Wilson plot (Å2) | 68.7 |
| Mean atomic B factor (Å2) | 65.6 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.36 |
| Coordinate precision, Cruickshank DPI (Å) | 0.27 |
| Correlation coefficient between Fo and Fc, free | 0.92 |
| Molprobity scores | |
| Rotamer outliers (%) | 2.83 |
| Ramachandran outliers (%) | 0.03 |
| Ramachandran favored (%) | 97.29 |
| Clash score (percentile) | 11.3 (97th) |
| Overall quality score (percentile) | 2.05 (98th) |
a Rmerge = ΣhΣl|Ihl − 〈Ih〉|/ΣhΣl〈Ih〉 where Ihl is the lth observation of reflection h and 〈Ih〉 is the weighted average intensity for all observations l of reflection h.
b Rcryst = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated as for Rcryst but using the test set of reflections (5% of the diffraction data, not used during refinement, and chosen to match the test set of reflections used in the determination of the wild-type structure (Protein Data Bank code 2WDQ)).
Structure Determination and Refinement
Phases for the mutant structure were initially determined by rigid body refinement in Refmac (22) of the protein part of the WT/carboxin model (Protein Data Bank code 2WDQ). Non-protein atoms were omitted from the search model and initial phase calculation. Difference maps (mFo − DFc) calculated from the initial phases confirmed the location of the mutation. Density for carboxin was clearly visible in the Q-site in 2mFo − DFc maps. Manual model building was performed using the program Coot (23), and refinement was done using phenix.refine (24) and Refmac (22). Strict non-crystallographic symmetry restraints were applied to the protein part of the model during refinement. Coordinates and restraints for carboxin were downloaded from the PRODRG server (25). Ideal coordinates for the malate-like intermediate TEO were obtained from the HIC-Up data base (26) and input to the PRODRG server to generate final coordinate and restraint files for modeling. The ligand building procedure within ARP/wARP (27) was used to fit carboxin into the density map. Water molecules were fitted using phenix.refine and Coot and validated using Coot. The model was refined to Rcryst = 21.7% and Rfree = 25.3%. TLS refinement (28) was implemented in the final stages of refinement, defining each chain in the asymmetric unit as a separate TLS group. This produced a model with a final Rcryst of 19.0% and Rfree of 22.2%. The TLS model was validated using TLSANL (29). The side chain of SdhD Trp-113 was truncated at the Cβ atom because density for the side chain was poor, and it was not possible to model it as a common rotamer without introducing clashes with symmetry-related side chains. Molprobity (30) was used to validate the final models and check for Asn/Gln/His flips. Refinement statistics are shown in Table 1. The root mean square deviation of superposed structures was determined using the CCP4 program LSQKAB. Figs. 1–4 and 7 were prepared using PyMOL (31).
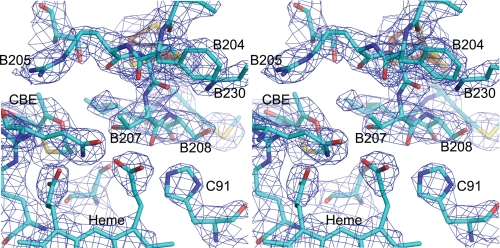
FIGURE 2.
Crystal structure of SdhB His207Thr mutant of E. coli SQR. A stereoview of the area around the mutation is shown. The density, shown in blue mesh, is a 2mFo − DFc map contoured at 1σ. The side chains of Pro-B160 and Ser-B161 have been removed to allow a clear view of the site of the mutation. The quinone-binding site inhibitor (carboxin) is labeled as CBE in the figure.
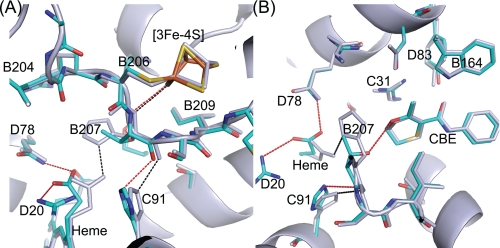
FIGURE 3.
Two views of E. coli wild-type and SdhB H207T mutant enzymes. A and B show different views of a superposition of the carboxin (CBE)-bound wild-type (in gray; Protein Data Bank code 2WDQ) and mutant structures (colored). Chains A–D of the structures were superimposed by minimizing the root mean square deviations of the Cα atoms using the CCP4 program LSQKAB. As in Fig. 2, side chains of Pro-B160 and Ser-B161 have been removed to allow a clear view of the site of the mutation. Hydrogen bonds involving His-B207 in the wild-type structure are shown as black dotted lines. Hydrogen bonds involving Thr-B207 in the mutant and new interactions found in the mutant between the protein and one of the heme propionates are shown as red dotted lines.
FIGURE 4.
Comparison of wild-type QFR FrdB subunit and SdhB H207T structures near their [3Fe-4S] cluster. Wild-type QFR FrdB is shown in pink (Protein Data Bank code 1KF6), and the mutant SdhB H207T mutant is shown in cyan. Backbone atoms of the loop covering the iron-sulfur cluster (equivalent to residues SdhB Cys-206 to Cys-212 and FrdB Cys-204 to Cys-210) are superimposed. Potential hydrogen bonds between protein backbone amines and sulfur atoms of the [3Fe-4S] cluster are shown as dotted lines. An additional hydrogen bond of the side chain of SdhB Thr-223 and cluster atom S4 is shown as a dotted line; the equivalent FrdB non-bonding Ala-221 is also shown. Also presented is a comparison of the amino acid sequence of the SdhB/FrdB subunit in this region from bacterial and mammalian complex II enzymes. The cysteine residues ligating the [3Fe-4S] cluster are underlined. The labeling of the sulfur atoms of the cluster is consistent with Protein Data Bank codes 2WDQ and 2WP9.
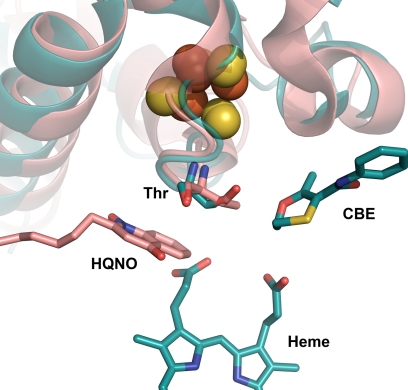
FIGURE 7.
Comparison of wild-type QFR and SdhB H207T mutant enzymes near their quinone-binding site. Wild-type QFR (Protein Data Bank code 1KF6) is shown in pink, and the SdhB H207T mutant is shown in cyan. 2-n-Heptyl-4-hydroxyquinoline-N-oxide (HQNO) in pink is shown at the site where menaquinone binds in QFR, and carboxin (CBE) is shown at the ubiquinone-binding site in SQR.
Potentiometric Titrations and EPR Spectroscopy
To obtain data as representative as possible of the in vivo enzymes, all EPR data reported herein were obtained from preparations of cytoplasmic membranes that had been activated with malonate as described (32). Potentiometric titrations were carried out at 25 °C as described previously (32, 33). Titrations of the heme and [3Fe-4S] cluster signals were carried out at pH 7.0 in a buffer containing 100 mm MOPS, 5 mm EDTA, and 1 mm malonate. Titrations of ubisemiquinone radical signals were carried out at pH 8.0 in an identical buffer except that Tricine was substituted for MOPS. The following redox mediators were used at a concentration of 25 μm: 2,6-dichlorindophenol, 1,2-naphthoquinone, toluylene blue, phenazine methosulfate, thionine, methylene blue, resorufin, indigotrisulfonate, indigocarmine, anthraquinone-2-sulfonic acid, and neutral red. EPR spectra were recorded using a Bruker ESP300E spectrometer. [3Fe-4S]1+,0 cluster and Fe3+ heme EPR spectra were recorded at 12 K (20-milliwatt microwave power and 100-kHz modulation frequency) using an Oxford Instruments ESR900 flowing helium cryostat. [3Fe-4S]1,+ and Fe3+ heme spectra were recorded at modulation amplitudes of 10 and 20 Gpp, respectively. Flavin and ubisemiquinone radical spectra were recorded at 150 K (microwave power of 20 milliwatts and a modulation amplitude of 2 Gpp at 100 kHz) using a Bruker liquid nitrogen cryostat (an ER4111 VT Variable Temperature Unit). Five scans were accumulated for each sample. Em values were representative of two to three independent titrations with a standard deviation of approximately ±10 mV. Potentiometric titration data were analyzed as described previously (34–36).
Measurement of Enzyme Activity
The standard assay medium contained 30 mm Bistris propane (pH range, 6.0–8.5), 0.1 mm EDTA, 0.006% detergent C12E9, and 3 mm potassium cyanide at 30 °C.
Succinate Oxidation
The succinate oxidase reaction of SQR was monitored by the decrease of the absorbance at 600 nm in the presence of succinate, 1.5 mm phenazine ethosulfate, and 50 μm 2,6-dichloroindophenol (ϵ600 = 21.8 mm−1 cm−1 (pH 7.8)). The succinate-ferricyanide reductase activity of QFR was determined at 420 nm with 0.5 mm potassium ferricyanide (ϵ420 = 1 mm−1 cm−1) and 20 mm succinate.
Fumarate Reduction
Fumarate reduction activities of QFR and SQR with quinol analogues were determined in a reaction coupled to DT diaphorase (NADH:quinone reductase) as described previously (7) with MQ1 (menaquinone) and UQ1 (ubiquinone) for QFR and SQR, respectively (MQ1 and UQ1 were kindly provided by Eisai Co. Ltd, Tokyo, Japan). All kinetic data are within 5% error.
Analytical Methods
FAD content was determined as described previously (37). Protein concentration was determined by the bicinchoninic acid (BCA) method (Pierce) with BSA as a standard in the presence of 1% (w/v) SDS. Heme b was determined by the pyridine hemochromogen method (38).
RESULTS
To investigate the effect of the redox potential of the [3Fe-4S] cluster on catalytic activity of complex II enzymes, two mutant proteins were constructed. The [3Fe-4S]1+,0 cluster in SQR has positively charged amino acid residues adjacent to one of the Cys ligands, whereas the lower redox potential cluster in QFR has uncharged polar side chains. Therefore, SdhB His-207 was mutated to a Thr residue, and the equivalent Thr residue in FrdB was mutated to His to mimic the SQR protein. The resulting SdhB H207T and FrdB T205H substituted enzymes were expressed in E. coli, and the yield and stability of the mutant proteins were equivalent to those of wild-type enzyme.
Crystal Structure of SdhB H207T Substitution of E. coli SQR at 2.7-Å Resolution
To determine the structural effects of the SdhB H207T substitution, we determined the x-ray structure of the protein in the presence of the quinone-binding site inhibitor carboxin. The crystal structure shows that the SQR SdhB H207T substituted enzyme is folded in a stable conformation and that the folds of the individual subunits are essentially identical to those of the wild-type enzyme. SQR molecules form a tightly packed trimeric arrangement in the asymmetric unit as described for the wild-type enzyme (18). The overall root mean square deviations for Cα atoms following superposition of chains A–D of the wild-type and mutant structures are 0.15 Å for SdhA, 0.15 Å for SdhB, 0.17 Å for SdhC, and 0.19 Å for SdhD. These values are within the estimated coordinate error of the two structures (0.27 Å for the SdhB H207T substituted structure and 0.19 Å for wild-type SQR-binding carboxin; see Table 1 here and supplemental Table 1 in Ref. 18), confirming that the mutation does not cause large conformational changes involving the Cα backbone. Analyzing the deviations for Cα atoms following superposition of individual chains does reveal some small but interesting changes around the site of the mutation. These residues lie within good electron density in both wild-type (Protein Data Bank code 2WDQ) and mutant structures (Fig. 2). Within the region of the mutation, the largest deviation is for B207 itself (0.52 Å).
The protein environment around the site of the mutation reveals localized structural changes compared with the wild type as shown in Fig. 3, left and right. In the wild-type structure, residue B207 forms a hydrogen bond interaction with one of the heme propionates with the Nδ atom of the imidazole ring lying 2.6 and 3.4 Å from the oxygen atoms of the heme propionate. The SdhB H207T substitution breaks this interaction, allowing the heme propionate to adopt a different conformation where it can form hydrogen bond interactions with SdhD Arg-20 (3.1 Å away) and SdhD Gln-78 (3.0 Å away) (Fig. 3). In the wild-type structure, the Nϵ atom of SdhC His-91 forms hydrogen bonds to the backbone carbonyl oxygen atom of SdhB His-207 (3.0 Å distant) and a heme propionate (3.1 Å away). The mutant structure shows a small movement of the side chain of SdhC His-91. This movement seems to be a consequence of the deviation in the Cα backbone at the SdhB 207 position, weakening the interaction between the substituted SdhB H207T and SdhC His-91 (3.4 Å apart) and the shift in the position of the heme propionate (4.5 Å away) (Fig. 3, left and right).
As shown in Fig. 3, there are no large changes to the protein structure around the [3Fe-4S] cluster. However, there is a small shift in the main chain position around residues SdhB 207 to SdhB 210. The wild-type structure shows that three of the sulfur atoms of the [3Fe-4S] cluster can interact with the protein amide backbone via hydrogen bonding (Fig. 4). Tables 2 and 3 compare donor-acceptor distances for these potential hydrogen bonds in the mutant structure and wild-type structures of SQR and QFR, respectively. Comparing wild-type SQR structures, it seems that the donor-acceptor distances of some of these hydrogen bonds can change without affecting the redox potential of the [3Fe-4S] cluster. However, three of the possible interactions have very similar donor-acceptor distances in both wild-type structures. These are from cluster atom S1 to B207N, S1 to B208N, and B212Sγ to B223N (we note that the latter two have poor geometry for forming a strong hydrogen bond). Furthermore, these donor-acceptor distances are all greater in the structure of the Thr-substituted enzyme compared with wild type. These distance changes would weaken the hydrogen-bonding interactions. Other studies have shown that removing hydrogen bonds to FeS clusters can lower midpoint potentials (e.g. Refs. 12–14); hence, this factor may contribute to the observed decrease in redox potential.
TABLE 2.
Donor-acceptor distances between amide nitrogen atoms and sulfur atoms of [3Fe-4S] cluster in x-ray structures of wild-type and mutant E. coli SQR (comparing SdhB subunit)
The labeling of cluster sulfur atoms is consistent with Protein Data Bank code 2WDQ.
| Interaction | Protein Data Bank code (resolution) |
||
|---|---|---|---|
| BH207T 2WP9 (2.7 Å) | Wild type |
||
| 2WDQ (2.4 Å) | 1NEK (2.6 Å) | ||
| Å | |||
| S1-H207N | 3.5 | 3.3 | 3.3 |
| S1-S208Na | 4.1 | 3.9 | 3.9 |
| S1-I209Na | 3.9 | 3.8 | 4.0 |
| S2-N211N | 3.1 | 3.3 | 3.5 |
| S2-C212N | 3.2 | 3.1 | 3.6 |
| S4-M210N | 3.3 | 3.4 | 3.2 |
| C159Sγ-S161N | 3.4 | 3.6 | 3.7 |
| C159Sγ-F162N | 3.5 | 3.5 | 3.8 |
| C206Sγ-S208N | 3.9 | 3.6 | 3.5 |
| C212Sγ-T223Na | 4.3 | 4.0 | 4.0 |
a These residues show poor geometry for a strong hydrogen bond.
TABLE 3.
Donor-acceptor distances between amide nitrogen atoms and sulfur atoms of [3Fe-4S] cluster in E. coli QFR FrdB subunit
The labeling of cluster sulfur atoms is consistent with Protein Data Bank code 2WDQ.
| Interaction | Protein Data Bank code 1KF6 (2.7-Å resolution) |
|---|---|
| Å | |
| S1-T205N | 3.5 |
| S1-F206Na | 3.7 |
| S1-V207N | 3.4 |
| S2-Y209N | 3.4 |
| S2-C210N | 3.4 |
| S4-G208N | 3.0 |
| C158Sγ-Q160N | 3.4 |
| C158Sγ-F161N | 3.5 |
| C204Sγ-F206N | 3.4 |
| C210Sγ-A221Na | 3.8 |
a These residues show poor geometry for a strong hydrogen bond.
Redox Properties of [3Fe-4S] Cluster in SQR/QFR
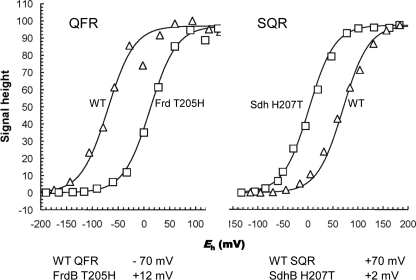
To gain insight into the role of the side chain in control of the redox potential of the [3Fe-4S] cluster, redox titrations of the mutant proteins were undertaken. The potentiometric EPR characterization of the complex II enzymes was performed using isolated membranes because of high expression levels of the proteins. In the wild-type QFR, the redox potential of the [3Fe-4S] center is near that for its electron donor, menaquinol QFR (Em[3Fe-4S] = −68 mV and EmMQ/MQH2 = −74 mV). Similarly, in SQR, the redox potential of the [3Fe-4S] cluster donating electrons to ubiquinone is thermodynamically favorable for this reaction (Em[3Fe-4S] = +70 mV and EmUQ/UQH2 = +90 mV). The midpoint potential of the same iron-sulfur cluster in the substituted enzymes demonstrates remarkable shifts; introducing His in the FrdB 205 position (FrdB T205H) increases the Em by 80 mV to +12 mV. A similar magnitude change is seen in the opposite direction when SdhB His-207 is changed to Thr. Here, the Thr substitution (SdhB H207T) lowers the Em by 70 mV to +2 mV (Fig. 5).
FIGURE 5.
Potentiometric titrations of [3Fe-4S] cluster in wild-type and mutant QFR and SQR. The titrations of the [3Fe-4S] signal for wild-type and the FrdB T205H mutant membranes are shown in the left panel. The titrations of the [3Fe-4S] signal for wild-type SQR and SdhB H207T mutant membranes are shown in the right panel. The g = 2.01 signals were plotted against Eh and normalized to 100% signal intensity. Below the plots, calculated Em,8 values for the [3Fe-4S] cluster in wild-type and mutant QFR and SQR are indicated. In both cases, wild-type enzymes are shown with open triangles (△), and the respective mutant enzymes are shown with open squares (□).
Properties of Heme b and Semiquinone Radical in SQR Enzymes
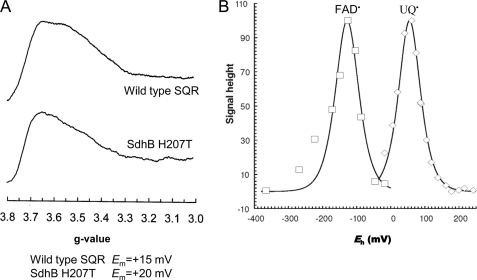
The most prominent structural changes for the His to Thr substitution in SQR are near the heme propionates (Fig. 3). The optical spectrum, however, of dithionite-reduced minus oxidized SQR as well as the midpoint potential of the heme b (Em = +15 and +20 mV for wild type and the H207T substitution, respectively) shows no difference between wild type and H207T (data not shown). This is consistent with the observed EPR spectra of oxidized membranes (Fig. 6A). A broad peak between g = 3.5 and g = 3.6 is characteristic for the low spin heme b in overexpressed SQR (37). The membranes containing the SdhB H207T enzyme exhibit a low spin signal of an amplitude similar to that of the wild-type enzyme but with a slightly modified line shape (an increase in the relative intensity of the g = 3.6 peak compared with the g = 3.5 peak). In wild-type SQR, the heme is reducible by succinate because of its favorable potential, and UQ is required for fast heme reduction (39). Similar rates of the heme reduction were observed in the SdhB H207T substitution (data not shown).
FIGURE 6.
EPR study of wild-type SQR and SdhB H207T mutant enzyme. A, low spin heme EPR spectra of SQR-enriched E. coli membranes. The Em,7 for the b heme is shown below the figure. B, potentiometric titration of the g = 2.005 EPR signal of E. coli membranes enriched for the SdhB H207T mutant. Peak troughs are plotted against Eh and normalized to 100% signal intensity. Both flavosemiquinone (□) and ubisemiquinone (△) are visible.
EPR titration of SdhB H207T membranes demonstrates the presence of two semiquinone radicals: one attributed to FAD and the other attributed to ubiquinone (USQ˙̄) with an Em for USQ radical of +55mV (Fig. 6B), similar to that of the wild-type SQR. The semiquinone radical of wild-type (36) and FrdB T205H QFR (data not shown) is too unstable for study by EPR spectroscopy. Thus, these data show that neither the properties of the heme b nor the semiquinone radical are significantly altered in SdhB H207T or FrdB T205H mutant enzymes. Also shown in Fig. 6B is the titration of the FAD semiquinone of the SdhB H207T substitution that has an Em of −128 mV in close agreement with that of the wild-type enzyme (32).
Catalytic Activity of Wild-type and Mutant Complex II Enzymes
Both SQR and QFR can reversibly catalyze the interconversion of succinate and fumarate (1, 2, 7). Both enzymes can also bind benzoquinones, naphthoquinones, and their derivatives and catalyze quinol oxidation and quinone reduction (2, 18, 39). The turnover numbers for their catalytic reaction with quinones are different, however, in that SQR is more proficient in reactions with ubiquinone and QFR is more proficient in reactions with menaquinone. Thermodynamically this makes sense in that the immediate electron donor/acceptor for the reaction with the quinone, the [3Fe-4S] cluster, is isopotential with it. It might therefore be expected that changing the redox potential of the [3Fe-4S] cluster, as shown above, would affect the propensity of SQR/QFR in reactions with quinones. Table 4 compares the activity of the SdhB H207T substitution in reactions of ubiquinone-1 reduction, ubiquinol oxidation, and menaquinol oxidation at different pH values. As seen in Table 4, there is a relatively minor effect on catalytic activity with quinones in the mutant enzyme. Thus, for SQR, substitution of the positively charged His residue with a neutral residue such as Thr does not significantly affect catalytic activity. The data also suggest that even though the [3Fe-4S] cluster redox potential has been lowered by ∼70 mV this does not increase the reactivity of the enzyme with the lower potential naphthoquinones such as MQ1.
TABLE 4.
Comparison of catalytic turnover of succinate-oxidase and fumarate-reductase reactions catalyzed by isolated wild-type SQR and SdhB H207T mutant
ND, not determined.
| pH | Turnover rates |
|||||
|---|---|---|---|---|---|---|
| Succinate-UQ1 |
UQ1H2-fumarate |
MQ1H2-fumarate |
||||
| WT | H207T | WT | H207T | WT | H207T | |
| s−1 | ||||||
| 6.0 | 9.7 | 12.4 | 0.6 | 0.4 | 1.7 | 1.2 |
| 7.0 | 44.1 | 47.1 | 1.7 | 1.4 | 3.7 | 2.9 |
| 8.0 | 101.8 | 89 | ND | ND | ND | ND |
There is, however, a relatively greater effect on binding of quinones as shown in Table 5. It had been suggested previously (40) that ubiquinone can bind in two different positions in the quinone binding pocket of SQR. It was further suggested that SdhB His-207 aided in movement of the quinone into the position deeper within the pocket where catalysis occurred (40). The two positions in the quinone binding pocket were confirmed by recent structural studies using different SQR quinone site inhibitors such as carboxin and pentachlorophenol (18). It can be seen in Table 5 that ubiquinone and carboxin, both of which bind more deeply in the quinone-binding pocket (18), showed reduced affinity in the SdhB H207T mutant, whereas there is no change for pentachlorophenol, which binds near the entrance of the quinone-binding site. The data in Table 5 also show that the substitution has similar effects on the relative affinity for ubi- and naphthoquinols.
TABLE 5.
Apparent affinity of quinones and quinone site inhibitors in SQR enzymes
PCP, pentachlorophenol.
| KmUQ1 | KiPCP | Kicarboxin | KmUQ1H2 | KmMQ1H2 | |
|---|---|---|---|---|---|
| μma | μma | μma | μmb | μmb | |
| WT SQR | 3 | 13 | 30 | 4 | 3.6 |
| SdhB H207T | 8.8 | 10 | 160 | 7.2 | 8 |
a Assay was done at pH 7.0.
b Assay was done at pH 8.0.
The side chain of FrdB Thr-205, like that for SdhB His-207, is oriented toward the quinone binding pocket (Fig. 7). Therefore, it is of interest that the FrdB T205H substitution does show significant effects on catalytic activity with quinones (Table 6), which is in contrast to the SdhB His-207-substituted enzyme. Both SQR and QFR show similar pH profiles for succinate oxidation with either ubiquinone or artificial electron acceptors with the activity increasing at higher pH (32). The FrdB T205H variant shows no difference in succinate-ferricyanide reductase activity as compared with wild-type enzyme (Table 6) in an assay that monitors the hydrophilic catalytic subunits of complex II. In contrast, succinate-ubiquinone reductase activity is severely compromised at higher pH with about a 2- and 4-fold reduction in activity at pH 7 and 8, respectively. It should be noted that at pH 6 the activity is only reduced ∼10% from wild-type enzyme. It can also be seen in Table 6 that in the mutant there is an increased Km for ubiquinone, suggesting that binding of the quinone is impaired. The FrdB T205H substitution also shows an increased Km for menaquinone and about a 2-fold reduction in menaquinol-fumarate reductase activity between pH 6 and 8. These data suggest that the catalytic quinone-binding (QP) site of QFR has been compromised in the variant enzyme.
TABLE 6.
Comparison of catalytic turnover of succinate-oxidase and fumarate-reductase reactions catalyzed by isolated wild-type QFR and FrdB T205H mutant
| Succinate-K3Fe(CN)6 |
Succinate-UQ2 |
MQ1H2-fumarate |
||||
|---|---|---|---|---|---|---|
| WT | T205H | WT | T205H | WT | T205H | |
| Turnover rates (s−1) | ||||||
| pH 6.0 | 3.2 | 3.5 | 1.9 | 1.7 | 144 | 64 |
| pH 7.0 | 7 | 7.6 | 7.6 | 4.2 | 220 | 121 |
| pH 8.0 | 14.6 | 15 | 12 | 2.8 | 177 | 119 |
| Km values (μm), pH 7.0 | ||||||
| KmUQ2 | 2 | 8.5 | ||||
| KmMQ1H2 | 4 | 22 | ||||
DISCUSSION
In this study, we have compared in two complex II homologues the effect of substitution of an amino acid residue distal to the middle Cys residue (in the primary sequence) coordinating the [3Fe-4S] cluster of the complex. This amino acid, SdhB His-207 in SQR and FrdB Thr-205 in QFR, appears important for controlling the redox potential of the [3Fe-4S] center. Replacing the natural amino acid in SQR/QFR with the one appearing in the homologue has an approximately equal effect on lowering/raising the redox potential (−68 mV lower for SQR and +82 mV higher for QFR). The His residue in this position is highly conserved in SQR enzymes as is the Thr residue in QFR enzymes, although a Met residue is found in the well studied Wolinella succinogenes QFR (41). X-ray structures for SQR and QFR all show that the side chain of the His or Thr residue is oriented toward the quinone-binding cavity in the enzymes, suggesting some involvement with the quinone catalytic site (Fig. 7) (18, 39, 41–45).
We have also determined the x-ray structure of the SdhB H207T enzyme to 2.7-Å resolution. The structure shows that there is a minimal perturbation of the overall structure of the SQR complex. There are, however, localized structural changes around the site of the mutation as shown in Figs. 2 and 3. The lowering of the redox potential of the cluster is most likely due to two factors. First, the mutation leads to an alteration of the electrostatic environment due to removal of the charge of the His residue. The wild-type structures clearly show that the imidazole side chain of the His is oriented away from the [3Fe-4S] cluster so that the charged side chain would lie about 5.5 Å away from the cluster. However, hydrophobic residues predominantly surround the cluster, so removal of the charged side chain would be expected to have an effect on the redox potential. Introduction of a histidine residue where the imidazole side chain is oriented toward the cluster in Azotobacter vinelandii ferredoxin I has been shown to have a significant effect on raising the redox potential of the [4Fe-4S] cluster in that protein (46). By contrast, in an E. coli SdhB I150H substitution, a decrease in the potential of the [4Fe-4S] cluster was observed (47); however, examination of the x-ray structure of wild-type SQR suggests that the imidazole side chain in this enzyme would be oriented away from the cluster.
A second factor that may contribute to the altered redox potential of the FeS cluster is changes in the hydrogen bonding network. A strengthening and increasing number of hydrogen bonds were noted in ferredoxin I (46). The wild-type SQR structure shows that three of the sulfur atoms of the [3Fe-4S] cluster can interact with the protein via hydrogen bonding (Fig. 4 and Table 2). Comparison of the available wild-type structures solved at better than 3-Å resolution shows that small movements of the main chain around the [3Fe-4S] cluster appear to be tolerated without causing a shift in redox potential. However, the distance between the S1 atom and the amide nitrogen of His-207 seems to be conserved. As a consequence of the mutation, there is a slight shift (∼0.2 Å) of the amide nitrogen of His-207 away from the S1 atom of the cluster, which would weaken the hydrogen-bonding interactions. This weakening of hydrogen bond interactions may contribute to the lower redox potential of the cluster in the SdhB H207T mutant as has been suggested for other proteins (11–14).
E. coli SQR SdhB H207T and wild-type QFR structures show similar hydrogen-bonding interactions of the [3Fe-4S] cluster within the immediate protein environment (Tables 2 and 3). The hydroxyl group is positioned closer to the cluster in QFR (4.2 Å) compared with SdhB H207T (4.9 Å) (Fig. 4). Interestingly, despite the similar fold of SdhB/FrdB loops near the [3Fe-4S] center, there is no sequence similarity; however, there is a similar hydrogen-bonding frame from backbone amide groups to the sulfur atoms. One significant difference between the SQR mutant and QFR wild-type proteins is that the side chain hydroxyl of SdhB Thr-223 is within 3 Å from the S4 atom of the cluster (Fig. 4), whereas in QFR, a non-hydrogen-bonding Ala residue is at the equivalent position. Thus, in SQR, the hydrogen bond from SdhB Thr-223 may be partially responsible for the overall 140-mV higher potential of the [3Fe-4S] center compared with QFR. In QFR, the x-ray structure of the wild-type enzyme suggests that a reason for the lower redox potential of the cluster is that the residue equivalent to SdhB Thr-223 in FrdB is Ala and that FrdB Phe-208 is present in place of SdhB Ser-208, making the environment around the cluster more hydrophobic in QFR. Although an x-ray structure is not available for the QFR FrdB T205H substitution, it is tempting to speculate that addition of the positive charge from the added His residue and the resulting more hydrophilic environment may be responsible for the +82-mV rise in the redox potential of the [3Fe-4S] cluster.
It is interesting to note that although the sequence identity between mammalian and E. coli SdhB proteins is only 55%, the structure of the loop shown in Fig. 4 is highly conserved. The x-ray structures of the avian (Protein Data Bank code 2H88) and porcine (Protein Data Bank code 1ZOY) SQR structures reveal a similar environment around the [3Fe-4S] cluster as compared with the E. coli enzyme (44, 45). The Em value for the [3Fe-4S] cluster in these enzymes is not known; however, the bovine enzyme is well studied with an Em similar to that of the E. coli enzyme (∼+60 to +70 mV) (48). There is one notable difference in the hydrogen bonding pattern in the porcine and avian complex II as compared with the E. coli SQR enzyme. In the porcine enzyme, the amino acid residue equivalent to SdhB Thr-223 that is hydrogen-bonded to the S4 atom of the [3Fe-4S] cluster is replaced by a non-bonding Gly residue. However, in the porcine enzyme, SdhB Tyr-178 is in a position homologous to SdhB Phe-169 of E. coli, and the porcine Tyr-178 is hydrogen-bonded to the sulfur of the FeS cluster. Thus, this hydrogen bond from the other side of the FeS cluster appears to maintain the number of hydrogen bonds to the [3Fe-4S] cluster as seen in other SQRs. The low potential [3Fe-4S] cluster of E. coli QFR has the non-hydrogen-bonding residues FrdB Phe-167 and FrdB Ala-221 at positions homologous to the Tyr and Thr residues found in SQR. It is possible to suggest that in addition to the presence of a His at the SdhB 207 position introduction of a hydrogen bond from either Thr-223 in E. coli SQR or Tyr-178 in the mammalian or avian complex II to the [3Fe-4S] cluster causes a further positive shift in the redox potential.
In addition to examining the effects the mutation in SdhB/FrdB had on the redox properties of the [3Fe-4S] cluster, we determined the effect on the quinone-binding site. As noted previously, the side chains of both SdhB His-207 and FrdB Thr-205 point into the quinone-binding cavity of each enzyme (Fig. 7). These residues are also part of a wall of amino acids that form the cavity where the SdhB/FrdB subunits interact with the hydrophobic C and D transmembrane peptides that form the rest of the quinone-binding site. As shown in Fig. 3, carboxin sits in essentially the same position within the Q-site for both wild-type and mutant structures. The structures rule out the possibility that the decreased affinity for carboxin is due to the substitution altering the structure of the quinone-binding site. As observed in the wild-type structure, the carbonyl group of carboxin interacts via hydrogen bonds with Tyr-D83 (2.7 Å) and Trp-B164 (2.9 Å). In both wild-type and mutant structures, carboxin interacts with other residues in the Q-site predominantly via hydrophobic interactions, including Phe-C20, Ile-B209, Pro-B260, Ile-C28, Ser-C27, and Arg-C31. Although carboxin lies close to His-B207 in the wild-type structure, the orientation of the ring was modeled to allow hydrogen bonding from the imidazole ring to one of the heme propionates and to a water molecule (18) rather than to the oxygen atom of the methyl-oxathiin ring of carboxin. The orientation of the imidazole ring seen in the structure would lead to poor hydrogen bonding geometry with carboxin. There is the possibility of a polar interaction between the oxygen atom of the methyl-oxathiin ring and the protonated His-B207. In the mutant structure, this would be replaced by a weak hydrogen bond interaction between the side chain oxygen atom of Thr-B207 and the oxygen atom in the methyl-oxathiin ring of carboxin (donor-acceptor distances are 3.4 Å with reasonable geometry). The difference in affinity for carboxin may be due to the altered interactions with B207 as a consequence of the substitution. Previous studies have suggested that SdhB His-207 is hydrogen-bonded to the O3 methoxy group of ubiquinone (43) and that rotation of the imidazole side chain is important for orienting the quinone in the binding site (40). Either a His or Thr residue at the SdhB 207 position provides a polar contact for carboxin and possibly to UQ. It will be of interest to determine whether non-polar substitutions at the SdhB 207 position lead to distinct differences in UQ binding and/or discrimination between reactions with UQ versus MQ. Although the data in Table 5 support the contention that SdhB His-207 plays some role in quinone binding, it is apparently not essential for this purpose. The data in Tables 4 and 5 also indicate that SdhB His-207 is not essential for electron transfer or protonation reactions with quinones. This is evidenced by the small change in enzyme activity with either ubiquinone or menaquinone even though the redox potential of the [3Fe-4S] cluster is lowered in the mutant enzyme.
In contrast to the rather small change in enzyme activity seen in the SQR mutant, the FrdB T205H substitution had more significant effects (Table 6). Using the electron transfer pathway program HARLEM (49), it is suggested that the backbone of FrdB Thr-205 is part of the electron transfer pathway from the [3Fe-4S] cluster to the QP site (not shown). Replacing the Thr-205 with a bulky charged residue such as His may have several effects on reactions with quinones in addition to alteration of the structural basis for electron transfer. The rate-limiting step for catalytic turnover with quinones in complex II is the two-electron/two-proton transfer between substrate and protein. Menaquinol oxidation by QFR is a pH-dependent reaction with a pKa of 7.4, and FrdC Glu-29 is a primary proton acceptor (15, 17). The 2-fold reduction in menaquinol oxidation in the FrdB T205H mutant enzyme regardless of pH along with the decreased affinity for MQH2 (Table 6) is consistent with an altered quinone-binding site rather than an increase in Em of the [3Fe-4S] cluster. Electron transfer models suggest that a change in Em of a redox group in a protein would not affect the electron transfer rate as long as distances between redox centers are conserved (50). As seen in Table 6, there is a more dramatic effect on the rate of ubiquinone reduction than menaquinol oxidation. At pH 6.0, the ubiquinone reductase activity is similar to that of the wild-type and mutant QFR enzymes. When electron transfer to the quinone site is faster with an increase of pH, protonation of the quinone may become rate-limiting either due to rearrangement of the hydrogen-bonded water network or to a change of local pKa due to His deprotonation. Previously, it was suggested that FrdC Tyr-25 was part of the protonation pathway to the quinone in QFR (15). Also the x-ray structures for E. coli QFR (42) have not modeled water in the molecule, although a chain of waters has been hypothesized to be present (15). This is consistent with previous suggestions that ubiquinone and menaquinone bind at slightly different places within the QP site and that different proton donors/acceptors are used for reduction/oxidation of the quinones (15, 42). It is also consistent with the possible effects of the mutation on the structure as described above.
CONCLUSION
Site-directed mutagenesis, x-ray structural analysis, kinetics, and spectroscopic methods have revealed the role of two residues that are part of the quinone-binding site of SQR and QFR. These residues are also proximal to the [3Fe-4S] cluster of the enzymes, and substitution affects the redox properties of the cluster. The altered redox state of the [3Fe-4S] cluster has relatively minor effects on steady state turnover kinetics of the mutant enzymes. The data do show, however, that SdhB His-207 aids in tight binding of quinone in SQR and that mutation of FrdB Thr-205 has significant effects on catalytic activity with quinones and binding in QFR.
This work was supported, in whole or in part, by National Institutes of Health Grant GM61606. This work was also supported by the Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Division; Biotechnology and Biological Sciences Research Council Grant B17935 with contributions from the Human Receptor Crystallography Project, Exploratory Research for Advanced Technology (ERATO), Japan Science and Technology Agency; and Canadian Institutes of Health Research Grant MOP89735.
The atomic coordinates and structure factors (code 2WP9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SQR
- succinate-quinone oxidoreductase
- UQ
- ubiquinone
- MQ
- menaquinone
- QFR
- menaquinol-fumarate oxidoreductase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Gpp
- gauss peak to peak
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- USQ
- ubisemiquinone
- Q
- quinone
- TLS
- translation libration screw-motion.
REFERENCES
- 1. Hägerhäll C. (1997) Biochim. Biophys. Acta 1320, 107–141 [DOI] [PubMed] [Google Scholar]
- 2. Cecchini G. (2003) Annu. Rev. Biochem. 72, 77–109 [DOI] [PubMed] [Google Scholar]
- 3. Lancaster C. R. D. (2002) Biochim. Biophys. Acta 1553, 1–6 [DOI] [PubMed] [Google Scholar]
- 4. Cecchini G., Schröder I., Gunsalus R. P., Maklashina E. (2002) Biochim. Biophys. Acta 1553, 140–157 [DOI] [PubMed] [Google Scholar]
- 5. Tran Q. M., Rothery R. A., Maklashina E., Cecchini G., Weiner J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18007–18012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oyedotun K. S., Sit C. S., Lemire B. D. (2007) Biochim. Biophys. Acta 1767, 1436–1445 [DOI] [PubMed] [Google Scholar]
- 7. Maklashina E., Cecchini G. (1999) Arch. Biochem. Biophys. 369, 223–232 [DOI] [PubMed] [Google Scholar]
- 8. Unden G., Bongaerts J. (1997) Biochim. Biophys. Acta 1320, 217–234 [DOI] [PubMed] [Google Scholar]
- 9. Kowal A. T., Werth M. T., Manodori A., Cecchini G., Schröder I., Gunsalus R. P., Johnson M. K. (1995) Biochemistry 34, 12284–12293 [DOI] [PubMed] [Google Scholar]
- 10. Morningstar J. E., Johnson M. K., Cecchini G., Ackrell B. A., Kearney E. B. (1985) J. Biol. Chem. 260, 13631–13638 [PubMed] [Google Scholar]
- 11. Stephens P. J., Jollie D. R., Warshel A. (1996) Chem. Rev. 96, 2491–2514 [DOI] [PubMed] [Google Scholar]
- 12. Adman E., Watenpaugh K. D., Jensen L. H. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 4854–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Breiter D. R., Meyer T. E., Rayment I., Holden H. M. (1991) J. Biol. Chem. 266, 18660–18667 [DOI] [PubMed] [Google Scholar]
- 14. Jang S. B., Seefeldt L. C., Peters J. W. (2000) Biochemistry 39, 641–648 [DOI] [PubMed] [Google Scholar]
- 15. Maklashina E., Hellwig P., Rothery R. A., Kotlyar V., Sher Y., Weiner J. H., Cecchini G. (2006) J. Biol. Chem. 281, 26655–26664 [DOI] [PubMed] [Google Scholar]
- 16. Maklashina E., Berthold D. A., Cecchini G. (1998) J. Bacteriol. 180, 5989–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Westenberg D. J., Gunsalus R. P., Ackrell B. A., Sices H., Cecchini G. (1993) J. Biol. Chem. 268, 815–822 [PubMed] [Google Scholar]
- 18. Ruprecht J., Yankovskaya V., Maklashina E., Iwata S., Cecchini G. (2009) J. Biol. Chem. 284, 29836–29846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kabsch W. (1993) J. Applied Crystallogr. 26, 795–800 [Google Scholar]
- 20. Evans P. R. (1993) in Proceedings of CCP4 Study Weekend on Data Collection and Processing, Warrington, January 29–30, 1993 (Sawyer L., Isaacs N., Bailey S. eds) pp. 114–122, SERC Daresbury Laboratory, Warrington, UK [Google Scholar]
- 21. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Afonine P. V., Grosse-Kunstleve R. W., Adam P. D. (2005) CCP4 Newsletter 42, contribution 8 [Google Scholar]
- 25. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 26. Kleywegt G. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Evrard G. X., Langer G. G., Perrakis A., Lamzin V. S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 29. Howlin B., Butler S. A., Moss D. S., Harris G. W., Driessen H. P. C. (1993) J. Appl. Crystallogr. 26, 622–624 [Google Scholar]
- 30. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 32. Maklashina E., Iverson T. M., Sher Y., Kotlyar V., Andréll J., Mirza O., Hudson J. M., Armstrong F. A., Rothery R. A., Weiner J. H., Cecchini G. (2006) J. Biol. Chem. 281, 11357–11365 [DOI] [PubMed] [Google Scholar]
- 33. Rothery R. A., Weiner J. H. (1996) Biochemistry 35, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 34. Hastings S. F., Kaysser T. M., Jiang F., Salerno J. C., Gennis R. B., Ingledew W. J. (1998) Eur. J. Biochem. 255, 317–323 [DOI] [PubMed] [Google Scholar]
- 35. Ohnishi T., King T. E., Salerno J. C., Blum H., Bowyer J. R., Maida T. (1981) J. Biol. Chem. 256, 5577–5582 [PubMed] [Google Scholar]
- 36. Hägerhäll C., Magnitsky S., Sled V. D., Schröder I., Gunsalus R. P., Cecchini G., Ohnishi T. (1999) J. Biol. Chem. 274, 26157–26164 [DOI] [PubMed] [Google Scholar]
- 37. Maklashina E., Rothery R. A., Weiner J. H., Cecchini G. (2001) J. Biol. Chem. 276, 18968–18976 [DOI] [PubMed] [Google Scholar]
- 38. Berry E. A., Trumpower B. L. (1987) Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 39. Tran Q. M., Rothery R. A., Maklashina E., Cecchini G., Weiner J. H. (2006) J. Biol. Chem. 281, 32310–32317 [DOI] [PubMed] [Google Scholar]
- 40. Horsefield R., Yankovskaya V., Sexton G., Whittingham W., Shiomi K., Omura S., Byrne B., Cecchini G., Iwata S. (2006) J. Biol. Chem. 281, 7309–7316 [DOI] [PubMed] [Google Scholar]
- 41. Lancaster C. R., Kröger A., Auer M., Michel H. (1999) Nature 402, 377–385 [DOI] [PubMed] [Google Scholar]
- 42. Iverson T. M., Luna-Chavez C., Cecchini G., Rees D. C. (1999) Science 284, 1961–1966 [DOI] [PubMed] [Google Scholar]
- 43. Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., Iwata S. (2003) Science 299, 700–704 [DOI] [PubMed] [Google Scholar]
- 44. Huang L. S., Sun G., Cobessi D., Wang A. C., Shen J. T., Tung E. Y., Anderson V. E., Berry E. A. (2006) J. Biol. Chem. 281, 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. (2005) Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 46. Chen K., Bonagura C. A., Tilley G. J., McEvoy J. P., Jung Y. S., Armstrong F. A., Stout C. D., Burgess B. K. (2002) Nat. Struct. Biol. 9, 188–192 [DOI] [PubMed] [Google Scholar]
- 47. Cheng V. W., Ma E., Zhao Z., Rothery R. A., Weiner J. H. (2006) J. Biol. Chem. 281, 27662–27668 [DOI] [PubMed] [Google Scholar]
- 48. Ohnishi T., Lim J., Winter D. B., King T. E. (1976) J. Biol. Chem. 251, 2105–2109 [PubMed] [Google Scholar]
- 49. Kurnivkov I. V. (2006) HARLEM, Northwestern University, Evanston, IL [Google Scholar]
- 50. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]