Abstract
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) plays an important role in DNA double-strand break (DSB) repair as the underlying mechanism of the non-homologous end joining pathway. When DSBs occur, DNA-PKcs is rapidly phosphorylated at both the Thr-2609 and Ser-2056 residues, and such phosphorylations are critical for DSB repair. In this study we report that, in addition to responding to DSBs, DNA-PKcs is activated and phosphorylated in normal cell cycle progression through mitosis. Mitotic induction of DNA-PKcs phosphorylation is closely associated with the spindle apparatus at centrosomes and kinetochores. Furthermore, depletion of DNA-PKcs protein levels or inhibition of DNA-PKcs kinase activity results in the delay of mitotic transition because of chromosome misalignment. These results demonstrate for the first time that DNA-PKcs, in addition to its role in DSB repair, is a critical regulator of mitosis and could modulate microtubule dynamics in chromosome segregation.
Keywords: Cell Cycle, Chromosomes, DNA Damage, Microtubules, Mitosis, Spindle Pole Body
Introduction
DNA-PKcs,3 the catalytic subunit of DNA-dependent protein kinase (DNA-PK), is known to play an essential role in non-homologous end joining-mediated DNA double-strand break (DSB) repair in mammalian cells. In response to DSBs, the Ku70/80 subunits of DNA-PK immediately bind to the broken DSB ends and subsequently recruit and activate DNA-PKcs kinase activity (1). The intrinsic kinase activity of DNA-PKcs is essential for its role in DSB repair (2), likely through phosphorylation and regulation of non-homologous end joining components, including DNA-PKcs itself. DNA-PKcs is rapidly autophosphorylated in vitro upon activation and is also phosphorylated in vivo after exposure to ionizing radiation (IR). Among all the phosphorylation sites identified (3–6), phosphorylation in vivo was clearly detected at the Ser-2056 residue and at the Thr-2609 cluster region (3, 7, 8). Whereas IR-induced DNA-PKcs phosphorylation at Ser-2056 is clearly mediated by the autophosphorylation of DNA-PKcs (7), IR-induced DNA-PKcs phosphorylation at the Thr-2609 cluster is mainly dependent on ataxia telangiectasia mutated (ATM) kinase but not DNA-PKcs itself (8). In addition, the Thr-2609 cluster region can be phosphorylated by ATR kinase in response to UV irradiation or replication stresses (9). Although the precise mechanism of DNA-PKcs phosphorylation remains to be clarified, like its kinase activity, DNA-PKcs phosphorylation at Ser-2056 and the Thr-2609 cluster are required for DSB repair.
In addition to its roles in DNA damage response, DNA-PKcs-mediated DSB repair is required for V(D)J recombination during T- and B-cell maturation (10). Recently, it has also been reported that DNA-PKcs phosphorylates and activates the upstream stimulatory factor, which in turn regulates the expression of fatty acid synthase in response to feeding (11). In contrast to the well studied role of DNA-PKcs in DSB repair, not much is known about the involvement of DNA-PKcs kinase and its phosphorylations in other cellular activities, particularly under normal conditions. While analyzing DNA-PKcs phosphorylation in response to IR, we observed that a small fraction of non-irradiated cells displayed positive staining with DNA-PKcs phosphorylation at the Thr-2609 cluster region. In this report, we present evidence that DNA-PKcs is activated during mitosis and is both physically and functionally associated with the mitotic spindle apparatus. As a result of this evidence, DNA-PKcs may be recognized for the first time as an important regulator in mitosis, one that is critical for mitotic spindle function in chromosome segregation.
EXPERIMENTAL PROCEDURES
Cell Culture, Synchrony, and siRNA Transfection
All cell cultures, including human cervical cancer HeLa cells, normal human skin fibroblasts (HSF), and human HCT116 DNA-PKcs−/− cells (12) were maintained in α-minimum Eagle's medium supplemented with 10% fetal bovine serum. HCT116 DNA-PKcs−/− cells were also complemented with a full-length DNA-PKcs cDNA, and the expression of DNA-PKcs was confirmed by Western blotting. A stable HeLa cell line expressing EGFP-α-tubulin was generated after transfection of pEGFP-Tub (Clontech) and was maintained in α-minimum Eagle's medium containing 200 μg/ml G418. A stable HeLa cell line expressing GFP-H2B was a kind gift from Dr. Hongtau Yu (13). HeLa cell synchronization was carried out by double thymidine blockage at the G1/S boundary and released in culture medium containing 50 ng/ml nocodazole for subsequent cell cycle arrest at mitosis (14). Small interfering RNA (siRNA) oligonucleotides designed against DNA-PKcs (15), ATM, and ATR (9) were transfected with RNAiMax (Invitrogen) as described (9).
Immunoblotting and Antibodies
Whole cell lysate preparation and Western blotting were performed as described (3, 7). For immunofluorescent (IF) staining, cells were fixed in 4% paraformaldehyde for 10 min, permeabilized in 0.5% Triton X-100 for 10 min, and blocked in 5% normal goat serum or bovine serum albumin for 1 h at room temperature. The cells were incubated with primary antibodies for 1 h, washed three times in PBS, and then incubated with Alexa Fluor 568- and Alexa Fluor 488-conjugated secondary antibodies for 30 min (Molecular Probes). Cells were then washed three times in PBS and mounted in Vectashield mounting medium with 4,6-diamidino-2-phenylindole (Vector Laboratories). Phospho-specific anti-DNA-PKcs antibodies were described previously (3, 7, 8). Anti-DNA-PKcs mouse monoclonal antibody (NeoMarkers), anti-phospho Histone H3, anti-cyclin A (Upstate), anti-ATR, anti-ATM, anti-PLK (Bethyl Laboratories), anti-α-tubulin, anti-γ-tubulin mAb (Sigma) were commercially available from the indicated vendors. Anti-Bub1 was a kind gift from Dr. Hongtau Yu.
Flow Cytometry Analysis
Flow cytometry analysis was performed as described before (16). In brief, cells were harvested and fixed in 70% ethanol. Fixed cells were washed with PBS three times and resuspended in propadium iodide solution (0.1 mg/ml RNase A, 0.1% Triton X-100, and 20 mg/ml PI in PBS). After incubation of the PI solution at 4 °C for 30 min in darkness, DNA content was measured by a FS500 flow cytometer, and cell cycle compartments were analyzed using the CXP cytometry analysis program (Beckman Coulter).
Mitotic Index
Cells were harvested by both shake-off and trypsin digestion in sequence. Harvested cells were swelled in a hypotonic solution containing 75 mm KCl for 15 min at 37 °C. Cells were fixed by dropwise addition of fixative (3:1 solution of methanol:glacial acetic acid). To make a mitotic spread, fixed cells were dropped onto prechilled slides. To visualize mitotic cells, dried slides were stained with Giemsa stain solution (Sigma). The mitotic index was generated after counting over 500 cells in each slide.
Time-lapse Live Cell Imaging
For live cell imaging, GFP-H2B and EGFP-Tub cells were cultured in CO2-independent medium (Invitrogen) with 10% FBS and were maintained at 37 °C in a temperature-controlled container in glass-bottom dishes (MatTek). Time-lapse images of GFP-H2B cells were acquired at 10-min intervals from an Axiovert 200 m microscope (Carl Zeiss) using a Plan-Apochromat 40×/NA 1.40 objective with an AxioCam HRm CCD camera. For the metaphase spindle repolymerization assay (17), GFP-Tub cells were treated with 10 μm NU7441b or dimethyl sulfoxide as mock treatment for 2 h. Nocodazole (100 ng/ml) was added into Nu7441-treated GFP-Tub cells for 15 min at 37 °C, given three quick washes with prewarmed PBS in 37 °C, and reintroduced into prewarmed fresh medium. Images were acquired at 5-min intervals from a LSM 510 Meta confocal microscope (Carl Zeiss) using a 1.4 NA plan-Apo 63× oil immersion objective.
RESULTS
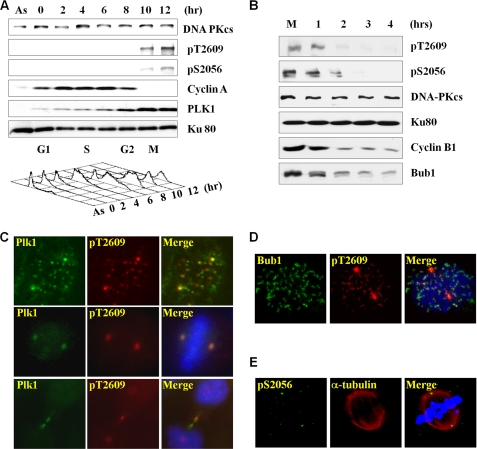
DNA-PKcs phosphorylations at both Thr-2609 and Ser-2056 are rapidly elicited upon DNA damage, particularly after IR. These phosphorylation events are critical for the function of DNA-PKcs in DNA DSB repair and radioresistance (3, 7, 8, 18). In addition, we found that DNA-PKcs phosphorylation occurs in a small fraction of asynchronous and non-irradiated cell populations. To determine whether the spontaneous induction of DNA-PKcs phosphorylation occurs at a specific cell cycle phase, HeLa cells were synchronized with a double-thymidine block at the G1/S boundary and were released into culture medium containing nocodazole for a subsequent cell cycle arrest at mitosis. Upon release, HeLa cells reenter the cell cycle immediately and reach mitosis 8–10 h afterward (Fig. 1A). Cell cycle progression was confirmed by Western blotting, showing the sharp decrease in cyclin-A protein levels (19, 20) and the increase of Polo-like kinase 1 (PLK1) expression in mitosis (21, 22). DNA-PKcs phosphorylations at the Thr-2609 and Ser-2056 residues were undetected in asynchronous HeLa cells and synchronized HeLa cells at the G1/S boundary and the S and G2 phases. Upon entering mitosis, DNA-PKcs phosphorylations at both residues increased drastically without changing the steady-state protein levels of DNA-PKcs (Fig. 1A). In addition, mitotic induction of DNA-PKcs phosphorylations were found in normal human skin fibroblasts (supplemental Fig. 1A) as well as in human colon cancer HCT116 cells (supplemental Fig. 1B), suggesting that spontaneous DNA-PKcs phosphorylation in mitosis is a general phenomenon. Furthermore, DNA-PKcs phosphorylations were detected in the HCT116 cells but not in DNA-PKcs−/− cells (12), thus demonstrating the specificity of DNA-PKcs phosphorylation detection in the Western blot analysis (supplemental Fig. 1B).
FIGURE 1.
Mitotic induction of DNA-PKcs phosphorylations. A, thymidine-synchronized HeLa cells were released into normal medium containing nocodazole (50 ng/ml) and were harvested at different time points for Western blotting and FACS. Asynchronous HeLa cells (As) were also processed. B, mitotically arrested HeLa cells were released for cell cycle reentry and were harvested at the indicated time points for Western blotting. C, asynchronous and exponentially growing HeLa cells were IF stained with the indicated antibodies. Mitosis-specific DNA-PKcs Thr-2609 phosphorylation overlapped with PLK1 at centrosomes and kinetochores in prometaphase and at the midbody during cytokinesis. D, DNA-PKcs Thr-2609 phosphorylation overlapped with Bub1 at prometaphase kinetochores. E, mitosis-specific DNA-PKcs Ser-2056 phosphorylation was detected at prometaphase centrosomes.
The induction of mitosis-specific DNA-PKcs phosphorylations was further examined in HeLa cells upon mitotic exit. Nocodazole-arrested HeLa cells were released for cell cycle reentry and were harvested at different time points afterward. Upon mitotic exit, as evidenced from the decrease in protein levels of Cyclin-B1 and Bub1 (23, 24), we observed a rapid decrease of DNA-PKcs phosphorylations at both Thr-2609 and Ser-2056 (Fig. 1B). This result thus demonstrated a temporary induction pattern of DNA-PKcs phosphorylation in mitosis.
To determine the spatial distribution of DNA-PKcs phosphorylation during mitosis, exponentially growing HeLa cells were subjected to IF staining. As shown in Fig. 1C, DNA-PKcs Thr-2609 phosphorylation occurred at both centrosomes and kinetochores during prometaphase and was completely overlapped with PLK1 staining. A similar overlapping pattern of Thr-2609 phosphorylation and PLK1 was also detected in the HCT116 cells (supplemental 1D). The presence of Thr-2609 phosphorylation at the prometaphase kinetochores was further demonstrated by the overlapping pattern of pT2609 with spindle checkpoint kinase Bub1 (24) and anti-kinetochore CREST antibody staining (25). Both Bub1 (Fig. 1D) and CREST (supplemental Fig. 2A) staining appeared only at kinetochores but not at centrosomes. On the other hand, the overlapping between DNA-PKcs Thr-2609 phosphorylation and PLK1 could be detected in prometaphase as well as in metaphase and cytokinesis. DNA-PKcs Thr-2609 phosphorylation and PLK1 appeared primarily at centrosomes during metaphase and was significantly reduced or undetectable at kinetochores. Similar to the pattern of Thr-2609 phosphorylation, DNA-PKcs phosphorylation at Ser-2056 could be detected at the centrosomes during metaphase (Fig. 1E). During cytokinesis, both DNA-PKcs Thr-2609 phosphorylation and PLK1 appeared at centrosomes as well as at the midbody structure (Fig. 1C). At this final phase of mitosis, Bub1 staining decreased to the background level because of proteasome-mediated degradation, whereas CREST staining remained at kinetochores (supplemental Fig. 2B).
We have previously reported that DNA-PKcs kinase autophosphorylates itself at Ser-2056 in response to IR (7), whereas ATM and ATR kinases are required for DNA-PKcs phosphorylation at Thr-2609 (8, 9). To determine whether ATM is involved in mitotic DNA-PKcs phosphorylation, ATM-proficient 1BR3 and deficient AT5 human fibroblasts were arrested in mitosis and were analyzed for DNA-PKcs phosphorylation. As shown in Fig. 2A, mitotic induction of DNA-PKcs Ser-2056 and Thr-2609 phosphorylations were clearly detected in both cell lines without a significant difference. Furthermore, depletion of either ATM or ATR protein levels with siRNA did not affect mitotic DNA-PKcs phosphorylation (Fig. 2B), suggesting that autophosphorylation of DNA-PKcs might be responsible for Ser-2056 and Thr-2609 phosphorylations in mitosis.
FIGURE 2.
DNA-PKcs autophosphorylation mediates mitotic induction of DNA-PKcs phosphorylations. A, ATM-deficient human AT5 cells and the control 1BR3 cells were treated with nocodazole (50 ng/ml for 16 h). Asynchronous (As) and mitotically arrested (M) cells were subjected to Western blot analysis. B, HeLa cells were transfected with small interfering RNA against ATM or ATR for 48 h, followed by nocodazole treatment. Asynchronous and mitotically arrested cells were analyzed. C, thymidine-synchronized HeLa cells were released into normal medium. At 6 h (late S/G2), cells were mock-treated or treated with 10 μm Nu7441 or Ku55933 for an additional 2 h. Cells harvested at 6 h and 8 h were subjected to FASC and Western blotting analyses.
To test this hypothesis, HeLa cells were synchronized with a double thymidine block and released for cell cycle reentry. At 6 h, when the majority cell population remained in late S phase, HeLa cells were treated with DNA-PKcs inhibitor Nu7441 or ATM inhibitor Ku55933 for an additional 2 h. As shown in Fig. 2C, Ser-2056 and Thr-2609 phosphorylations were both induced at 8 h in mock- and Ku55933-treated cells. In contrast, treatment with Nu7441 attenuated both Ser-2056 and Thr-2609 phosphorylations effectively. Such inhibition was not due to an alteration in cell cycle progression, as neither Nu7441 nor Ku55933 blocked mitosis entry.
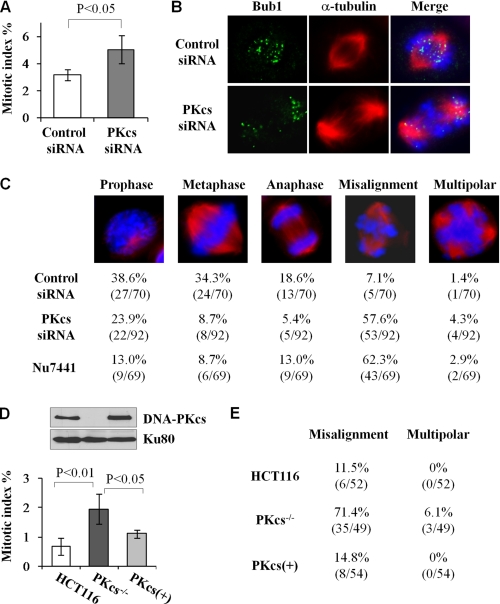
The induction of mitotic DNA-PKcs phosphorylations and their association with the spindle apparatus strongly indicate a critical function of DNA-PKcs in mitosis. Consistent with this notion, an increase of the mitotic index was detected upon siRNA depletion of DNA-PKcs in HeLa cells (Fig. 3A) and in DNA-PKcs-deficient human M059J glioma cells (supplemental Fig. 3B) in contrast with the control cells. In addition, we observed that depletion of DNA-PKcs resulted in chromosome misalignment (Fig. 3B), a possible cause and explanation for the increase of the mitotic index in the absence of DNA-PKcs. In the DNA-PKcs-depleted cell population, more than half of mitotic cells (57%) displayed misaligned chromosomes as compared with 7% in the control siRNA-transfected cells (Fig. 3C). The induction of chromosome misalignment was also found in HeLa cells treated with DNA-PKcs inhibitor Nu7441 (Fig. 3C) and in HCT116 DNA-PKcs−/− cells (E). Furthermore, complementation with wild-type DNA-PKcs rescued the mitotic defects in HCT116 DNA-PKcs−/− cells (Fig. 3, D and E). This rules out the possible off-target effect from siRNA depletion of DNA-PKcs and provides strong evidence for the function of DNA-PKcs in mitosis.
FIGURE 3.
DNA-PKcs is required for normal mitosis progression. A, HeLa cells were transfected with control siRNA or DNA-PKcs siRNA for 48 h and stained with Hoechst 33258 to determine the percentage of mitosis cell population. The results were generated from four independent experiments, and more than 500 cells were counted for each experiment. B, the same HeLa cells were analyzed by IF staining with the indicated antibodies. C, HeLa cells transfected with siRNA or treated with Nu7441 (10 μm for 16 h) were IF stained with anti-α-tubulin antibody. The percentages of each mitotic stage among mitotic cells were determined with a given phenotype. D, human HCT116, HCT116 DNA-PKcs−/−, and DNA-PKcs−/− cells complemented with DNA-PKcs (PKcs+) were analyzed for the mitotic index. The results were generated from four independent experiments. The upper panel shows the protein levels of DNA-PKcs. E, the same HCT116 cell lines were IF stained and analyzed for chromosome misalignment in mitosis.
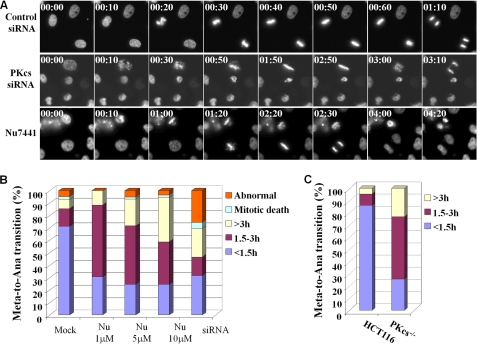
To further investigate the role of DNA-PKcs in mitotic transition, HeLa cells expressing the GFP-H2B fusion protein were subjected to live cell imaging to capture chromosome condensation and segregation during mitosis. The live cell imaging analysis revealed that upon siRNA depletion or kinase inhibition of DNA-PKcs a large portion of mitotic cells displayed misaligned chromosomes and prolonged mitotic transition (Fig. 4A). The time duration from initial chromosome condensation (prometaphase) to chromosome segregation (anaphase) was completed within 1.5 h in the majority of control GFP-H2B cells, and only a small fraction of mitotic cells displayed abnormal spindle organization or died during mitosis (Fig. 4B). Upon DNA-PKcs siRNA transfection at 48 h, the prometaphase-to-anaphase transition was significantly prolonged together with an increase of abnormal spindle structure (Fig. 4B). Treatment with DNA-PKcs kinase inhibitor Nu7441 also led to a delay in the prometaphase-to-anaphase transition in a dose-dependent manner. However, no clear increase in abnormal spindle or mitotic cell death was found in the Nu7441-treated cell population. The prometaphase-to-anaphase transition was further analyzed in the HCT116 and DNA-PKcs−/− cells expressing the GFP-H2B fusion protein. A similar delay in mitotic transition was also found in DNA-PKcs−/− cells as compared with the control HCT116 cells (Fig. 4C).
FIGURE 4.
DNA-PKcs is required for proper chromosome segregation during mitosis. A, HeLa GFP-H2B cells were transfected with control siRNA or DNA-PKcs siRNA or treated with DNA-PKcs kinase inhibitor Nu7441 (10 μm) prior to live cell image recording. Live cell images were recorded at 10-min intervals for 12–16 h (supplemental movies 1–3). Representative images of the prometaphase-to-anaphase transition are shown 2 days after siRNA transfection or treatment with Nu7441. B, the durations of the prometaphase-to-anaphase transitions were determined from >100 mitotic cells of each experimental group. C, the GFP-H2B fusion protein was stably expressed in HCT116 and DNA-PKcs−/− cells. The durations of prometaphase-to-anaphase transitions were determined from >50 mitotic cells in each cell line.
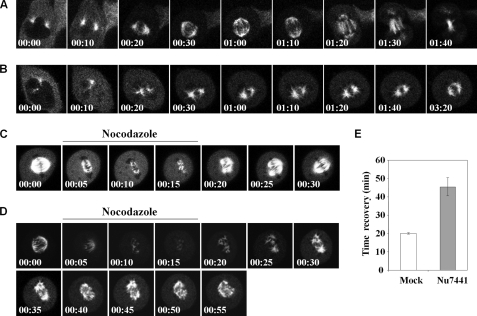
The dysfunction in chromosome alignment upon DNA-PKcs inhibition revealed that DNA-PKcs could influence microtubule dynamics and the formation of the spindle apparatus. To investigate the role of DNA-PKcs in modulating mitotic spindle formation, HeLa cells expressing the EGFP-α-tubulin fusion protein were subjected to live cell imaging analysis. EGFP-α-tubulin cells in early mitosis, with microtubule fibers projecting from the newly separated centrosomes, were identified for continuous live cell image recording in a confocal microscope setup. In a control group of siRNA-transfected cells, 9 of 10 cells analyzed were able to progress through mitosis within 3 h without any apparent abnormality in spindle formation (Fig. 5A). Upon DNA-PKcs siRNA transfection, microtubule growth and spindle formation were significantly attenuated. In 10 mitotic cells monitored, only two cells were able to complete the mitosis. The remaining cells were either unable to assemble the spindle structure or were unable to complete the mitosis within 200 min (Fig. 5B). In addition, EGFP-α-tubulin cells in metaphase were treated with nocodazole to destabilize the microtubule fibers and spindle apparatus. Upon nocodazole removal, the microtubule fibers recovered immediately and reassembled into the spindle apparatus (Fig. 5C). It took an average of 20 min for spindle reassembly in mock-treated EGFP-α-tubulin cells (Fig. 5E). In contrast, treatment with Nu7441 significantly attenuated microtubule repolymerization (Fig. 5, D and E), and more than half of the Nu7441-treated cells displayed dysfunction in spindle formation (data not shown). Taken together, our results demonstrate that DNA-PKcs can modulate microtubule dynamics in vivo and that DNA-PKcs kinase activity is involved in such regulation.
FIGURE 5.
DNA-PKcs and its kinase activity affect microtubule dynamics in vivo. HeLa cells expressing the EGFP-α-tubulin fusion protein were transfected with control siRNA (A) or DNA-PKcs siRNA (B) for 48 h and were subjected to live cell image recording at 10-min intervals for the indicated time (hr:min). C, EGFP-α-tubulin cells with a visible spindle apparatus were pulsed with nocodazole (100 ng/ml) for 15 min to monitor mitotic spindle disassembly and reassembly. Dimethyl sulfoxide was used as a mock control treatment. D, the same experiment was carried out in the presence of Nu7441 (10uM). E, the average time of spindle recovery after nocodazole withdrawal. Ten mitotic cells were analyzed for each treatment.
DISCUSSION
In this study, we report for the first time that DNA-PKcs, a well established DNA DSB repair factor, plays an important role during normal cell cycle progression through mitosis. Our results demonstrate that DNA-PKcs phosphorylations at both Thr-2609 and Ser-2056 are spontaneously and transiently elicited during mitosis. Furthermore, mitotically induced DNA-PKcs phosphorylations are associated with the spindle apparatus, specifically at centrosomes (the microtubule organization centers) and kinetochores (where microtubule fibers attach to chromosomes). The temporal and spatial patterns of mitotic DNA-PKcs phosphorylations suggest that DNA-PKcs is functionally involved in the spindle apparatus. Indeed, depleting DNA-PKcs protein levels with siRNA or inhibiting DNA-PKcs kinase activity with small molecule inhibitor Nu7441 resulted in delay and dysfunction in mitotic transition, including abnormal spindle formation and chromosome misalignment. Consequently, the possibility of chromosome segregation infidelity or uneven chromosome segregation during mitosis will lead to aneuploidy, the loss or gain of a whole chromosome, which is the most common chromosomal instability associated with human cancers (26). In agreement with this assertion, the DNA-PKcs-deficient M059J cell line was first derived from a malignant human glioma autopsy sample (27). It is plausible that the lack of a functional DNA-PKcs in human cells will cause chromosomal instability and induce tumorigenesis.
It was demonstrated previously that the kinase activity of DNA-PKcs is essential for DSB repair in response to IR (2). Likewise, DNA-PKcs kinase activity is also required for its critical function during mitosis, as DNA-PKcs kinase inhibitor Nu7441 confers chromosome misalignment, prolonged prometaphase-to-anaphase transition, and abnormal spindle formation (Figs. 3–5). The necessity of DNA-PKcs kinase activity in mitotic transition is due in part to DNA-PKcs autophosphorylation, as treatment with Nu7441 attenuated mitotic Ser-2056 phosphorylation, which is a genuine autophosphorylation event of DNA-PKcs (7). In addition, Nu7441 but not the ATM kinase inhibitor Ku55933 attenuated mitotic DNA-PKcs phosphorylation at Thr-2609. We have reported previously that DNA-PKcs Thr-2609 phosphorylation is mediated by ATM and ATR kinases in response to IR and UV-induced replication stress, respectively (8, 9). The result in Fig. 2C thus suggests a preferential regulation of Thr-2609 phosphorylation by DNA-PKcs itself during mitosis. In agreement, siRNA depletion of either ATM or ATR did not affect mitotic Thr-2609 phosphorylation (Fig. 2B), although we do not rule out the possibility that additional mitotic kinases could activate or phosphorylate DNA-PKcs directly at Thr-2609.
The requirement of DNA-PKcs activity in mitosis is closely associated with microtubule dynamics and the development of the spindle apparatus. These results also imply that DNA-PKcs is involved in modulating centrosome activity. Indeed, IF staining analyses revealed that DNA-PKcs phosphorylations occur at centrosomes throughout different mitotic phases. In addition, our sucrose gradient analysis also revealed that Thr-2609 phosphorylated DNA-PKcs is co-fractionated with the critical centrosomal component γ-tubulin (supplemental Fig. 1C). Consistently, it has been reported the presence of DNA-PKcs staining in centrosomes (29) and identification of the Ku70/80 heterodimer from a proteomic profiling of the human centrosome (28). Furthermore, Shang et al. (30) recently reported that inactivation of DNA-PKcs resulted in spindle disruption and centrosome instability.
The presence of DNA-PKcs at the spindle apparatus is consistent with the function of DNA-PKcs in spindle formation and/or microtubule attachment to chromosomes, both of which are critical for proper chromosome alignment during metaphase. Indeed, attenuation of DNA-PKcs activity via siRNA or kinase inhibitor resulted in a higher incidence of chromosome misalignment and consequently caused a delay in mitotic transition (Figs. 3 and 4). The current model suggests that kinetochores are initially captured by microtubules that extend from the opposite spindle poles and pull the kinetochores toward them. Microtubule fibers extending from the secondary spindle pole subsequently reach to the unattached sister kinetochores to achieve a bi-orientation (31). It is plausible that DNA-PKcs is required for the long stretch of microtubules originating from the opposite pole to catch the unattached kinetochores, as we observed that the projection of microtubule fibers is attenuated in the absence of functional DNA-PKcs (Fig. 5). Taken together, our results reveal that the catalytic/kinase activity of DNA-PKcs is required for chromosome segregation and normal cell cycle progression through mitosis. In addition, our results also demonstrate that DNA-PKcs is a versatile regulator of genomic integrity, not only in DSB repair but also in chromosomal stability maintenance.
Acknowledgment
We thank Dee Hill for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA50519. This work was also supported by National Aeronautics and Space Administration Grant NNX07AP84G and National Science Council, Taiwan Grant NSC96-2321-B002-008.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and movies 1–3.
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- DSB
- double-strand break
- IR
- ionizing radiation
- ATM
- ataxia telangiectasia mutated
- EGFP
- enhanced green fluorescent protein
- IF
- immunofluorescent
- PLK1
- Polo-like kinase 1
- ATR
- ataxia telengiectasia and rad3 related.
REFERENCES
- 1. Smith G. C., Jackson S. P. (1999) Genes Dev. 13, 916–934 [DOI] [PubMed] [Google Scholar]
- 2. Kurimasa A., Kumano S., Boubnov N. V., Story M. D., Tung C. S., Peterson S. R., Chen D. J. (1999) Mol. Cell Biol. 19, 3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Douglas P., Sapkota G. P., Morrice N., Yu Y., Goodarzi A. A., Merkle D., Meek K., Alessi D. R., Lees-Miller S. P. (2002) Biochem. J. 368, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M. R. (2005) J. Biol. Chem. 280, 33839–33846 [DOI] [PubMed] [Google Scholar]
- 6. Douglas P., Cui X., Block W. D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S. P., Meek K. (2007) Mol. Cell Biol. 27, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen B. P., Chan D. W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D. J. (2005) J. Biol. Chem. 280, 14709–14715 [DOI] [PubMed] [Google Scholar]
- 8. Chen B. P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Löbrich M., Shiloh Y., Chen D. J. (2007) J. Biol. Chem. 282, 6582–6587 [DOI] [PubMed] [Google Scholar]
- 9. Yajima H., Lee K. J., Chen B. P. (2006) Mol. Cell Biol. 26, 7520–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gellert M. (2002) Annu. Rev. Biochem. 71, 101–132 [DOI] [PubMed] [Google Scholar]
- 11. Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruis B. L., Fattah K. R., Hendrickson E. A. (2008) Mol. Cell Biol. 28, 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Z., Shu H., Oncel D., Chen S., Yu H. (2004) Mol. Cell 16, 387–397 [DOI] [PubMed] [Google Scholar]
- 14. Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. (2002) Nat. Cell Biol. 4, 358–366 [DOI] [PubMed] [Google Scholar]
- 15. Peng Y., Zhang Q., Nagasawa H., Okayasu R., Liber H. L., Bedford J. S. (2002) Cancer Res. 62, 6400–6404 [PubMed] [Google Scholar]
- 16. Yajima H., Lee K. J., Zhang S., Kobayashi J., Chen B. P. (2009) J. Mol. Biol. 385, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong J., Fang G. (2006) J. Cell Biol. 173, 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding Q., Reddy Y. V., Wang W., Woods T., Douglas P., Ramsden D. A., Lees-Miller S. P., Meek K. (2003) Mol. Cell Biol. 23, 5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geley S., Kramer E., Gieffers C., Gannon J., Peters J. M., Hunt T. (2001) J. Cell Biol. 153, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. den Elzen N., Pines J. (2001) J. Cell Biol. 153, 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Golsteyn R. M., Mundt K. E., Fry A. M., Nigg E. A. (1995) J. Cell Biol. 129, 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee K. S., Yuan Y. L., Kuriyama R., Erikson R. L. (1995) Mol. Cell Biol. 15, 7143–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu H., King R. W., Peters J. M., Kirschner M. W. (1996) Curr. Biol. 6, 455–466 [DOI] [PubMed] [Google Scholar]
- 24. Taylor S. S., McKeon F. (1997) Cell 89, 727–735 [DOI] [PubMed] [Google Scholar]
- 25. Moroi Y., Peebles C., Fritzler M. J., Steigerwald J., Tan E. M. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1627–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lengauer C., Kinzler K. W., Vogelstein B. (1998) Nature 396, 643–649 [DOI] [PubMed] [Google Scholar]
- 27. Lees-Miller S. P., Godbout R., Chan D. W., Weinfeld M., Day R. S., 3rd, Barron G. M., Allalunis-Turner J. (1995) Science 267, 1183–1185 [DOI] [PubMed] [Google Scholar]
- 28. Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 29. Zhang S., Hemmerich P., Grosse F. (2007) J. Cell. Biochem. 101, 451–465 [DOI] [PubMed] [Google Scholar]
- 30. Shang Z. F., Huang B., Xu Q. Z., Zhang S. M., Fan R., Liu X. D., Wang Y., Zhou P. K. (2010) Cancer Res. 70, 3657–3666 [DOI] [PubMed] [Google Scholar]
- 31. Tanaka T. U., Stark M. J., Tanaka K. (2005) Nat. Rev. Mol. Cell Biol. 6, 929–942 [DOI] [PubMed] [Google Scholar]