Abstract
The mammalian circadian clock component PERIOD2 (PER2) plays a critical role in circadian rhythm entrainment. Recently, a missense mutation at a putative phosphorylation site in hPER2, Ser-662, was identified in patients that suffer from familial advanced sleep phase syndrome (FASPS). Patients with FASPS display abnormal sleep-wake patterns characterized by a lifelong pattern of sleep onset in the early evening and offset in the early morning. Although the phosphorylation of PER2 is strongly implied from functional studies, it has not been possible to study the site-specific phosphorylation of PER2 on Ser-662, and the biochemical functions of this residue are unclear. Here, we used phospho-specific antibodies to show that PER2 is phosphorylated on Ser-662 and flanking casein kinase (CK) sites in vivo. The phosphorylation of PER2 was carried out by the combined activities of casein kinase 1δ (CK1 δ) and casein kinase 1ϵ (CK1ϵ) and was antagonized by protein phosphatase 1. PER2 phosphorylation was rapidly induced in response to circadian entrainment of mammalian cell lines and occurred in both cytosolic and nuclear compartments. Importantly, we found that the pool of Ser-662-phosphorylated PER2 proteins was more stable than the pool of total PER2 molecules, implying that the FASPS phosphorylation cluster antagonizes PER2 degradation. Consistent with this idea, a Ser-662 → Ala mutation that abrogated PER2 phosphorylation significantly reduced its half-life, whereas a phosphomimetic Ser-662 → Asp substitution led to an elevation in half-life. Our combined findings provide new insights into PER2 regulation and the biochemical basis of FASPS.
Keywords: DNA Damage, PP1, Protein Phosphorylation, Serine-Threonine Protein Kinase, Serine-Threonine Protein Phosphatase, CK1, PERIOD 2, Circadian Rhythms
Introduction
Circadian rhythms refer to the roughly 24-h periodicity of biochemical and physiological processes in light-sensitive organisms. These rhythms are synchronized by external environmental cues such as the light/dark cycle or a temperature cycle and serve to regulate processes as diverse as sleep-wake cycles, nutrient metabolism, immunity, and cellular division (reviewed in Refs. 1–3). The biochemical machinery regulating rhythm generation in vertebrates is conserved across evolution and, at its core, is comprised of an oscillatory transcriptional feedback circuit (1). In Drosophila, this feedback circuit consists of two positive regulators from the basic helix-loop-helix family of transcription factors, CYCLE (CYC)3 and CLOCK (CLK), and negative regulators PERIOD (PER) and TIMELESS. Heterodimeric complexes of CYC·CLK bind to E-box promoter sequences and drive the expression of a variety of genes regulating the circadian clock, including those of per and tim. Newly synthesized PER·TIMELESS complexes accumulate in the cytoplasm before translocating to the nucleus, where PER inhibits CYC·CLK activity and completes the feedback loop (1). In mammals, structurally conserved proteins play similar, if only more complex roles in regulating the central feedback circuit (1–3). In mammals, CLOCK and BMAL1 transcription factors play a role analogous to the CYC·CLK complex, driving expression of Cryptochrome (Cry) and Period (Per) genes, the products of which inhibit CLOCK·BMAL activity. The existence of functionally overlapping homologous genes for Per (Per1, Per2, and Per3) and Cry (Cry1 and Cry2) imparts additional complexity to the mammalian circadian oscillator (2).
Reversible protein phosphorylation events also play an essential role in the regulation of the circadian cycle (4–6). The initiation of a new circadian cycle in Drosophila and mammals is accomplished through phosphorylation-dependent degradation of inhibitory PER proteins (5, 7, 8). Phosphorylation of the PER proteins is carried out by members of the casein kinase 1 (CK1) family, including DOUBLE-TIME (DBT) in Drosophila and casein kinase 1δ (CK1δ) and casein kinase 1ϵ (CK1ϵ) in mammals (5, 6). In mammals, the CK1ϵ- or CK1δ-dependent phosphorylation of PER1 and PER2 recruits the F-box protein β-TRCP, which stimulates the ubiquitylation and proteasome-dependent degradation of both proteins (7–9). On the other hand, phosphorylation-mediated degradation of PER1/PER2 is antagonized by the PP1 and PP2A families of protein phosphatases (10, 11). The significance of post-translational modifications in the regulation of clock timing has been clearly revealed by the use of CK1 and proteasome inhibitors, which increase circadian periods in cell culture (12). In addition, genetic studies in Drosophila identified dbt alleles imparting either short or long periods. Interestingly, both sets of DBT mutants display decreased kinase activity in vitro (5, 13, 14).
Genetic evidence supporting protein phosphorylation as an essential component of the circadian oscillation has also been obtained through study of mammalian sleep disorders. The tau hamster, which was the first mammalian circadian mutant identified, contains a missense mutation in CK1ϵ that leads to a reduction in circadian period (15). More recently, studies of the inherited disorder familial advanced sleep phase syndrome (FASPS), have implicated site-specific phosphorylation of PER2 as a crucial event in the circadian oscillation (16, 17). FASPS patients are “morning larks” that display a markedly advanced sleep phase and a shortened circadian period. In the first study identifying a genetic link to the syndrome, Toh et al. (16) identified a Ser-to-Gly mutation at position 662 in hPER2 that segregated with FASPS-affected members in a large pedigree. The authors subsequently showed that the mutation led to hypophosphorylation of a PER2 polypeptide in vitro (16). Subsequent studies have attempted to gain a greater understanding of the molecular impact of the hPER2 mutation and its role in the FASPS pathophysiology. Xu et al. (18) showed that PER2-deficient mice genetically reconstituted with an hPer2 BAC clone harboring the FASPS S662G mutation displayed marked phase advancement. Interestingly, these mice exhibited reduced levels of Per2 gene transcription, suggesting that PER2 regulates its own expression (18). On the other hand, Vanselow et al. (19) demonstrated that the mPER2S659G protein was less stable than wild-type mPER2 and proposed that reduced protein stability was a consequence of impaired nuclear import. While providing important insights into hPER2 regulation, neither study directly analyzed PER2 proteins site-specifically phosphorylated at Ser-662 in vivo.
To better elucidate the mechanisms and functional consequences of PER2 phosphorylation, we generated a phosphospecific antibody that detects Ser-662 phosphorylation in vivo. We have used this reagent to answer several outstanding questions about PER2 regulation. We show that Ser-662 and adjacent CK1 sites at Ser-665 and Ser-668 are coordinately phosphorylated in response to circadian entrainment and confirm that CK1δ, CK1ϵ, and PP1 are key regulators of the PER2 phosphorylation state. In addition, we demonstrate for the first time that Ser-662/665/668-phosphorylated hPER2 possesses increased stability versus the unphosphorylated hPER2. Increased stability of Ser-662/665/668-phosphorylated PER2 occurs in the absence of nuclear retention and is recapitulated in PER2 proteins harboring phosphomimetic amino acids at codon 662. These results provide new insights into the biochemical mechanisms of PER2 phosphorylation, and the phospho-PER2 antibody described here will be a useful tool for interrogating mechanisms of PER2 regulation in response to circadian and noncircadian cues.
EXPERIMENTAL PROCEDURES
DNA Constructs
pcDNA3.1Myc-hPER2(zeo) was constructed by cloning hPer2 (BC111453 clone from Open Biosystems) into the KpnI and NotI sites of a modified pcDNA3.1zeo (with an N-terminal Myc tag). Site-directed mutagenesis was performed using the QuikChange method (Stratagene) to make the following hPER2 mutants using the indicated primers: hPER2S662A (5′-CCGGGCAAGGCAGAGGCTGTGGCGTCGCTCACC-3′ and its reverse complement), hPER2S665A (5′-GCAGAGAGTGTGGCGGCGCTCACCAGCCAGTGC-3′ and its reverse complement), hPER2S668A (5′-GTGGCGTCGCTCACCGCCCAGTGCAGCTACAGC-3′ and its reverse complement), and hPER2A664V (5′-CAAGGCAGAGAGTGTGGTGTCGCTCACCAGCCAG-3′ and its reverse complement). The various hPER2 C-terminal truncation mutants were generated by introducing a STOP codon at the desired position in the coding sequence by the QuikChange method using the indicated primers: hPER2(1–1157) (5′-GCTGCCTTCCCGAAATTAAGAAGCGGTTTTGAAGG-3′ and its reverse complement), hPER2(1–806) (5′-GGGTCAAACCTCGAGACTAATCTGAGAGCACCGG-3′), and hPER2(1–682) (5′-CATGTGGGAGACAAGTAGCCGCAGCCGGAGTTAG-3′). hPER2(401–806) was generated by cloning the hPer2 fragment into the NotI and KpnI sites of pcDNA3.1zeo-myc using the following primers: 5′-GCCGGGCGGACAGCGGCCGCCCAGATCCGGTGCTC-3′ and (5′-TCACCTACATGGTACCCGCGCCCGGAACGGAGAG-3′. QuikChange mutagenesis of V5-tagged mPER1 was performed to generate mPER1V716A,V718L using 5′-GGCAGAGAGCGTGGCGTCCCTCACCAGTCAGTGTAGC-3′ and its reverse complement). The dominant-negative PP1 plasmid harboring the D95N mutation was a kind gift from Dr. David Virshup at the Duke-NUS Graduate Medical School. Epitope (FLAG)-tagged CK1ϵ was generated by cloning hCK1ϵ cDNA into the pFLAG-CMV 6b vector. Myc-tagged wild-type CK1δ was a kind gift from Dr. Wade Harper.
Cell Culture, Antibodies, and Inhibitors
HEK 293T cells an U-2 OS cells were purchased from ATCC and maintained in DMEM containing 5% FBS. NIH-3T3 and 293T cells stably expressing hPer2 were maintained in DMEM containing 10% FBS supplemented with 300 μg/ml Zeocin (Invitrogen). NIH-3T3 (Per:luc) cells were a kind gift from the Dr. Achim Kramer. HEK 293T cells and NIH-3T3 cells were transfected with pcDNA3.1(zeo)Myc-hPer2 using the calcium phosphate method, followed by selection in 300 μg of Zeocin (Invitrogen). Individual clones were selected and propagated in medium containing antibiotic. The pPER2(FASPS) antibody was generated by immunizing rabbits with a triply phosphorylated hPER2 peptide (KAEpSVApSLTpSQC) (Cocalico Biologicals, Reamstown, PA). Peptide synthesis and purification of antisera were performed as described before for the pCREB-108/111/114 antibody (20). Other antibodies used in this study include: α-PER2 (Novus), α-Myc (SCBT), α-FLAG-M2 (Sigma). α-CK1ϵ (SCBT), α-CK1δ (Bethyl), α-CREB (Millipore), and α-PP1 (SCBT). CK1 inhibitor D4476 (4-(4-(2,3-dihydrobenzo[1,4] dioxin-6-yl)-5-pyridin-2-yl-1H-imidazol-2-yl)benzamide), CK1 inhibitor IC261 (3-[2,4,6-(trimethoxyphenyl)methylidenyl]-indolin-2-one), and okadaic acid (OA) were used at 75 μm, 10 μm, and 100 nm, respectively. All of the inhibitors were obtained from EMD Biosciences and added to culture medium for 4 h. Cycloheximide (Sigma) was used at a final concentration of 20 μg/ml for the indicated times. Dexamethasone (Dex) was obtained from Sigma and used as described below.
Transfections and Immunoblotting
Transfections were performed using the calcium phosphate DNA precipitation procedure as described. siRNA SMARTpools against hCK1δ and hCK1ϵ were obtained from Dharmacon Inc. The cells were harvested 48 h later, and the extracts were prepared as described previously (20). Standard Western blotting procedures were followed, as described before (20). Where indicated, band pixel intensities were determined using the density function of Quantity One software (Bio-Rad).
Dexamethasone Shock and Nucleocytoplasmic Fractionation
NIH-3T3-hPer2 cells washed with PBS were first suspended in cytoplasmic extract buffer (10 mm HEPES, pH 7.9, 50 mm NaCl, 1 mm DTT, 0.1 mm EDTA + protease and phosphatase inhibitors). The cytoplasmic extracts were clarified by sedimentation at 5000 × g. This was followed by treatment with equal volume nuclear extract buffer (20 mm HEPES, pH 7.9, 400 mm NaCl, 1 mm DTT, 1 mm EDTA, 1 mm EGTA + inhibitors) and sedimentation at 15,000 × g. The extracts were assayed by immunoblotting using the indicated antibodies. Dexamethasone shock was performed as described before (19). Briefly, NIH-3T3-hPer2 cells were grown to confluence for 4 days before the experiment. At time 0, the culture medium was exchanged with serum-free medium containing 100 nm Dex for 2 h. After 2 h, the medium was once again exchanged with serum-free medium (lacking Dex), and the cells were harvested at the indicated times.
RESULTS
Generation and Characterization of a pPER2(FASPS) Antibody
Previous work in our lab defined a cluster of DNA damage-inducible phosphorylation sites in the cAMP response element-binding protein (CREB) that are collaboratively phosphorylated by CK1, CK2, and ataxia-telangiectasia mutated (ATM) protein kinases (20). In the CREB phosphorylation paradigm, ATM-dependent phosphorylation of Ser-111 creates consensus phosphorylation sites for CK1 and CK2 on Ser-108, Ser-114, and Ser-117. An in silico screen for proteins encoding a consensus ATM Ser-Gln phosphorylation site flanked by putative CK1/CK2 sites identified several circadian rhythm proteins, including PER1 and PER2 (Fig. 1A). The candidate ATM-CK1/CK2 phosphorylation cluster in PER2 spans codons 662–673 and contains the Ser-662 phosphorylation site implicated in FASPS. Given that PER2 (and PER1) was identified as a tumor suppressor protein and previously linked to cellular DNA damage responses, we hypothesized that PER2 was a direct phosphorylation target of ATM (21–23).
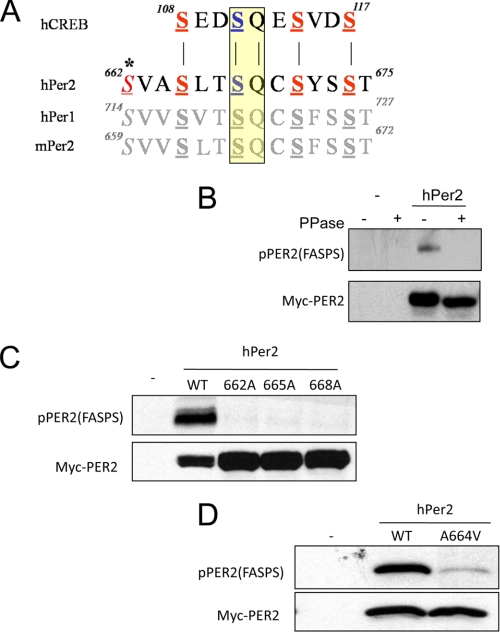
FIGURE 1.
Characterization hPER2 phospho-FASPS cluster antibodies. A, sequence overlay between mammalian CREB and PER proteins. Homologous putative phosphorylation sites are shown in bold and underlined. Putative ATM phosphorylation sites in the PER proteins are highlighted in yellow. The residue Ser-662, mutated in FASPS, is italicized and marked with an asterisk. B, phosphatase sensitivity of pPER2(FASPS) antisera. HEK 293T cells were transfected with vector DNA (−) or a plasmid encoding Myc epitope-tagged hPER2 for 24 h. The cell extracts were prepared and treated with λ phosphatase prior to analysis by SDS-PAGE and immunoblotting with α-Myc and α-pPER2(FASPS) antibodies. C, phosphorylation site requirements. HEK 293T cells were transfected with plasmids encoding Myc-tagged hPER2WT or the indicated hPER2 phosphorylation site mutants. The cell extracts were then prepared and analyzed by immunoblotting using α-Myc and α-pPER2(FASPS) antibodies. D, α-pPER2(FASPS) antibodies are selective for hPER2. HEK 293T cells were transfected with plasmids encoding Myc-tagged hPER2WT or the hPER2A664V mutant. The cell extracts were then made and analyzed by immunoblotting using α-Myc and α-pPER2(FASPS) antibodies.
To begin testing whether PER2 was phosphorylated by ATM and CK1/CK2 in response to DNA damage, we raised a phospho-specific antibody against a human PER2 peptide triply phosphorylated on Ser-662, Ser-665, and Ser-668, using the logic that processive phosphorylation by CK1 would result in the phosphorylation of all three sites in vivo (see “Experimental Procedures” for details). We refer to this purified antisera as the α-pPER2(FASPS) antibody and the phosphorylated PER2 region spanning amino acids 662–668 as the FASPS cluster.
The α-pPER2(FASPS) antibody was first tested in immunoblotting experiments for immunoreactivity against overexpressed PER2 using HEK 293T cell extracts. The α-pPER2(FASPS) antibody displayed reactivity with transfected hPER2 in HEK 293T cells, which otherwise do not express the PER2 protein (Fig. 1B). Immunoreactivity was completely abolished upon phosphatase treatment of the cell extracts, indicating that the α-pPER2(FASPS) antibody was phospho-specific (Fig. 1B). We further tested the specificity of the α-pPER2(FASPS) antibody by assessing the effects of single Ser → Ala substitutions at the Ser-662, Ser-665, and Ser-668 sites on immunoreactivity. Mutation of Ser-662, Ser-665, or Ser-668 abolished immunoreactivity of overexpressed hPER2 proteins with α-pPER2(FASPS) (Fig. 1C). Thus, the pPER2(FASPS) antibody requires that all three sites be phosphorylated. These findings provide strong evidence that the FASPS site Ser-662 is phosphorylated in vivo. Our data also strongly suggest that, as postulated, Ser-665 and Ser-668 are obligatorily phosphorylated in parallel with Ser-662.
We next tested whether the pPER2(FASPS) antibody recognized mouse PER2 protein or human PER1 protein, which are nearly identical to PER2 throughout the region surrounding the FASPS cluster (Fig. 1A). Lacking a mouse Per2 cDNA, we introduced a single A664V mutation into a hPer2 cDNA that converted the human FASPS cluster to the corresponding murine sequence. This single, conservative change severely attenuated pPER2(FASPS) antibody reactivity (Fig. 1D). Similarly, α-pPER2(FASPS) antisera did not detect overexpressed hPER1, which differed from hPER2 at two amino acids (supplemental Fig. S1A). These data demonstrated the high specificity of α-pPER2(FASPS) for hPER2 versus closely related hPER1 and mPER2 proteins. Interestingly, a hPER1V696A/V698L mutant that converted the PER1 FASPS cluster to the homologous hPER2 sequence reacted with α-pPER2(FASPS) antibody (supplemental Fig. S1A). These data also suggest that the analogous sites in PER1 are possibly phosphorylated in mammalian cells, as has been previously suggested (24).
We next proceeded to test whether pPER2(FASPS) immunoreactivity was enhanced upon exposure to DNA-damaging agents that are known activators of ATM. HEK 293T cells stably overexpressing Myc-tagged hPER2 were exposed to 10 grays of ionizing radiation, 50 J/m2 UV light, or 200 μm H2O2 for a period of 2 h. No changes in pPER2(FASPS) immunoreactivity were observed. In comparison, phosphorylation of CREB on Ser-108, Ser-111, and Ser-114 was clearly induced by these stimuli (supplemental Fig. S1B). Thus, even though the FASPS cluster contains a consensus ATM phosphorylation site, these findings suggest that Ser-662 of PER2 is not a direct substrate of ATM in vivo.
CK1δ/ϵ Cooperatively Phosphorylate the PER2 FASPS Cluster
Although indirect evidence had suggested a role for the CK1 kinase family in regulating hPER2 FASPS cluster phosphorylation in intact cells, definitive evidence was lacking. To address this issue, we first examined the effects of specific small molecule inhibitors of CK1, D4476, and IC261, on FASPS cluster phosphorylation (25, 26). Although D4476 is a pan-CK1 inhibitor, IC261 is selective for CK1δ/ϵ when used at a concentration of 10 μm. HEK293T cells expressing Myc-hPER2 were exposed to solvent, 75 μm D4476, or 10 μm IC261 for 4 h (26). D4476 treatment almost completely abrogated FASPS cluster phosphorylation, strongly supporting a role for CK1 proteins as bona fide hPER2 kinases (Fig. 2A). IC261 also strongly inhibited FASPS cluster phosphorylation, suggesting that CK1δ/ϵ are the relevant CK1 isoforms (Fig. 2A). To substantiate the inhibitor findings, we used siRNA to silence CK1δ or CK1ϵ in Myc-hPER2-expressing HEK 293T cells. Although CK1δ and CK1ϵ protein levels were strongly suppressed by the siRNA transfection, knockdown of either CK1δ or CK1ϵ alone had no effect on phosphorylation of the hPER2 FASPS cluster (supplemental Fig. S1C). However, knockdown of both isoforms together led to a more than 50% reduction in phosphorylation (Fig. 2B), suggesting that CK1δ and CK1ϵ cooperatively phosphorylate hPER2 in mammalian cells. Overexpression of CK1ϵ also strongly induced FASPS cluster phosphorylation in HEK 293T cells cotransfected with Myc-hPER2 (Fig. 2C). The overexpression of CK1ϵ imparted a phosphatase-sensitive reduction in hPER2 electrophoretic mobility and caused a reduction in total hPER2 expression level, which is consistent with a previous conclusion that CK1ϵ phosphorylates PER2 and targets hPER2 for degradation (compare third and fifth lanes in Fig. 2C) (18). It is important to note that FASPS cluster phosphorylation was observed even in the absence of the electrophoretic mobility shift (Fig. 2C, third lane), which must therefore be due to modification of different CK1ϵ-dependent sites. Consistent with this, an hPER2S662A mutant defective for phosphorylation of the FASPS cluster still undergoes an electrophoretic mobility shift upon the coexpression of the CK1ϵ mutant tau (Fig. 2D). The tau mutant has been shown to be a hypermorphic CK1 variant that hyperphosphorylates PER proteins with faster kinetics than the wild-type protein (27). Together the RNAi and overexpression experiments show that CK1δ and CK1ϵ redundantly regulate hPER2 FASPS cluster phosphorylation and that CK1ϵ targets at least two distinct motifs: the FASPS cluster and a distinct motif that reduces hPER2 electrophoretic mobility and stability.
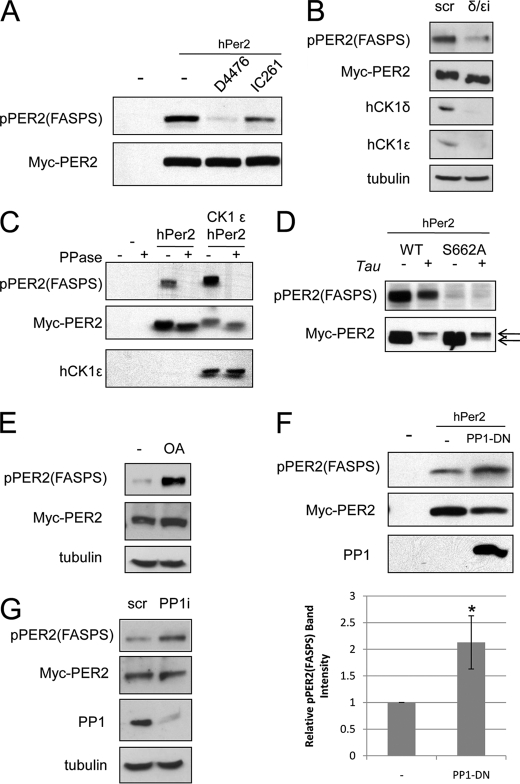
FIGURE 2.
A CK1δ, CK1ϵ, and PP1 regulate hPER2 FASPS phosphorylation. A, inhibition of hPER2 FASPS cluster phosphorylation by CK1 inhibitors. HEK 293T cells overexpressing Myc-tagged hPER2 were treated with D4476 or IC261 for 4 h. The cell extracts were then made and analyzed by immunoblotting using α-Myc and α-pPER2(FASPS) antibodies. B, CK1δ and CK1ϵ redundantly phosphorylate the hPER2 FASPS cluster. HEK 293T-Myc-hPER2 cells were transfected with scrambled siRNA (scr) or siRNA directed against the CK1δ and CK1ϵ isoforms (δ/ϵi) for 48 h. The cell extracts were then prepared and analyzed by immunoblotting using α-Myc, α-pPER2(FASPS), and α-tubulin antibodies. C, effect of CK1ϵ overexpression on FASPS cluster phosphorylation. HEK 293T cells were transfected with vector DNA (−) or a plasmid encoding Myc-tagged hPER2 alone or plasmids for both hPER2 and FLAG-tagged hCK1ϵ for 24 h. The cell extracts were prepared and treated with λ phosphatase prior to analysis by immunoblotting with α-Myc, α-pPER2(FASPS), and α-FLAG antibodies. D, CK1tau targets the FASPS mutant for degradation. HEK 293T cells were transfected with a plasmid encoding Myc-tagged hPER2WT or hPER2S662A alone or plasmids for both hPER2 and FLAG-tagged hCK1tau for 24 h. The cell extracts were prepared and analyzed by immunoblotting with α-Myc and α-pPER2(FASPS) antibodies. The arrows indicate hyperphosphorylated (top arrow) and hypophosphorylated (bottom arrow) forms of PER2. E, okadaic acid induces FASPS cluster phosphorylation. HEK 293T-Myc-hPER2 cells were treated with solvent (−) or 100 nm OA. The cell extracts were analyzed by immunoblotting using α-Myc, α-pPER2(FASPS), and α-tubulin antibodies. F, dominant-negative PP1α catalytic subunit (PP1D95N) induces hPER2 FASPS cluster phosphorylation. Top panels, HEK 293T cells were transfected with plasmids encoding Myc-tagged hPER2WT alone or plasmids for both hPER2 and Myc-tagged PP1D95N for 24 h. The cell extracts were analyzed by immunoblotting as described for E. Bottom panel, graphical representation of immunoblotting data. The data are depicted as relative mean band intensities ± S.E. The asterisk indicates p < 0.05 (n = 3). G, knockdown of the PP1α catalytic subunit induces hPER2 FASPS cluster phosphorylation. HEK 293T-Myc-hPER2 cells were transfected with scrambled siRNA (scr) or siRNA directed against PP1α for 48 h. The cell extracts were then prepared and analyzed by immunoblotting using α-Myc, α-pPER2(FASPS), α-PP1α, and α-tubulin antibodies.
PP1 Is a Negative Regulator of PER2 FASPS Cluster Phosphorylation
There is conflicting evidence concerning the identity of the hPER2 phosphatase (10, 11). Although both PP1 and PP2A have been implicated, the absence of appropriate phospho-specific antibodies has precluded definitive studies. To clarify this issue, we first used the small molecule protein phosphatase inhibitor OA, which can be used to distinguish OA-sensitive PP1, PP2A, and PP5 phosphatases from OA-insensitive PP2B/calcineurin and PP7 phosphatases. Treatment of Myc-PER2-expressing HEK 293T cells with a concentration of OA (100 nm) that inhibits PP1, PP2A, and PP5 strongly induced FASPS cluster phosphorylation, suggesting a role for one or more of these phosphatases (Fig. 2E and Ref. 28). Furthermore a dominant-negative mutant of the PP1α catalytic subunit (PP1D95N) (10) caused a significant increase in FASPS cluster phosphorylation of cotransfected hPER2 in HEK 293T cells, strongly supporting an important role for PP1 (Fig. 2F). PP1α knockdown also increased hPER2 FASPS cluster phosphorylation (Fig. 2G). Taken together, our findings show that PP1 antagonizes CK1δ/ϵ-dependent phosphorylation of the FASPS cluster; however, these studies do not rule out a supportive role for PP2A or other OA-sensitive phosphatases.
Domain Requirements for hPER2(FASPS) Phosphorylation
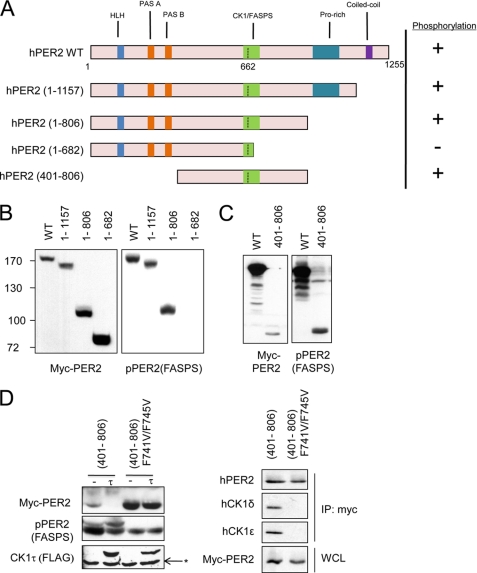
The availability of the α-hPER2(FASPS) antibody allowed us to map hPER2 structural determinants that are required for FASPS cluster phosphorylation. Specifically, hPER2 contains several protein-protein interaction domains and several identified binding partners (29), and we tested whether deletion of these domains affected phosphorylation on Ser-662/665/668 (Fig. 3A). Deletion of HLH, PAS-A, PAS-B, Pro-rich, or coiled-coil domains did not prevent FASPS cluster phosphorylation in transiently transfected HEK 293T cells. On the other hand, a fragment of hPER2 spanning amino acids 1–682 was defective for phosphorylation, indicating that amino acids C-terminal to the FASPS cluster are required for its phosphorylation (Fig. 3B). Finally, we found that a minimal fragment of hPER2 spanning amino acids 401–806, hPER2(401–806), was efficiently phosphorylated on the FASPS cluster in HEK 293T cells (Fig. 3C). We used this hPER2 miniprotein to further evaluate sequence elements regulating hPER2 phosphorylation. Previous studies have identified two conserved Phe residues in mPER1, Phe-793 and Phe-797, responsible for CK1 binding, and we sought to test the effects of mutating the homologous residues in hPER2, Phe-741 and Phe-745, on FASPS cluster phosphorylation (8, 30). Mutation of Phe-741 and Phe-745 attenuated hPER2 phosphorylation on the FASPS cluster while simultaneously inhibiting phosphorylation-mediated degradation of the miniprotein, clearly identifying the CK1-binding region of hPER2 as an important structural element in the regulation of PER2 stability (Fig. 3D). We verified that mutation of these residues abrogated binding of PER2 to both CK1δ and CK1ϵ (Fig. 3D, right panel). A role for Phe-741 and Phe-745 is also in agreement with our initial truncation studies implicating the region spanning amino acids 682–806 as determinants of hPER2 FASPS cluster phosphorylation. This miniprotein can be further used to identify other sequence elements regulating PER2 stability.
FIGURE 3.
Domain requirements for hPER2 FASPS cluster phosphorylation. A, schematic depiction of the various functional domains of hPER2 and the various hPER2 truncation mutants made. HLH indicates the helix-loop-helix domain; PAS A and PAS B are protein interaction domains; CK1/FASPS indicates the CK1 interaction domain and FASPS phosphorylation cluster; Pro-rich indicates a proline-rich sequence of unknown functional significance and coiled-coil refers to the C-terminal domain that plays a role in Cry binding. B, C-terminal truncation of hPER2. HEK 293T cells were transfected with vector DNA (−) or plasmids encoding various Myc-tagged hPER2 truncation mutants. The cell extracts were prepared and analyzed by immunoblotting with α-Myc, α-pPER2(FASPS) antibodies. C, an hPER2(401–806) miniprotein is competent for hPER2 FASPS cluster phosphorylation. HEK 293T cells were transfected with plasmids encoding Myc-tagged hPER2WT or Myc-tagged hPER2(401–806). The cell extracts were prepared and analyzed by immunoblotting with α-Myc and α-pPER2(FASPS) antibodies. D, mutation of putative CK1-binding domain attenuates hPER2 FASPS cluster phosphorylation. Left panel, HEK 293T cells were transfected with plasmids encoding the Myc-tagged hPER2(401–806) or a Myc-tagged hPER2(401–806) F741V/F745V mutant. The cell extracts were prepared and analyzed by immunoblotting with α-Myc, α-pPER2(FASPS), and α-FLAG antibodies. Where indicated, the cells were cotransfected with FLAG-tagged CK1tau (τ). The asterisk indicates endogenous CK1. Right panel, HEK 293T cells were transfected with plasmids encoding the Myc-tagged hPER2(401–806) or a Myc-tagged hPER2(401–806) F741V/F745V mutant. The cell extracts were subjected to immunoprecipitation (IP) with α-Myc antibody. Precipitates and whole cell lysates (WCL) were analyzed by immunoblotting with α-Myc, α-pPER2(FASPS), α-CK1δ, and α-CK1ϵ antibodies.
Phosphorylation of hPER2(FASPS) Occurs during the Circadian Cycle
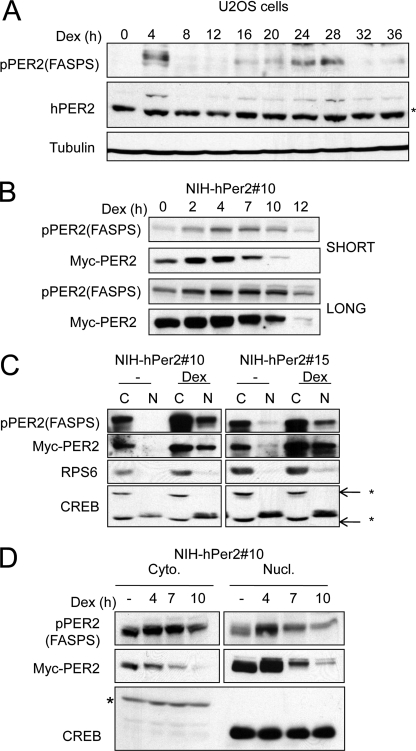
It has not been possible, until now, to observe the site-specific phosphorylation of hPER2 during a circadian oscillation. We sought to use the pPER2(FASPS) antisera to delineate the temporal regulation of PER2 phosphorylation. We used a well established Dex shock entrainment protocol to induce circadian oscillations in U-2 OS cells (31). The Dex shock induced a robust yet transient up-regulation of endogenous PER2 protein levels at 4 h followed by a reduction in PER2 levels at subsequent times (Fig. 4A). PER2 levels again rose at 24 h post-Dex shock and disappeared at 32 h post-entrainment, consistent with the circadian regulation of its expression. Interestingly, PER2 FASPS cluster phosphorylation almost completely mirrored that seen with the total PER2 protein (Fig. 4A). To eliminate the confounding effects of Per2 transcription on our studies with PER2 post-translation modifications, we generated two independent NIH-3T3 cell lines stably expressing Myc-tagged hPER2 (19). We refer to these cells as NIH-3T3-myc-hPER2 10 and 15, respectively. These cells were then subjected to the Dex shock protocol, and cell extracts were harvested at various times. As seen in Fig. 4B and supplemental Fig. S1D, Dex shock initially induced transient stabilization of hPer2 accompanied by an increase in FASPS cluster phosphorylation. At later time points (7, 10, and 12 h), hPER2 electrophoretic mobility was impeded, and the expression level of the protein was dramatically reduced (Figs. 4B and supplemental Fig. S1D). This finding is consistent with coupled phosphorylation and degradation of hPER2, as described by others (7, 10). The levels of FASPS cluster phosphorylation also declined at late time points after Dex shock; however, the rate of decline was lower than observed for total hPER2 (Figs. 4B and supplemental Fig. S1D). This finding provided initial evidence that phosphorylation of the FASPS cluster may correlate with enhanced hPER2 stability.
FIGURE 4.
FASPS cluster phosphorylation during a circadian cycle. A, temporal regulation of FASPS cluster phosphorylation during a circadian cycle. U-2 OS cells were Dex shocked as described under “Experimental Procedures.” The cell extracts were made at the indicated times and subjected to immunoblotting with α-PER2, α-tubulin, and α-pPER2(FASPS) antibodies. The asterisk indicates a nonspecific band detected by hPER2 antibody. B, temporal regulation of FASPS cluster phosphorylation in NIH-3T3 cells. NIH-3T3-hPer2 clone 10 cells were subjected to the Dex shock protocol. The cell extracts were made at the indicated times and subjected to immunoblotting with α-Myc and α-pPER2(FASPS) antibodies. Short and long exposures of the film are shown. C, nucleocytoplasmic localization of Ser-662/665/668-phosphorylated hPER2. NIH-3T3-hPer2 clones (clones 10 and 15) were either left untreated or subjected to a Dex shock. Nucleocytoplasmic fractionation of the cell extracts was then performed as indicated under “Experimental Procedures.” Immunoblotting analysis was performed with α-Myc, α-pPER2(FASPS), α-CREB, and α-ribosomal protein S6 antibodies (rpS6). CREB is a documented nuclear protein, whereas rpS6 is largely cytoplasmic. An asterisk indicates nonspecific bands detected by the α-CREB antibody. D, increased stability of phosphorylated hPER2 in nuclear (Nucl.) and cytoplasmic (Cyto.) fractions. NIH-3T3-hPer2 clone 10 cells were subjected to a Dex shock, and samples were taken at the indicated times. Nucleocytoplasmic fractionation of the cell extracts was then performed as described above. Immunoblotting was performed using α-Myc, α-pPER2(FASPS), α-CREB, and α-ribosomal protein S6 antibodies (rpS6). The asterisk indicates a cross-reactive nonspecific band detected by the α-CREB antibody.
Nucleocytoplasmic Shuttling of hPER2 Is Unaffected by FASPS Site Phosphorylation
The PER proteins are synthesized in the cytoplasm and translocated into the nucleus where they repress CLOCK/BMAL transcription. It has been proposed that PER2 phosphorylation on the FASPS sites affects its nucleocytoplasmic shuttling (19). To study this, we separated control or Dex-shocked NIH-3T3-hPer2 cell extracts into nuclear and cytoplasmic extracts and immunoblotted Myc-hPER2 and pPER2(FASPS). In untreated Myc-hPER2 NIH-3T3 cell lines, virtually all of the Myc-hPER2 and pPER2(FASPS) immunoreactivity was observed in the cytoplasmic fraction (Fig. 4B). Upon Dex stimulation, both total Myc-hPER2 and Ser-662/665/668-phosphorylated hPER2 accumulated in the nucleus (Fig. 4C). In no instance did we observe enrichment of Ser-662/665/668-phosphorylated hPER2 in the nuclear fraction. The simplest interpretation of these findings is that phosphorylation of the FASPS cluster does not influence hPER2 nucleocytoplasmic shuttling. To test the possibility that the nuclear fraction of PER2 alone may be more stable over the course of the circadian cycle, we performed a Dex time course as described above and fractionated the cell extracts into nuclear and cytoplasmic extracts. Immunoblotting with Myc-hPER2 and α-pPER2(FASPS) revealed that the rate of reduction of Ser-662/665/668-phosphorylated hPER2 is lower than the rate of reduction of the total hPER2 protein in both the nuclear and cytoplasmic extracts (Fig. 4D). These findings suggested that increased nuclear retention is not responsible for delayed degradation of PER2 that is phosphorylated on the FASPS cluster as has been previously proposed (19).
FASPS Cluster Phosphorylation Regulates hPER2 Stability
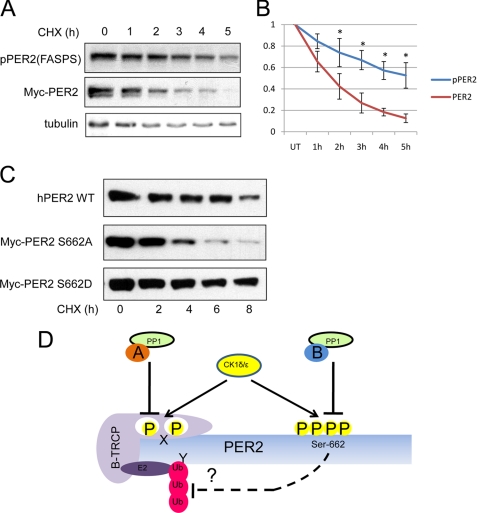
The above data suggested that phosphorylation of the FASPS cluster stabilized hPER2. To test whether this was the case, we used the protein synthesis inhibitor cycloheximide (CHX) to evaluate the half-lives of phosphorylated and total hPER2. HEK 293T-hPER2 cells were treated with CHX for the indicated lengths of time and subjected to immunoblotting analyses with the α-Myc and α-pPER2(FASPS) antibodies (Fig. 5A). Densitometric quantification of unsaturated band intensities was used to plot fractional decay of Myc-hPER2 and pPER2(FASPS) for the duration of the experiment (Fig. 5B). In agreement with our previous findings in Fig. 3, the rate of decay of the phospho-hPER2 was much slower than that for the total hPER2 (Fig. 5, A and B). Although the total protein had a half-life of ∼2 h, the phosphorylated species had a half-life of nearly 5 h. This was also observed over the course of a much longer time course (supplemental Fig. S2A), where the difference was more pronounced at later time points. A low level of phospho-hPER2 persisted throughout the 11-h duration of the experiment, whereas total Myc-PER2 was essentially undetectable by 9 h (supplemental Fig. S2A). This also illustrates that the α-pPER2(FASPS) antibody is highly sensitive and can detect Ser-662/665/668-phosphorylated PER2 proteins that are undetectable using the α-Myc antibody. To confirm a role for FASPS cluster phosphorylation in increasing PER2 stability, we transfected HEK 293T cells with PER2 mutated at the Ser-662 sites to alanine or aspartate (phosphomimetic) and subjected them to CHX treatment. In agreement with our previous findings, hPER2WT and hPER2S662D had significantly elevated half-lives in comparison with the hPER2S662A mutant (Fig. 5C). Together, these findings strongly suggest a model in which phosphorylation of the FASPS cluster stabilizes hPER2.
FIGURE 5.
FASPS site phosphorylation regulates hPER2 stability. A, increased half-life of Ser-662/665/668-phosphorylated hPER2. HEK 293T-hPER2 cells were treated with 20 μg/ml CHX for the indicated times. The cell extracts were made and subjected to immunoblotting with α-Myc, α-pPER2(FASPS), and tubulin antibodies. B, graphical representation of immunoblotting data. The data are depicted as relative mean band intensities ± S.E. An asterisk indicates p < 0.05 (n = 3). C, effects of FASPS phosphorylation cluster mutations on hPER2 stability. HEK 293T were transfected with hPER2WT, hPER2S662D, or hPER2S662A mutants and treated with 20 μg/ml CHX for the indicated times. The cell extracts were then made and subjected to immunoblotting with α-Myc, α-pPER2(FASPS), and α-CREB antibodies. D, model for hPER2 FASPS cluster phosphoregulation. CK1δ/ϵ phosphorylate both the degradation cluster (X) and the stabilizing FASPS cluster (Ser-662/665/668) on hPER2. Phosphorylation of the degradation cluster recruits the ubiquitination machinery (the ubiquitin ligase β-TrCP) and phosphorylation of FASPS cluster stabilizes the protein, perhaps by blocking Lys (Y) ubiquitination. In this model, different targeting subunits of PP1, denoted as A and B, may confer selectivity toward the X and FASPS phosphorylation clusters.
DISCUSSION
In this study we have exploited an α-pPER2(FASPS) antibody that recognizes hPER2 phosphorylated on Ser-662, Ser-665, and Ser-668—the FASPS phosphorylation cluster—to study dynamic PER2 phosphorylation and dephosphorylation. Our findings confirm several important, yet unproven, aspects of the hPER2 phosphorylation model and have provided new insights into the mechanisms of hPER2 regulation. We identify roles for CK1δ, CK1ϵ, and PP1 in the control FASPS cluster phosphorylation (Fig. 2). Although all three proteins have been implicated in the regulation of hPER2 phosphorylation, their respective contributions to site-specific modification of PER2 could not previously be ascertained (4–6). We now show that CK1δ and CK1ϵ are redundant with respect to hPER2 FASPS cluster phosphorylation and that only simultaneous inactivation of both proteins impairs phosphorylation (Fig. 2B). These data are consistent with the recent demonstration of functional overlap between the two kinases in the generation of mammalian circadian rhythms (32).
Several studies on PER2 phosphorylation have led to contradictory conclusions regarding the effects of CK-mediated phosphorylation on protein stability. Our data now provide compelling evidence that there are two distinct CK1/PP1-regulated phosphorylation clusters in PER2 that exert different influences on hPER2 stability and, possibly, function. We show that although one as yet unidentified cluster targets PER2 for phosphorylation-mediated degradation, phosphorylation of the FASPS cluster stabilizes the protein. Thus, CK1 appears to phosphorylate two different regions of hPER2 yielding opposing functional outcomes. The consequence of phosphorylation may be dictated by the distinct phosphorylation kinetics of the two clusters. Although pPER2(FASPS) levels rise almost immediately following entrainment with Dex, PER2 phosphorylation on the degradation cluster occurs only much later in the circadian cycle (Fig. 4B). The cell may exercise specificity and control on these phosphorylation events by regulating the phosphatase targeting subunits for PP1 (Fig. 5D). PP1 phosphatase targeting subunits have been shown to recruit and regulate the activities of PP1 toward its various substrates, and consistent with this idea, some PP1 subunits are regulated in a circadian fashion (5, 33, 34).
Since the identification of the PER2 FASPS mutation, two reports have put forward diverging models on the precise biochemical role of this site (18, 19). Both reports agree on a role for the mutation in reducing PER2 protein levels but differ on the precise biochemical mechanism. Vanselow et al. (19) postulated that the mutation led to defective nuclear retention of PER2, leading to its premature export and degradation. On the other hand, Xu et al. (18) suggested that PER2 phosphorylation at Ser-662 increased its own expression. Although our cell system does not allow the testing of the model proposed by Xu et al., our data argue against a role for FASPS cluster phosphorylation in regulating PER2 nuclear entry and retention. Our data instead point toward the existence of a stabilizing mechanism that is operational in both the nucleus and the cytosol. We hypothesize that FASPS cluster phosphorylation of hPER2 may in fact modulate hPER2 ubiquitination (Fig. 5D). Reduced recruitment of the PER2 ubiquitin ligase β-TrCP is one such plausible mechanism for increased stability of Ser-662/665/668-phosphorylated hPER2; however, we found that phosphorylated hPER2 retained interaction with β-TrCP in coimmunoprecipitation experiments (supplemental Fig. S2B). Among several alternative models, it is possible that FASPS cluster phosphorylation electrostatically inhibits ubiquitylation of proximal Lys residues. Further studies are required to test this model and better define the mechanisms of CK1-dependent PER2 stabilization.
An interesting next step in the study of hPER2 regulation will be to determine whether the FASPS cluster is phosphorylated in response to circadian-independent cues. Given the recently identified roles for the PER2 protein in tumor suppression (21), cardiovascular regulation (35), immune system function (36), and metabolic control (37), it is possible that this phosphorylation event may integrate other signals into the circadian oscillation network. These studies will provide much more in depth mechanistic insights into the working of the mammalian circadian clock.
Acknowledgments
We thank Dr. Gary Case (University of Wisconsin, Madison) for expert assistance with peptide synthesis. We also thank Dr. Achim Kramer, Dr. David Virshup, Dr. Wade Harper, and Dr. Vladimir Spiegelman for providing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants CA124722 (to R. S. T.) and T32ES007015 (to J. A. H.). This work was also supported by American Cancer Society Grant RSG-06-113-01 and a Shaw scientist award (to R. S. T.) from the Greater Milwaukee Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- CYC
- CYCLE
- CLK
- CLOCK
- CK
- casein kinase
- PP
- protein phosphatase
- PER
- PERIOD
- FASPS
- familial advanced sleep phase syndrome
- DBT
- DOUBLE-TIME
- OA
- okadaic acid
- Dex
- dexamethasone
- CREB
- cAMP response element-binding protein
- ATM
- ataxia-telangiectasia mutated
- CHX
- cycloheximide.
REFERENCES
- 1. Mackey S. R. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 7–19 [DOI] [PubMed] [Google Scholar]
- 2. Siepka S. M., Yoo S. H., Park J., Lee C., Takahashi J. S. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosbash M., Bradley S., Kadener S., Li Y., Luo W., Menet J. S., Nagoshi E., Palm K., Schoer R., Shang Y., Tang C. H. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 75–83 [DOI] [PubMed] [Google Scholar]
- 4. Mehra A., Baker C. L., Loros J. J., Dunlap J. C. (2009) Trends Biochem. Sci. 34, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Virshup D. M., Eide E. J., Forger D. B., Gallego M., Harnish E. V. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 413–420 [DOI] [PubMed] [Google Scholar]
- 6. Vanselow K., Kramer A. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 167–176 [DOI] [PubMed] [Google Scholar]
- 7. Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E. L., Giovanni A., Virshup D. M. (2005) Mol. Cell. Biol. 25, 2795–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shirogane T., Jin J., Ang X. L., Harper J. W. (2005) J. Biol. Chem. 280, 26863–26872 [DOI] [PubMed] [Google Scholar]
- 9. Reischl S., Vanselow K., Westermark P. O., Thierfelder N., Maier B., Herzel H., Kramer A. (2007) J. Biol. Rhythms 22, 375–386 [DOI] [PubMed] [Google Scholar]
- 10. Gallego M., Kang H., Virshup D. M. (2006) Biochem. J. 399, 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sathyanarayanan S., Zheng X., Xiao R., Sehgal A. (2004) Cell 116, 603–615 [DOI] [PubMed] [Google Scholar]
- 12. Eide E. J., Virshup D. M. (2001) Chronobiol. Int. 18, 389–398 [DOI] [PubMed] [Google Scholar]
- 13. Preuss F., Fan J. Y., Kalive M., Bao S., Schuenemann E., Bjes E. S., Price J. L. (2004) Mol. Cell. Biol. 24, 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suri V., Hall J. C., Rosbash M. (2000) J. Neurosci. 20, 7547–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ralph M. R., Menaker M. (1988) Science 241, 1225–1227 [DOI] [PubMed] [Google Scholar]
- 16. Toh K. L., Jones C. R., He Y., Eide E. J., Hinz W. A., Virshup D. M., Ptácek L. J., Fu Y. H. (2001) Science 291, 1040–1043 [DOI] [PubMed] [Google Scholar]
- 17. Xu Y., Padiath Q. S., Shapiro R. E., Jones C. R., Wu S. C., Saigoh N., Saigoh K., Ptácek L. J., Fu Y. H. (2005) Nature 434, 640–644 [DOI] [PubMed] [Google Scholar]
- 18. Xu Y., Toh K. L., Jones C. R., Shin J. Y., Fu Y. H., Ptácek L. J. (2007) Cell 128, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanselow K., Vanselow J. T., Westermark P. O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A. (2006) Genes Dev. 20, 2660–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shanware N. P., Trinh A. T., Williams L. M., Tibbetts R. S. (2007) J. Biol. Chem. 282, 6283–6291 [DOI] [PubMed] [Google Scholar]
- 21. Fu L., Pelicano H., Liu J., Huang P., Lee C. (2002) Cell 111, 41–50 [DOI] [PubMed] [Google Scholar]
- 22. Gery S., Komatsu N., Baldjyan L., Yu A., Koo D., Koeffler H. P. (2006) Mol. Cell 22, 375–382 [DOI] [PubMed] [Google Scholar]
- 23. Kondratov R. V., Antoch M. P. (2007) Trends Cell Biol. 17, 311–317 [DOI] [PubMed] [Google Scholar]
- 24. Takano A., Nagai K. (2006) Biochem. Biophys. Res. Commun. 346, 95–101 [DOI] [PubMed] [Google Scholar]
- 25. Rena G., Bain J., Elliott M., Cohen P. (2004) EMBO Rep. 5, 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brockschmidt C., Hirner H., Huber N., Eismann T., Hillenbrand A., Giamas G., Radunsky B., Ammerpohl O., Bohm B., Henne-Bruns D., Kalthoff H., Leithäuser F., Trauzold A., Knippschild U. (2008) Gut 57, 799–806 [DOI] [PubMed] [Google Scholar]
- 27. Meng Q. J., Logunova L., Maywood E. S., Gallego M., Lebiecki J., Brown T. M., Sládek M., Semikhodskii A. S., Glossop N. R., Piggins H. D., Chesham J. E., Bechtold D. A., Yoo S. H., Takahashi J. S., Virshup D. M., Boot-Handford R. P., Hastings M. H., Loudon A. S. (2008) Neuron 58, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swingle M., Ni L., Honkanen R. E. (2007) Methods Mol. Biol. 365, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albrecht U., Bordon A., Schmutz I., Ripperger J. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 95–104 [DOI] [PubMed] [Google Scholar]
- 30. Okamura H., Garcia-Rodriguez C., Martinson H., Qin J., Virshup D. M., Rao A. (2004) Mol. Cell. Biol. 24, 4184–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., Schütz G., Schibler U. (2000) Science 289, 2344–2347 [DOI] [PubMed] [Google Scholar]
- 32. Lee H., Chen R., Lee Y., Yoo S., Lee C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Connor J. H., Quan H. N., Ramaswamy N. T., Zhang L., Barik S., Zheng J., Cannon J. F., Lee E. Y., Shenolikar S. (1998) J. Biol. Chem. 273, 27716–27724 [DOI] [PubMed] [Google Scholar]
- 34. Udho E., Tedesco V. C., Zygmunt A., Krucher N. A. (2002) Biochem. Biophys. Res. Commun. 297, 463–467 [DOI] [PubMed] [Google Scholar]
- 35. Martino T. A., Tata N., Belsham D. D., Chalmers J., Straume M., Lee P., Pribiag H., Khaper N., Liu P. P., Dawood F., Backx P. H., Ralph M. R., Sole M. J. (2007) Hypertension 49, 1104–1113 [DOI] [PubMed] [Google Scholar]
- 36. Arjona A., Sarkar D. K. (2006) J. Interferon Cytokine Res. 26, 645–649 [DOI] [PubMed] [Google Scholar]
- 37. Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K., Chesham J., Odell M., Lilley K. S., Kyriacou C. P., Hastings M. H. (2006) Curr. Biol. 16, 1107–1115 [DOI] [PubMed] [Google Scholar]