Abstract
G protein-coupled receptor kinases (GRKs) phosphorylate activated G protein-coupled receptors (GPCRs) to initiate receptor desensitization. In addition to the canonical phosphoacceptor site of the kinase domain, activated receptors bind to a distinct docking site that confers higher affinity and activates GRKs allosterically. Recent mutagenesis and structural studies support a model in which receptor docking activates a GRK by stabilizing the interaction of its ~20-amino acid N-terminal region with the kinase domain. This interaction in turn stabilizes a closed, more active conformation of the enzyme. To investigate the importance of this interaction for the process of GRK activation, we first validated the functionality of the N-terminal region in rhodopsin kinase (GRK1) by site-directed mutagenesis and then introduced a disulfide bond to cross-link the N-terminal region of GRK1 with its specific binding site on the kinase domain. Characterization of the kinetic and biophysical properties of the cross-linked protein showed that disulfide bond formation greatly enhances the catalytic efficiency of the peptide phosphorylation, but receptor-dependent phosphorylation, Meta II stabilization, and inhibition of transducin activation were unaffected. These data indicate that the interaction of the N-terminal region with the kinase domain is important for GRK activation but does not dictate the affinity of GRKs for activated receptors.
G protein-coupled receptor kinases (GRKs)1 are serine/threonine protein kinases involved in the desensitization of GPCR signaling (1, 2). Similar to heterotrimeric G proteins and arrestins, they preferentially recognize activated GPCRs (3). Upon activation, a conformational change in the GPCR exposes a binding surface for GRKs, permitting the phosphorylation of the C-terminal tail and/or intracellular loop 3 (IL3) of the receptor. Although a detailed understanding of the molecular interactions between activated GPCRs and GRKs is lacking, it is expected that GPCRs bind to a docking site distinct from the active site that allosterically activates GRKs (Scheme 1). Evidence supporting this mechanism includes the much higher catalytic efficiency of GRKs for receptors than peptides derived from native receptors (4–7), and the enhancement of peptide phosphorylation in the presence of activated receptors (4, 8, 9).
Scheme 1.
Phosphorylation of GPCRs by GRKs, Using GRK1 and Rho as an Example
The seven GRKs in mammals contain a highly conserved N-terminal region of ~20 amino acids, followed by a regulator of G protein signaling (RGS) homology (RH) domain, into which is inserted a central protein kinase domain closely related to that of protein kinase A (PKA) (Figure 1A) (10). This kinase domain includes a C-terminal extension known as the C-tail, which plays a regulatory role in other AGC kinases (11, 12). The extreme C-terminal region is the most diversified region in GRKs and consists of structural elements that are important for membrane association and, in some cases, regulation of activity (1). As in other protein kinases, the active site is located between the two lobes of the kinase domain (the N- and C-lobes). However, the mechanism of allosteric activation of GRKs induced by receptor docking is poorly understood (13).
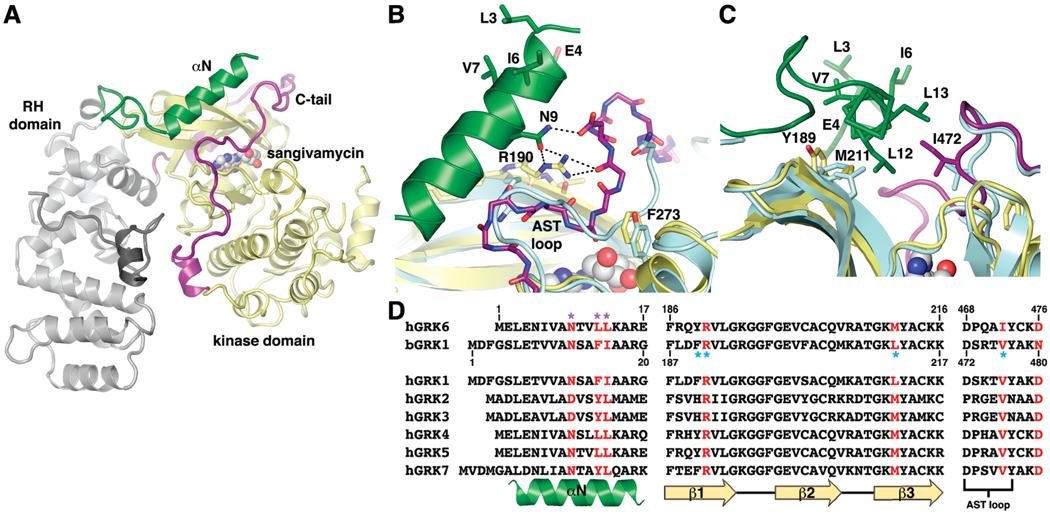
FIGURE 1.
Key interactions between αN and the kinase domain observed in GRK6 are conserved in GRK1. (A) Overall structure of GRK6 in the closed conformation (PDB entry 3NYN). The RH domain and C-terminal region, the kinase domain, αN, and the C-tail are colored gray, yellow, green, and purple, respectively. The nucleoside analogue sangivamycin bound in the active site is shown as a sphere model. (B) Hydrogen bonds formed among Arg190 of the kinase N-lobe, Asn9 of αN, and the C-tail of GRK6. The structure of GRK1 in a complex with ATP (PDB entry 3C4Z, light blue) was superimposed with GRK6 using the N-lobe. The AST loop (residues 472–477) of GRK1 adopts a conformation similar to that in GRK6. Phe273 of αD on the kinase large lobe of GRK6 interacts with the backbone of the C-tail, as does the analogous Tyr274 of GRK1. (C) van der Waals interactions formed between αN and the kinase domain of GRK6, including Leu12 and Leu13 of αN, Ile472 of the C-tail, and Met211 and Tyr189 of the kinases mall lobe. Corresponding positions in GRK1, including Val476, Leu212, and Phe190, are colored light blue. (D) Residues of GRKs involved in key interactions between αN and the kinase domain, and between the small lobe and the C-tail, are highly conserved. Conserved contact residues in GRK6 are colored red, and residues previously identified as being important for kinase activity in GRK1 and GRK6 are labeled with purple and blue asterisks, respectively. Note that bovine GRK1 is used in this study.
Current biochemical and structural data support a model in which the N-terminal region of GRKs and the kinase domain interact to activate the kinase (14, 15). In recent crystal structures of GRK6, the kinase domain adopts a conformation that closely resembles the active state of other characterized protein kinases (15), and the extreme N-terminal region forms a single α-helix (αN) that packs between the amino-terminal domain (N-lobe) and the C-tail of the kinase domain. Important interactions between αN and the kinase domain include a hydrogen bond network formed byArg190 on β1 of the N-lobe, Asn9 of αN, and backbone atoms of the C-tail (Figure 1B), and extensive van der Waals interactions involving residues from αN, the kinase N-lobe, and the C-tail, including Leu12, Leu13, Tyr189, Met211, and Ile472 (Figure 1C). All of these residues are highly conserved in GRKs (Figure 1D) and are among those that have been shown to be important for the kinase activity against both receptor and peptide substrates (14–19), strongly suggesting that the observed interactions between αN and the kinase domain represent a general mechanism for stabilization of the active conformation of GRKs.
These same structures also reveal a potential receptor-binding site. On the surface of αN facing away from the kinase domain, several conserved hydrophobic residues form a conspicuous hydrophobic patch (Figure 1B,C). Mutation of the residues in this patch results in a decreased reactivity toward receptor phosphorylation but does not affect peptide phosphorylation, consistent with the idea that these residues directly interact with receptors (15).
Three main events are therefore expected to occur during the activation of GRKs: receptor binding to the docking site, αN interacting with the kinase domain, and kinase domain closure. Although the sequence of these events is not known, kinase domain closure is likely the last prior to the chemical step. Thus, a working hypothesis is that both binding of the receptor to the docking site and binding of αN to the kinase domain stabilize the active GRK conformation. To further investigate the role of these two events in the catalytic mechanism of GRKs, we first used site-directed mutagenesis to confirm the interactions between αN and the kinase domain in GRK1 that we previously observed in structures of active GRK6 (15). We then introduced an intramolecular disulfide bond to cross-link αN and the C-tail of the GRK1 kinase domain and facilitate interactions between αN and the kinase domain. Our data demonstrate that cross-linking enhances kinase activity but does not contribute to binding of activated receptors to GRK1.
EXPERIMENTAL PROCEDURES
Reagents
[γ-32P]ATP was purchased from MP Biomedicals. Peptide C (DDEASTTVSKTETSQVARRR) was synthesized by solid-phase synthesis, and its concentration in solution was determined by quantitative amino acid analysis at the Protein Structure Facility, University of Michigan Medical Center (Ann Arbor, MI). Dark-adapted bovine retinas were purchased from W. L. Lawson Co.
Modeling
PyMOL was used to prepare structural figures.
Production of GRK1 Mutants
Double and single mutants of GRK1 were generated using the QuikChange Multi Site-Directed Mutagenesis and QuikChange Site-Directed Mutagenesis Kits (Stratagene) in a bovine GRK1 construct C-terminally truncated at residue 535 and tagged with hexahistidine (H6) (GRK1535H6) (20). All mutations were verified by DNA sequencing, and the resulting proteins were expressed in High Five insect cells using the Bac-to-Bac system (Invitrogen) and purified as described previously (20). Briefly, metal-chelating affinity chromatography (Ni-NTA, Qiagen) was followed by cation exchange chromatography (Source 15S, GE Healthcare). Some mutants were further purified by size-exclusion chromatography (Superdex 200,GE Healthcare). The purity of each GRK1535H6 variant was >90% as judged by SDS–PAGE. All the proteins were stored in small aliquots at −80 °C, and their concentrations were determined by absorbance at 280 nm using an estimated molar extinction coefficient of 63830 M−1 cm−1.
Production of K3Fe(CN)6-Treated GRK1535H6 Double Cysteine Mutants (CC mutants)
GRK1535H6 CC mutants (1–2 µM) were incubated with 1–2 mM K3Fe(CN)6 in 100 mM HEPES (pH 7.5), 0.15 M NaCl, 200 µM ADP, and 10 mM MgCl2 at room temperature for 1–2 h (21). Proteins were then concentrated, buffer-exchanged into 20 mM HEPES (pH 7.5) and 0.15 M NaCl, and purified by size-exclusion chromatography (Superdex 200, GE Healthcare). Fractions were analyzed via 10% SDS–PAGE under nonreducing conditions. Monomeric proteins were collected, concentrated, and stored in small aliquots at −80 °C for further characterization. To test for reversibility, 10 mM DTT was added to the K3Fe(CN)6-treated protein. After incubation at room temperature for 30 min, the sample was analyzed via SDS–PAGE.
Pepsin Digestion and Tandem Mass Spectrometry Analysis of K3Fe(CN)6-Treated GRK1535H6 T8C/N480C
For each experiment, 10 µg of K3Fe(CN)6-treated GRK1535H6 T8C/N480C was buffer-exchanged into 25 mM NH4HCO3 (pH 7.4). Remaining free cysteine residues were alkylated with 10 µL of 200 mM iodoacetamide (Sigma, St. Louis, MO) and incubated for 1 h in the dark at 25 °C. Pepsin (Worthington, Lakewood, NJ) was dissolved in Milli-Q H2O and equilibrated on ice for 10 min. Before incubation with pepsin, the pH of the protein solution was adjusted to ~2.5 via addition of 10 µL of a 20 mM NH4HCO3/formic acid solution. The reaction mixture was incubated at room temperature for 20 min using a 1:1 ratio of pepsin to K3Fe(CN)6-treated GRK1535H6 T8C/N480C (w/w). Following digestion, 100 µL of the peptic digest was loaded on a Mercury MS column (Phenomenex, Torrance, CA) previously equilibrated for at least 20 min in 2% 2-propanol with 0.1% formic acid and coupled to an HPLC system (Agilent Technologies 1100 Series). After the column was washed for 4 min with 2% 2-propanol and 0.1% formic acid (v/v), peptides were eluted by a 14 min linear gradient from 2 to 98%2-propanol in 0.1% formic acid (v/v) at a flow rate of 0.2 mL/min. The eluate from the HPLC column was injected onto a Finnigan LXQ (Thermo Scientific, Waltham, MA) mass spectrometer equipped with an electrospray ionization source. Identification of pepsin-generated fragments and the disulfide bound peptide was performed using Mascot (22) and MassMatrix (23). For cross-linked GRK1535H6 T8C/N480C treated with DTT, samples were analyzed in the same way except for incubation with 10 mM DTT for 1 h prior to alkylation.
Rod Outer Segment (ROS) and Peptide C Phosphorylation
Both assays were performed as described previously (14). Briefly, phosphorylation of photoactivated rhodopsin (Rho*) was assessed with a gel-based assay using urea-washed bovine ROS as the substrate. Reactions were run at a saturating (1 mM) [γ-32P]ATP concentration, with 1.5–40 µM ROS and 0.05–2 µM GRK1535H6 variants in 100 mM HEPES-NaOH (pH 7.5), 0.15 M NaCl, 10 mM MgCl2, and 1 mM EDTA (buffer A) at room temperature. At various time points up to 5 min, small aliquots of reaction mixtures were quenched with SDS–PAGE sample buffer. Formation of phosphorylated Rho* was analyzed by SDS–PAGE and quantified by phosphorimaging. Phosphorylation of peptide C was assessed by a phosphocellulose filter binding assay (8). The reaction mixtures contained 0.5–10 µM GRK1535H6 variants, 0.1 mM [γ-32P]ATP, and 0.2–2 mM peptide C in buffer A at room temperature. At various time points up to 2 h, small aliquots of reaction mixtures were quenched by 10–20% TCA. After centrifugation at 13000 rpm (16000g) for 10 min, supernatants were applied to phosphocellu-lose P-81 papers (2.5 cm circles, Whatman) and washed extensively with 75 mM H3PO4 (~20–30 mL). Filters were air-dried, and their radioactivity was quantified by liquid scintillation. Initial rates of Rho* and peptide C phosphorylation were calculated from the slope of phosphorylated product formation at various time points and fitted to the Michaelis–Menten equation to yield kcat and KM values using Prism version 4.0a. For cross-linked GRK1535H6 T8C/N480Ctreated with DTT, the protein (1 µM) was incubated with 10 mM DTT on ice for 1 h prior to the measurements.
Activation of Peptide C Phosphorylation with Asp-N-Cleaved Rho* (Asp-N Rho*)
Asp-N-treated rhodopsin (Rho) was produced as described previously (8). Briefly, 500 µL of ~7 mg/mL ROS in 20 mM HEPES-NaOH (pH 7.5) (buffer B) were incubated with 4 ng/µL endoproteinase Asp-N (Roche) in the dark at room temperature overnight (~16 h). ROS were pelleted, washed with 5Murea once, washed with buffer B three times, and then resuspended in buffer B. The concentration of Asp-N-treated Rho was determined by absorbance at 500 nm using the molar extinction coefficient of 40600 M−1 cm−1 (24). Digestion was verified by the reduced size of Rho via SDS–PAGE. Reaction mixtures contained 1–2 µM GRK1535H6 variants, 1 mM peptide C, 0.1 mM [γ-32P]ATP, and 5–80 µM Asp-N Rho* in buffer A at room temperature. After 10 min, reactions were stopped by addition of 20% TCA and mixtures were analyzed as described above. Ymax (maximal Vi/[E]) and KD,app (apparent affinity) values were obtained by fitting the kcat/KM(Y) at various Asp-NRho* (X) concentrations with eq 1:
| (1) |
Nucleotide Binding Assay
Dissociation constants (KD) of GRK1535H6 variants for ADP were determined using a competition fluorescence polarization assay as described previously (14). These assays monitored the decrease in millipolarization caused by the displacement of BODIPYTR-ADP (Invitrogen) (final concentration 0.2 µM) from each GRK1535H6 variant (final concentration 1 µM) by 0–10 µM ADP at 25 °C. See Supporting Information for a full description.
Transducin (Gt) Activation Assay
Gt was purified as described previously (25). Rho was purified from ROS isolated by Zn2+ extraction and had an A280/A500 absorbance ratio of 1.63 (26). ZnCl2 was removed by dialysis in the presence of 0.1% DDM to prevent protein precipitation. The effect of GRK1 on activation of transducin (Gt) was determined by fluorescence measurements (27, 28). Samples contained 25 nM Rho, 250 nM Gt, and 250 nM GRK1535H6 variants in 500 µL of buffer [20mM Bis-Tris propane (BTP) (pH 7.5), 100 mM NaCl, 1 mM MgCl2, and 2 mM n-dodecyl β-d-maltoside (DDM)]. GRK1 was omitted from the control experiment. Samples were incubated for 15 min in the dark at room temperature and then exposed to light for 15 s with a Fiber-light covered with a band-pass filter (480–525 nm). Intrinsic fluorescence was measured at excitation and emission wavelengths of 300 and 345 nm, respectively, with a Perkin-Elmer L55 luminescence spectrophotometer. While fluorescence was being recorded, the temperature in the cuvette chamber of the fluorometer was maintained at 20 °C using a thermostat and the sample was continuously stirred at low speed. After 300 s, GTPγS was added to the final concentration of 5 µM. The relative rate constant (k) was derived from the function A(t) = Amax(1 − e−kt), where Amax is the maximal Gt fluorescence enhancement, A(t) is the amplitude, and k is the relative rate constant of Gt activation in inverse seconds.
Meta II Decay
The effect of GRK1 on the stability of the Rho Meta II state was determined by fluorescence measurements (29). Samples were prepared using 50nMRho and 100 nM GRK1535H6 variants in 500 µL of buffer containing 20 mM BTP, 100 mM NaCl, 1 mM MgCl2, and 2 mM DDM (pH 6.5). GRK1 was excluded for the control experiment. Samples were incubated for 15 min in the dark at room temperature and then exposed to light for 15 s using a Fiber-light covered with a band-pass filter (480–525 nm). Intrinsic fluorescence was measured at excitation and emission wavelengths of 295 and 330 nm, respectively, using a Perkin-Elmer L55 luminescence spectrophotometer (30) with a cuvette maintained at 20 °C. Data were fitted to a first-order reaction equation to calculate relaxation times (τ).
RESULTS
Although we previously determined the structure of GRK6 in an active conformation (15), we chose to use GRK1 in this study for several reasons. First, there are fewer reactive surface cysteine residues in GRK1 (3–5 residues) than in GRK6 (> 12 residues), as determined by Ellman’s titration (data not shown) and by visual examination of representative crystal structures. Therefore, GRK1 is more likely to form an intramolecular disulfide bridge at the engineered positions. Second, GRK1 has higher catalytic activity toward Rho* than GRK6, making it easier to assess the effects of cross-linking on the catalytic efficiency. Third, GRK1 is the native kinase for Rho*, so the studies herein using Rho* and peptide C, which is derived from the C-terminus of Rho, as substrates are more physiologically relevant.
Functional Analysis of the αN Helix of GRK1
The residues involved in the interactions between αN and the kinase domain in the closed structures of GRK6 are highly conserved in GRKs (Figure 1D), and the portions of the C-tail observed in prior structures of GRK1 (20) that are expected to interact with αN are essentially the same as in GRK6 (Figure 1B,C) (15). Thus, we speculated that GRK1 would adopt an active conformation similar to that seen in closed structures of GRK6. To test this hypothesis, we mutated conserved residues in the GRK1 αN helix and assessed whether these mutations have the same activity profile for phosphorylating receptor and peptide substrates as mutations of their corresponding residues in GRK6. Unlike our previous study using human GRK6 isoform C where only the second-order rate constant kcat/KM could be obtained because of the high KM value (15), our use of GRK1 would allow us to obtain both kcat and KM parameters for these variants and thereby provide additional insights into the functional roles of individual residues.
Nine highly conserved residues of the GRK1 αN helix were chosen for mutagenesis studies: Leu6, Glu7, Val9, Val10, Ala11, Asn12, Phe15, Ile16, and Ala18 [corresponding to Leu3, Glu4, Ile6, Val7, Ala8, Asn9, Leu12, Leu13, and Ala15 of GRK6 (Figure 1B,C), respectively]. All residues were mutated to alanine with the exception of Ala11 and Ala18, which were mutated to glutamic acid. For receptor phosphorylation, all GRK1 αN alanine variants exhibited a decrease in kcat (2–21-fold) and an increase in KM (3–6-fold) compared to the values of the wild-type (wt) protein. Overall, a 7–130-fold decrease in kcat/KM was observed (Table 1). This same pattern was also observed for mutants of the GRK1 kinase domain, including F190A, R191A, L212A, Y274A, V476A, and V484A (14), whose equivalent residues in GRK6 make important contacts with αN (15). Because we observed defects in both kcat and KM, our data suggest that both receptor binding and αN interacting with the kinase domain, along with the associated kinase domain closure, are required for efficient receptor phosphorylation. The effect caused by any individual mutation is not as large as for the ΔN19 mutant of GRK1, which lacks the amino-terminal 19 residues and exhibits a > 3900-fold decrease in kcat/KM (14). This may reflect the cumulative contribution from all the amino acid side chains in αN.
Table 1.
Kinetic Parameters of ROS Phosphorylation for GRK αN Variantsa
| bGRK1 variants | kcat (×10−3 s−1) | KM (×10−6 M) | kcat/KM (×103 M−1 s−1) | fold decrease (kcat/KM) | hGRK6 variants | fold decreaseb (kcat/KM) |
|---|---|---|---|---|---|---|
| wt | 25 ± 4 | 3.4 ± 1.7 | 7.2 ± 2.7 | 1 | wt | 1 |
| L6A | 4.9 ± 0.4 | 12 ± 3 | 0.40 ± 0.05 | 18 | L3A | 2 |
| E7A | 7.7 ± 0.6 | 13 ± 2 | 0.62 ± 0.07 | 12 | E4A | 3 |
| V9A | 2.2 ± 0.3 | 18 ± 5 | 0.13 ± 0.02 | 55 | I6A | 12 |
| V10A | 1.2 ± 0.3 | 21 ± 9 | 0.057 ± 0.013 | 130 | V7A | 13 |
| A11E | 22 ± 1 | 8.8 ± 1.2 | 2.5 ± 0.2 | 3 | ||
| N12A | 1.7 ± 0.7 | 20 ± 15 | 0.083 ± 0.032 | 87 | N9A | 140 |
| F15A | 3.5 ± 0.6 | 11 ± 5 | 0.32 ± 0.08 | 23 | L12A | 1100 |
| I16A | 11 ± 2 | 9.5 ± 4.8 | 1.1 ± 0.4 | 7 | L13A | 6 |
| A18E | 8.7 ± 0.7 | 6.1 ± 1.4 | 1.4 ± 0.2 | 5 |
Numbers shown represent means ± the standard error of the fit calculated from three to six independent experiments. Reactions were performed in 100 mM HEPES-NaOH (pH 7.5), 0.15 M NaCl, 10 mM MgCl2, and 1 mM EDTA for <5 min at room temperature.
Data taken from ref 15.
If residues in αN are involved in only GPCR binding, then mutations at these positions should have little effect on phosphorylation of peptide C. Indeed, mutations of residues proposed to interact solely with the receptor (L6A, E7A, V9A, and V10A) showed no defects in peptide phosphorylation (Table 2). In contrast, mutations of residues involved in interacting with the kinase domain (N12A, F15A, and I16A) showed a 6–22-fold decrease in kcat and kcat/KM when compared to the values of wt. Therefore, the activity profile of GRK1 αN mutants is very similar to that of the analogous GRK6 mutants (Tables 1 and 2), and is consistent with the proposed function of αN in the closed GRK6 structures (15). Because there is no change in their KM values, the binding of peptide substrates is probably not highly dependent on the conformational state of the kinase domain, consistent with the fact that peptide substrates form most of their interactions with the C-terminal kinase domain (C-lobe) in the AGC kinase family.
Table 2.
Kinetic Parameters of pC Phosphorylation for GRK αN Variantsa
| bGRK1 variants | kcat (×10−6 s−1) | KM (×10−4 M) | kcat/KM (×10−2 M−1 s−1) | fold decrease (kcat/KM) | hGRK6 variants | fold decreaseb (kcat/KM) |
|---|---|---|---|---|---|---|
| wt | 33 ± 6 | 3.2 ± 1.4 | 11 ± 3 | 1 | wt | 1 |
| L6A | 58 ± 13 | 2.8 ± 1.7 | 21 ± 8 | 0.5 | L3A | 1 |
| E7A | 80 ± 21 | 4.1 ± 2.4 | 19 ± 6 | 0.6 | E4A | 2 |
| V9A | 91 ± 15 | 3.5 ± 1.3 | 26 ± 13 | 0.4 | I6A | 1 |
| V10A | 81 ± 12 | 3.9 ± 1.3 | 21 ± 4 | 0.5 | V7A | 1 |
| A11E | 13 ± 3 | 3.1 ± 1.6 | 4.3 ± 1.4 | 3 | ||
| N12A | 4.8 ± 1.4 | 5.0 ± 2.9 | 1.0 ± 0.3 | 11 | N9A | 8 |
| F15A | 1.7 ± 0.4 | 3.5 ± 2.1 | 0.50 ± 0.18 | 22 | L12A | 6 |
| I16A | 3.9 ± 0.6 | 2.2 ± 1.0 | 1.8 ± 0.5 | 6 | L13A | 5 |
| A18E | 5.0 ± 1.6 | 4.1 ± 2.9 | 1.2 ± 0.5 | 9 |
Numbers shown represent means ± the standard error of the fit calculated from three to six independent experiments. Reactions were performed in 100 mM HEPES-NaOH (pH 7.5), 0.15 M NaCl, 10 mM MgCl2, and 1 mM EDTA for <2 h at room temperature.
Data taken from ref 15.
The A11E and A18E mutants displayed modest defects in kcat (<3-fold decrease) and KM (<3-fold increase) for receptor phosphorylation and a <7-fold decrease in kcat for peptide phosphorylation. When a glutamate residue is modeled at position 8 or 15 of GRK6 (position 11 or 18 of GRK1, respectively), the longer side chain clashes with neighboring amino acids. Therefore, the observed effects could result from loss of productive interactions with the receptor or disruption of the interactions between αN and the kinase domain, leading to a relatively inactive conformation of the kinase domain.
Design of an Intramolecular Disulfide Bond between αN and the Kinase Domain
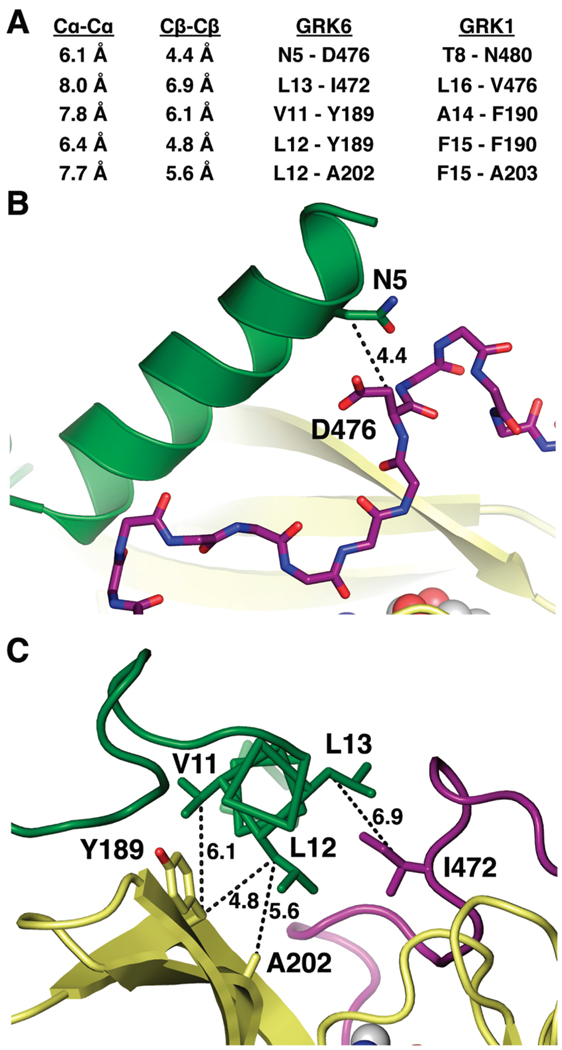
We used the GRK6 structure as a model to determine which positions on αN and the kinase domain of GRK1 could be mutated to cysteine residues with the goal of forming an intramolecular disulfide bond. The Sγ–Sγ, Cβ–Cβ, and Cα–Cα atomic distances are ~2, 3.4–4.2, and <6.5 Å, respectively, in disulfide bonds from high-resolution X-ray structures, and the Cβ–Sγ–Sγ angle is ~100° (31). Based on these criteria, five pairs of positions, with one on the αN helix and the other either on the N-lobe or on the C-tail of the kinase domain, were selected for mutation to cysteine residues (Figure 2): Thr8/Asn480, Ala14/Phe190, Phe15/Phe190, Phe15/Ala203, and Ile16/Val476 of GRK1 (corresponding to Asn5/Asp476, Val11/Tyr189, Leu12/Tyr189, Leu12/Ala202, and Leu13/Ile472 of GRK6, respectively). Although some interatomic distances for these pairs in the GRK6 structure are too long for ideal disulfide bonds (Figure 2A), slight adjustments might be accommodated without perturbing the kinase activity.
FIGURE 2.
Pairs of residues targeted for disulfide bridge formation. (A) Cα–Cα and Cβ–Cβ interatomic distances for chosen pairs. The Cα–Cα and Cβ–Cβ distances of typical disulfide bonds are <6.5 and 3.4–4.2 Å, respectively. (B and C) Locations of CC mutants on the GRK6 structure. The Cβ–Cβ distances are indicated. The N5C/D476C (B) and L13C/I472C (C) pairs were designed to cross-link αN with the C-tail, whereas the V11C/Y189C, L12C/Y189C, and L12C/A202C pairs (C) were designed to cross-link αN with the kinase N-lobe. The color scheme is the same as in Figure 1.
GRK1535H6 T8C/N480C Forms an Intramolecular Disulfide Bond between αN and the C-Tail
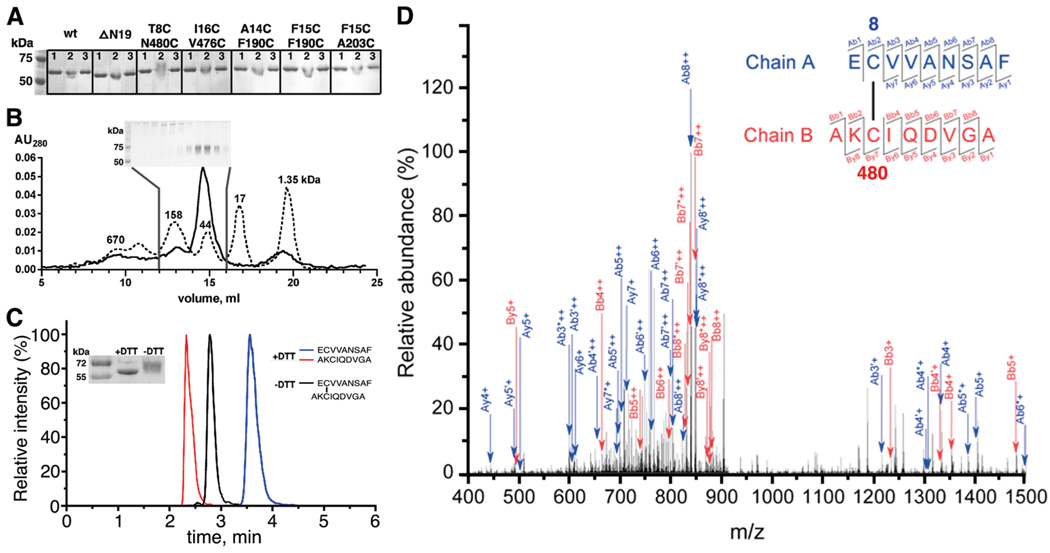
To test whether the various CC mutants of GRK1 are able to form disulfide bonds, we first used a gel-shift assay under nondenaturing conditions to detect cross-linked species after oxidation. After the protein (1–2 µM) had been treated with 1 mM K3Fe(CN)6 for 1 h at room temperature, a distinct upward shift was observed for the T8C/N480C mutant but not for wt, ΔN19, the other four CC mutants (Figure 3A), or single mutants at position 8 or 480 (T8C or N480C) (data not shown). The upward shift of T8C/N480C was reversed in the presence of the reducing agent DTT (Figure 3A). Collectively, these data show that the shifted species likely results from formation of a specific disulfide bond between positions 8 and 480.
FIGURE 3.
GRK1535H6 T8C/N480C forms a disulfide bond between positions 8 and 480 under oxidizing conditions. (A) K3Fe(CN)6-treated GRK1535H6 T8C/N480C, but not wt, ΔN19, or other CC mutants, displays a delayed migration on nonreducing 10% SDS–PAGE that can be reversed by DTT: lane 1, before K3Fe(CN)6 treatment; lane 2, after K3Fe(CN)6 treatment; lane 3, after K3Fe(CN)6 treatment and then incubation with DTT. (B) GRK1535H6 T8C/N480C is monomeric after K3Fe(CN)6 oxidation. The elution profile from size-exclusion chromatography (Superdex 200, 25 mL) is shown as a solid line, and that of molecular mass standards is shown as a dotted line with numbers indicating molecular masses in kilodaltons. Nonreducing 10% SDS–PAGE analyzed fractions from the column are shown in the inset. Fractions of monomers show an upward gel shift, as seen in panel A. (C) Overlays of HPLC runs of peptidic fragments from oxidized GRK1535H6 T8C/N480C in the absence (black) or presence (red and blue) of DTT. The inset shows the degree of reduction via SDS–PAGE by application of DTT before tandem mass spectrometry analysis. (D) Identification of the Cys8–Cys480 disulfide bond by tandem mass spectrometry. The specific disulfide bond was verified on the basis of the MS2 spectra of the doubly charged ion at m/z 920.39. Ions used to identify fragments of chain A (EC8VVANSAF) are colored blue, whereas ions used to identify chain B (AKC480IQDVGA) are colored red. Product ions generated due to neutral losses are labeled using primes (loss of water) and asterisks (loss of ammonia). Further statistics for the confirmation of the disulfide-linked peptide by mass spectroscopy are provided in Table S1 of the Supporting Information.
To test whether the shifted species corresponds to an intra- or inter-molecular disulfide linkage, K3Fe(CN)6-treated GRK1535H6 T8C/N480C was purified by size-exclusion chromatography. As shown in Figure 3B, the majority of K3Fe(CN)6-treated T8C/N480C eluted as a monomer and these fractions retained the same slowed migration upon nonreducing SDS–PAGE (Figure 3B, inset). Thus, cross-linked GRK1535H6 T8C/N480C forms an intramolecular disulfide bond.
To further confirm that the oxidized T8C/N480C mutant forms the disulfide bridge at the designed position, the cross-linked protein purified by size-exclusion chromatography was subjected to proteolysis with pepsin and the resulting peptide fragments were subjected to tandem mass spectrometry analysis. As shown in Figure 3C, peptides corresponding to the disulfide-linked regions of GRK1 elute as a single peak from an HPLC column, whereas addition of the reducing agent DTT before pepsin digestion results in two distinct peaks attributed to AKC8IQDVGA and EC480VVANSAF, with only trace amounts of the cross-linked peak. The cross-linked peptide shows an ion at m/z 920.39 on the MS2 spectrum. On the basis of the y and b ions, and the ions due to neutral losses of water and ammonia, the disulfide-linked peptides with chains A (EC8VVANSAF) and B (AKC480IQDVGA) were identified as having a disulfide bond joining positions 8 and 480 (Figure 3D and Table S1 of Supporting Information).
Cross-Linked GRK1535H6 T8C/N480C Exhibits a Profound Increase in kcat for Peptide Phosphorylation
To characterize the properties of the cross-linked T8C/N480C mutant, we measured its catalytic activities for ROS and peptide phosphorylation and its affinity for ADP (Table 3). Similar to those of ΔN19 and other key mutants of the kinase domain (14), the dissociation constant for ADP is unchanged for cross-linked T8C/N480C, suggesting that nucleotide binding is relatively independent of the conformation of the kinase domain and to any effects of the αN helix. For receptor phosphorylation, the cross-linked T8C/N480C mutant showed no significant differences in either kcat or KM values from that of wt. However, a robust 10-fold increase in kcat and kcat/KM was observed for peptide phosphorylation. The increase in kcat was not observed for either nonoxidized T8C/N480C or wt protein treated with K3Fe(CN)6 and was reversed by treatment with reducing agent DTT, confirming that the increase in catalytic efficiency resulted from formation of a disulfide bond between position 8 and 480. This indicates that cross-linking between αN and the C-tail favors formation of the active kinase and hence increases the reactivity toward peptides. Similar to those of mutants of αN and the kinase domain, the KM for peptide C phosphorylation of the cross-linked T8C/N480C mutant was unaltered, once again supporting the conclusion that binding of peptides does not strongly depend on the active state of the kinase.
Table 3.
Kinetic Parameters and ADP Binding of bGRK1535H6 T8C/N480Ca
| ROS phosphorylation | peptide C phosphorylation | ADP | |||||
|---|---|---|---|---|---|---|---|
|
kcat (×10−3 s−1) |
KM (×10−6 M) |
kcat/KM (×103 M−1 s−1) |
kcat (×10−6 s−1) |
KM (×10−4 M) |
kcat/KM (×10−2 M−1 s−1) |
KD (µM) |
|
| wt | 25 ± 4 | 3.4 ± 1.7 | 7.2 ± 2.7 | 33 ± 6 | 3.2 ± 1.4 | 11 ± 3 | 0.84 ± 0.15 |
| wt, K3Fe(CN)6b | 27 ± 2 | 3.2 ± 0.8 | 8.5 ± 1.6 | 39 ± 8 | 4.4 ± 1.8 | 8.7 ± 2.0 | |
| T8C/N480C | 17 ± 2 | 4.7 ± 1.4 | 3.6 ± 0.7 | 55 ± 18 | 5.2 ± 3.5 | 11 ± 4 | 0.88 ± 0.30 |
| T8C/N480C, K3Fe(CN)6b | 23 ± 4 | 3.1 ± 1.9 | 7.4 ± 3.5 | 410 ± 60 | 2.7 ± 1.0 | 152 ± 33 | 1.2 ± 0.2 |
| T8C/N480C, K3Fe(CN)6, DTTb | 23 ± 2 | 5.8 ± 1.5 | 3.9 ± 0.7 | 78 ± 8 | 4.6 ± 1.2 | 17 ± 3 | 1.4 ± 0.8 |
Numbers represent means ± the standard error of the fit calculated from three to six independent experiments. Reactions were performed in 100 mM HEPES-NaOH (pH 7.5), 0.15 M NaCl, 10 mM MgCl2, and 1 mM EDTA for <5 min for ROS phosphorylation and <2 h for peptide phosphorylation at room temperature.
Proteins were treated with 1 mM K3Fe(CN)6 at room temperature for 1 h, and then monomers were purified by size-exclusion chromatography. Some of the protein was then reduced with 10 mM DTT at 4 °C for 1 h before the experiments described above.
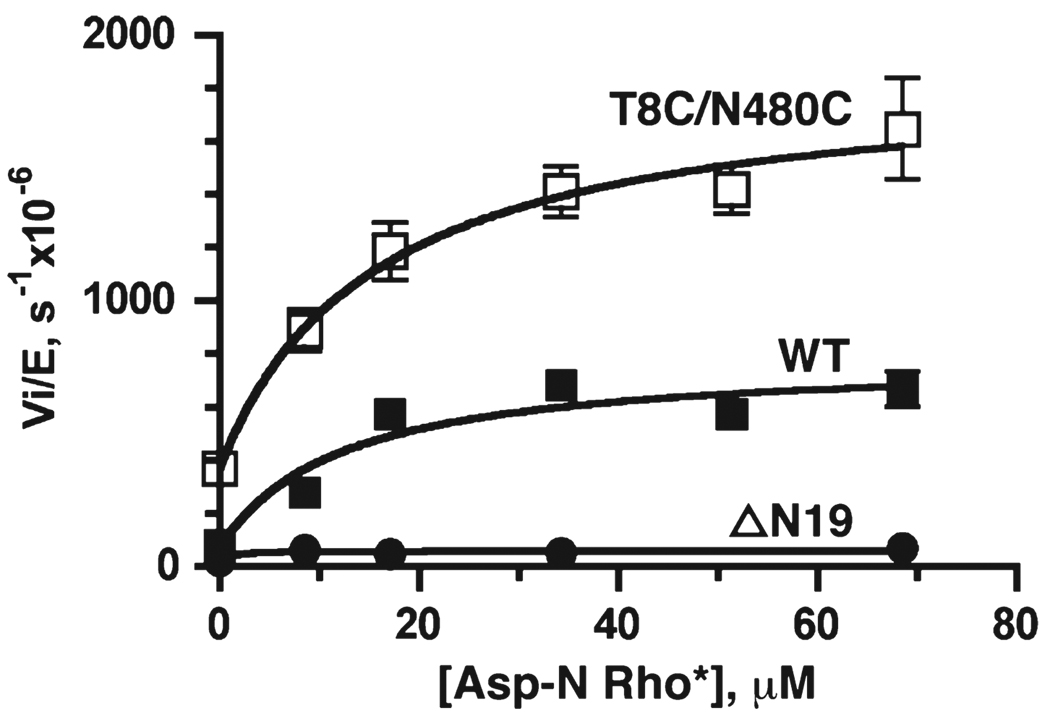
To investigate if cross-linking has an effect on direct interactions with an activated receptor, we measured the activity of peptide phosphorylation in the presence of Rho* treated with endoproteinase Asp-N, which cleaves Rho at position 329 and removes all the phosphorylation sites while leaving the GRK docking site intact. It was previously shown that Asp-N Rho* enhances the peptide phosphorylation of GRK1, presumably by directly stabilizing the active conformation of the kinase domain (8). In the presence of Asp-N Rho*, the kcat values for peptide C phosphorylation are enhanced ~10- and 5-fold for the wt protein and the cross-linked T8C/N480C mutant, respectively, at saturating Asp-N Rho* concentrations (Figure 4). As expected, the ΔN19 mutant does not show any increase in catalytic activity in the presence of Asp-N Rho*. The apparent binding constants, KD,app, are similar for the wt protein and the cross-linked T8C/N480C mutant (12 ± 5 and 15 ± 7 µM, respectively), suggesting that receptor binding is not affected by cross-linking of the N-terminal region of GRK1 to the C-tail. The fact that cross-linked T8C/N480C is still 2–3 times more active than the wt protein at saturating Asp-N Rho* concentrations suggests that both receptor docking and the interaction of αN with the kinase domain contribute to stabilization of the active conformation of the kinase domain.
FIGURE 4.
Effects of Asp-N-cleaved activated Rho (Asp-NRho*) on peptide C phosphorylation by GRK1535H6. Both the wt protein (■) and the cross-linked T8C/N480C mutant (□) show activation by Asp-N Rho* in a concentration-dependent fashion, while the ΔN19 mutant did not show activation by Asp-NRho* (●). Data were fitted to eq 1 from four independent experiments. Ymin, Ymax, and KD,app values are (79 ± 5) × 10−6 s−1, (780 ± 80) × 10−6 s−1, and 12 ± 5 µM for wt and (370 ± 40) × 10−6 s−1, (1900 ± 170) × 10−6 s−1, and 15 ± 6 µM for the cross-linked T8C/N480C mutant, respectively.
Both GRK1535H6 wt and Cross-Linked T8C/N480C Inhibit Transducin (Gt) Activation by Rho*
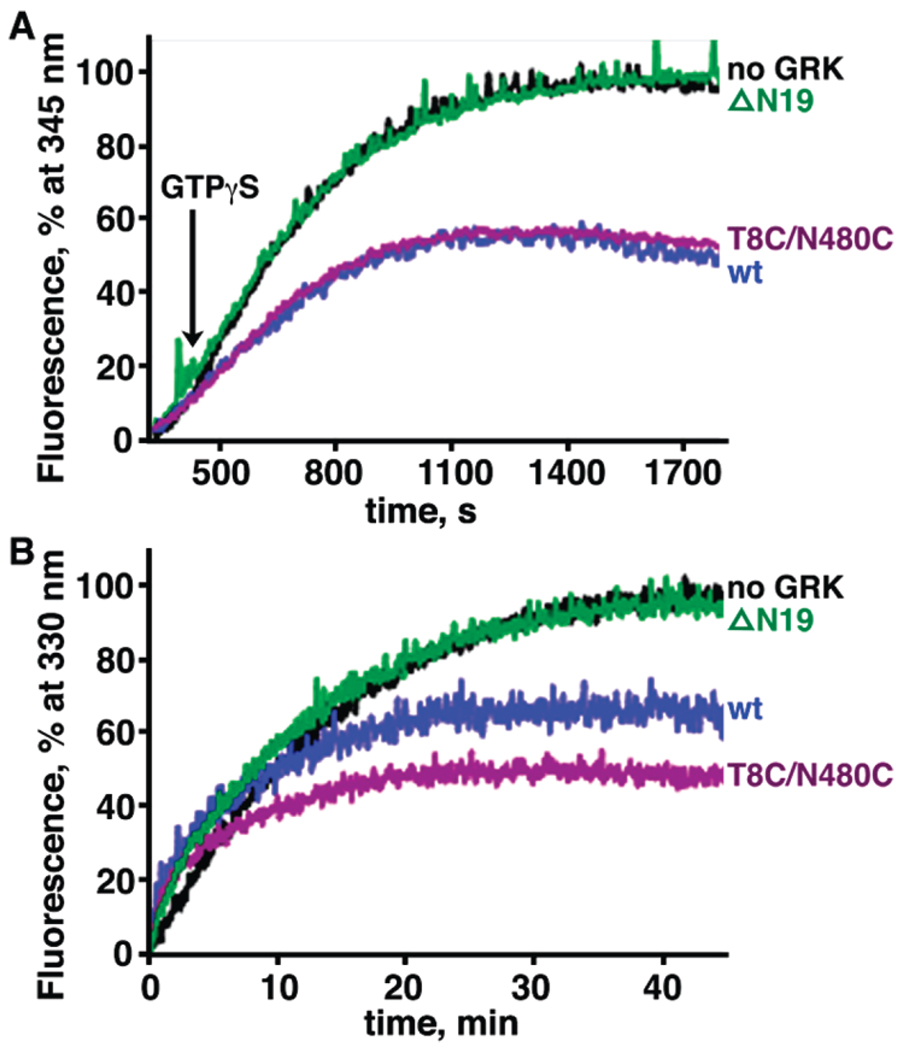
An indirect way to test binding affinity for activated receptors is to compete with binding of transducin (Gt) to activated Rho. Zn2+-extracted Rho* was used to activate Gt, and the change in the tryptophan fluorescence of Gαt upon GTP binding was monitored at 345 nm (Figure 5A) (28, 32, 33). Conditions for the assay were chosen so that the Gt activation rate was the same as that determined by GTPγS-induced complex dissociation (27). The relative activation rates were determined in the absence or presence of GRK1 proteins. Relative fluorescence intensity was decreased by ~50% for both wt and cross-linked T8C/N480C proteins, whereas no effect was observed for the ΔN19 mutant (Figure 5A). In addition, a slight increase in the rate constant was also observed for both GRK1 wt and cross-linked T8C/N480C proteins. These results strongly suggest that GRK1 directly competes with Gt for binding to Rho*. Because the same degree of fluorescence decrease was observed for wt and cross-linked T8C/N480C, these results are consistent with the idea that cross-linking of αN to the C-tail does not affect binding of the kinase to the receptor.
FIGURE 5.
Effects of GRK1535H6 variants on transducin (Gt) activation by Rho* and Meta II decay. Data for reactions without GRK1, with wt GRK1, with cross-linked T8C/N480C, and with ΔN19 are colored black, blue, purple, and green, respectively. Rate constants were calculated from three independent experiments, and data from a representative reaction are shown. (A) GRK1 inhibits activation of Gt by Rho*. The activation of Gt was monitored by the increase in its intrinsic tryptophan fluorescence because of nucleotide exchange catalyzed by Meta II (λexcitation = 300 nm, and λemission = 345 nm). GTPγS (5 µM) was added to initiate the reaction after data had been recorded for 300 s. (B) GRK1 promotes Meta II decay. Meta II decay was measured by the increase of the intrinsic tryptophan fluorescence of Rho due to the release of the retinal chromophore (λexcitation = 295 nm, and λemission = 330 nm).
Both GRK1535H6 wt and the Cross-Linked T8C/N480C Mutant Promote Meta II Decay
To investigate whether cross-linking of GRK1 T8C/N480C exerts a different level of Rho* stabilization, we measured the rate of Meta II decay, which corresponds to the release of the chromophore all-trans-retinal from Rho*, via the intrinsic tryptophan fluorescence change (30, 34, 35). Reactions were conducted in the presence or absence of GRK1 proteins using a λexcitation of 295 nm and a λemission of 330 nm (Figure 5B). The relaxation time was 8.4 ± 0.5 min in the presence of GRK1 wt and 7.2 ± 0.4min in the presence of the cross-linked T8C/N480C mutant, significantly shorter than that measured in the absence of any GRK1 (14.0 ± 0.9 min) or in the presence of the ΔN19 mutant (12.8 ± 0.7 min). Additionally, the magnitude of the fluorescence change is smaller in the presence of GRK1 by ~35% for wt and ~50% for the cross-linked T8C/N480C mutant. Therefore, unlike transducin, which stabilizes the Meta II intermediate (36), and arrestin, which affects the amplitude of Meta II decay but not the relaxation time (37), GRK1 binding to Rho* accelerates the rate of Meta II decay and influences the environment of tryptophan residues in opsin. Thus, it is possible that GRK1 stabilizes a unique conformation of Rho*. However, once again, the cross-linking between αN and the C-tail does not significantly affect interactions between GRK1 and the receptor.
DISCUSSION
Previous structural and biochemical studies suggested that activated receptor allosterically activates GRK by stabilizing an active configuration of the GRK kinase domain through interactions with the αN helix (14, 15). In this study, we further investigated the role of the interactions between the αN helix and the kinase domain by cross-linking αN and the C-tail at a specific position and determined the effect of this cross-linking on the catalytic activity of GRK1 and its direct interactions with Rho*, its physiological substrate. Formation of the T8C/N480C disulfide bond results in a 10-fold increase in kcat and kcat/KM for peptide substrate phosphorylation but has no effect on either receptor phosphorylation or nucleotide binding. Additionally, cross-linking does not enhance the binding of GRK to the receptor, as inferred from Asp-N Rho*-dependent peptide phosphorylation, inhibition of Gt activation, and Meta II decay.
The catalytic efficiency of rhodopsin phosphorylation and peptide phosphorylation by GRK1 is very different. The kcat value is ~103-fold higher and the KM value ~102-fold lower for rhodopsin phosphorylation, resulting in a total ~105-fold difference in kcat/KM. Therefore, although the chemical reaction is identical, the rate-limiting step is most likely different for rhodopsin and peptide phosphorylation. For peptide phosphorylation, the kcat value is so small (3.3 × 10−5 s−1) that neither product (ADP) release (koff ~ 10−1 s−1 assuming kon ~ 104M−1 s−1 and KD ~ 1 µM) nor the chemical step (≥kcat of rhodopsin phosphorylation, 2.5 × 10−2 s−1) is likely to be rate-limiting. Furthermore, GRK1 ΔN19 and cross-linked T8C/N480C mutants display a 10-fold decrease and a 10-fold increase in kcat, respectively, but show no change in either the KM for peptide or the KD for ADP, suggesting that both peptide substrate and nucleotide binding are independent of αN and its interactions with the kinase domain. Thus, the major factor in determining the catalytic efficiency of peptide phosphorylation is the equilibrium between active and inactive GRK. The ΔN19 mutant disfavors formation of the active GRK conformation, whereas cross-linked T8C/N480C favors it.
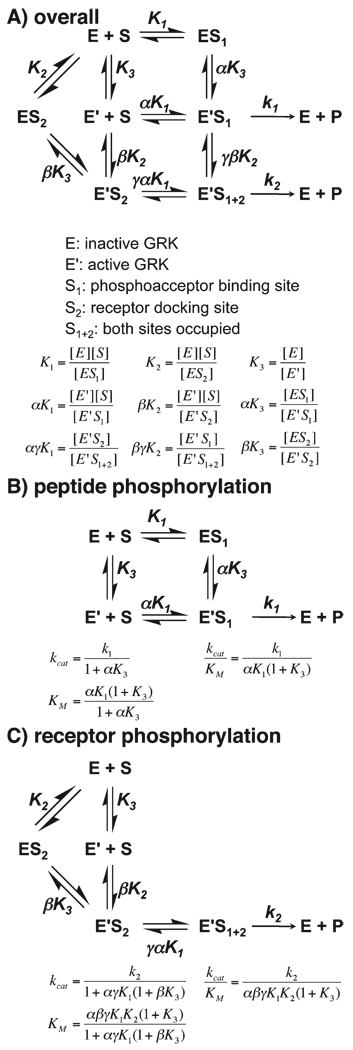
Therefore, a simplified kinetic scheme of GRK1 that is focused on the transition of GRK from an inactive to active state is described in Scheme 2A under the assumption of rapid equilibrium. All enzyme species also are assumed to be pre-occupied with ATP for simplification. Three steps are considered: (1) peptide binding to the active site, (2) receptor binding to αN and perhaps other structural elements of GRK1 that comprise the docking site, and (3) the conformational change of GRK1 from an inactive (E) to an active (E′) state, in which αN interacts with the kinase domain and stabilizes kinase domain closure. Equilibrium constants for these steps are described as K1, K2, and K3, respectively.
Scheme 2.
Reaction Diagrams for Phosphorylation of GPCRs and Peptides by GRKs
For peptide C phosphorylation, step 2 (receptor docking to the kinase) is absent. Therefore, a further simplified scheme can be derived (Scheme 2B). On the basis of this scenario, if an alteration stabilizes the active GRK conformation (E′), a shift in equilibrium toward E′ will result in a decreased K3 and hence an increased kcat, and vice versa for alterations that destabilize the active state. For the cross-linked T8C/N480C mutant, which was designed to facilitate the docking of αN to the kinase domain, a 10-fold increase in kcat was observed, consistent with the situation in which E′ is stabilized. In contrast, for GRK1 mutants of αN and the kinase domain that impair the docking of αN to the kinase domain, a decreased kcat was observed, consistent with the destabilization of E′. Thus, the docking of αN to the kinase domain appears to be important for formation of an active GRK and is closely linked to kinase domain closure. However, the ΔN19 mutant is still able to catalyze peptide phosphorylation, albeit with decreased reactivity, indicating that interaction of αN with the kinase domain is not absolutely required for kinase domain closure. Instead, αN facilitates the formation of the active state. Additionally, because the KM value is unaffected, the binding of the peptide substrate appears to be independent of the conformational state of GRK1 (α = γ = 1, and KM = K1).
Asp-N Rho* is known to enhance GRK-catalyzed peptide phosphorylation, presumably by stabilizing GRK1 in its active conformation (shift E toward E′). The cross-linked T8C/N480C mutant exhibits the same phenomenon. Peptide phosphorylation in the presence of Asp-N Rho* for the cross-linked T8C/N480C mutant allows us to evaluate individual contributions to GRK activation of receptor docking to the kinase (step 2) and of αN interacting with the kinase domain (step 3). If the mechanism used by cross-linked T8C/N480C is the same as that used by Asp-N Rho*, then the activity of the cross-linked T8C/N480C mutant will be insensitive to Asp-N Rho* and will be similar to that of the wt protein at saturating Asp-N Rho* concentrations, where almost all GRK1 is bound to Asp-N Rho*. The fact that the cross-linked T8C/N480C mutant is still activated by Asp-NRho* suggests that this mutant is not fully active despite the disulfide linkage. In other words, the introduced cross-link between the N-terminal region and the C-tail only partially favors the activated conformation observed in the GRK6 · sangivamycin structure. Furthermore, the activity is higher for the cross-linked T8C/N480C mutant than wt at saturating Asp-N Rho* concentrations, implying that the interaction of αN with the kinase domain (step 3) and receptor binding to the docking site (step 2) are two distinct steps contributing to GRK1 activation.
When considering receptor phosphorylation, we must take an additional step of receptor docking to GRK (step 2) into account. Because the formation of active GRK in the presence of the receptor is the dominant pathway (k2 ≫ k1, and K2 ≪ K1), Scheme 2C is sufficient to describe the reaction of receptor phosphorylation. For GRK1 mutants that perturb both the interaction of αN with the kinase domain (step 3) and receptor binding to the docking site (step 2), a decrease in kcat and an increase in KM were observed. This is consistent with the idea that impairment of either step will disfavor the active kinase conformation and/or receptor docking. However, it was unexpected that the cross-linked T8C/N480C mutant did not have any effect on receptor phosphorylation. On the basis of the current scheme, an improved kcat, KM, or both should have been observed. Several possibilities could account for this discrepancy. The first is that other steps not considered here, for example, release of the product ADP, are rate-limiting for receptor phosphorylation. Another is that once the receptor docks to the kinase, it may act processively and not dissociate before additional rounds of phosphorylation occur (there are seven potential phosphorylation sites in the C-terminal region of rhodopsin).
To directly probe the importance of the interactions of αN with the kinase domain on the binding of GRK1 to Rho, we assessed both wt and cross-linked T8C/N480C GRK1 for their effects on inhibition of transducin activation by Rho* and Meta II decay. In either assay, a robust effect was observed upon addition of GRK1, but no significant difference from that of wt was observed for the cross-linked T8C/N480C mutant. These data are consistent with peptide phosphorylation in the presence of Asp-N Rho*, in which wt and the cross-linked T8C/N480C mutant have similar apparent affinities for Asp-N Rho*. All three pieces of data suggest that cross-linking the N-terminal region of GRK1 to the C-tail does not promote the interactions between GRK1 and rhodopsin. It is possible that receptor docking to GRK is independent of the interaction formed between the N-terminal region and the kinase domain. It is also likely that even when this intramolecular contact is enforced by a disulfide bond, the αN helix still fails to form and productively interact with the small lobe without the assistance of an activated receptor. Such action would be consistent with the crystal structure of the cross-linked T8C/N480C mutant, wherein the kinase domain retains an open conformation and the αN helix remains disordered (Figures S1 and S2 of the Supporting Information). Although the preponderance of current data suggests that the αN helix is the most important region for interacting with rhodopsin, it is possible that other regions of GRKs also contribute to the binding surface (38). It should be noted that the N-terminal region of GRKs has also been postulated to promote interactions with phospholipids (18), but the fact that GRK1 can efficiently phosphorylate detergent-solubilized Rho* appears to argue that phospholipid binding is not the most important function of the αN helix with respect to its activity.
In conclusion, we have used site-directed mutagenesis and an engineered disulfide linkage to provide additional evidence that the interaction between the αN helix and the kinase domain of GRK1 is important for formation of the catalytically competent conformation of GRK1 and, by extension, other GRKs. Our assays provide further insights into receptor binding affinity and suggest that receptor binding is not highly dependent on contacts formed between the N-terminal region of GRK1 and the C-tail of the kinase domain. This could imply that αN adopts a helical conformation only after it docks to the activated GPCR, after which stabilization of the kinase domain occurs rapidly through contacts formed on the opposite side of the αN helix. However, receptor docking to GRKs and the interactions of αN with the kinase domain are both clearly important for stabilizing the active conformation and constitute a unique mechanism for allosteric regulation of an AGC kinase.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health (NIH) Grants HL071818 and HL086865 (to J.J.G.T.) and EY008061, GM079191, and P30EY11373 (to K.P.). Our research used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center, supported by NIH Grant DK20572.
Abbreviations: AGC, protein kinase A, G, and C; AST, active site tether; BTP, 1,3-bis[tris(hydroxymethyl)methylamino]propane; CC mutants, double cysteinyl mutants; C-tail, C-terminal tail; DDM, n-dodecyl β-d-maltoside; DTT, d,l-dithiothreitol; EDTA, (ethylenedinitrilo)tetraacetic acid; Gt, transducin; Gαt, transducin α subunit; GRK, G protein-coupled receptor kinase; GPCR, G protein-coupled receptor; GTPγS, guanosine 5′-[γ-thio]triphosphate; HEPES, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid; HPLC, high-performance liquid chromatography; H6, hexahistidine; Meta II, Meta II intermediate of rhodopsin; PDB, Protein Data Bank; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; mP, millipolarization; peptide C, DDEASTTVSKTETSQVARRR; PKA, protein kinase A; RH, regulator of G protein signaling homology; Rho, rhodopsin; Rho*, photo-activated rhodopsin; ROS, rod outer segments; TCA, trichloroacetic acid; wt, wild-type.
SUPPORTING INFORMATION AVAILABLE
Supplemental methods, a table containing statistics describing the identification of the T8C/N480C disulfide bond by tandem mass spectrometry (Table S1), and a table and figures describing the crystal structure of oxidized T8C/N480C GRK1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu. Rev. Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski K, Benovic JL. G-protein-coupled receptor kinases. Trends Biochem. Sci. 1991;16:387–391. doi: 10.1016/0968-0004(91)90157-q. [DOI] [PubMed] [Google Scholar]
- 3.Huang CC, Tesmer JJ. Recognition in the face of diversity: The interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J. Biol. Chem. 2011 doi: 10.1074/jbc.R109.051847. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowles C, Sharma R, Akhtar M. Mechanistic studies on the phosphorylation of photoexcited rhodopsin. FEBS Lett. 1988;238:56–60. doi: 10.1016/0014-5793(88)80224-2. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, McDowell JH, Hargrave PA. Rhodopsin kinase: Substrate specificity and factors that influence activity. Biochemistry. 1988;27:2306–2313. doi: 10.1021/bi00407a010. [DOI] [PubMed] [Google Scholar]
- 6.Kelleher DJ, Johnson GL. Characterization of rhodopsin kinase purified from bovine rod outer segments. J. Biol. Chem. 1990;265:2632–2639. [PubMed] [Google Scholar]
- 7.Benovic JL, Onorato J, Lohse MJ, Dohlman HG, Staniszewski C, Caron MG, Lefkowitz RJ. Synthetic peptides of the hamster β2-adrenoceptor as substrates and inhibitors of the β-adrenoceptor kinase. Br. J. Clin. Pharmacol. 1990;30 Suppl. 1:3S–12S. doi: 10.1111/j.1365-2125.1990.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. Mechanism of rhodopsin kinase activation. J. Biol. Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 9.Chen CY, Dion SB, Kim CM, Benovic JL. β-Adrenergic receptor kinase. Agonist-dependent receptor binding promotes kinase activation. J. Biol. Chem. 1993;268:7825–7831. [PubMed] [Google Scholar]
- 10.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: A complex between G protein-coupled receptor kinase 2 and Gβγ. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 11.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauge C, Antal TL, Hirschberg D, Doehn U, Thorup K, Idrissova L, Hansen K, Jensen ON, Jorgensen TJ, Biondi RM, Frodin M. Mechanism for activation of the growth factor-activated AGC kinases by turn motif phosphorylation. EMBO J. 2007;26:2251–2261. doi: 10.1038/sj.emboj.7601682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palczewski K. GTP-binding-protein-coupled receptor kinases: Two mechanistic models. Eur. J. Biochem. 1997;248:261–269. doi: 10.1111/j.1432-1033.1997.00261.x. [DOI] [PubMed] [Google Scholar]
- 14.Huang CC, Yoshino-Koh K, Tesmer JJ. A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J. Biol. Chem. 2009;284:17206–17215. doi: 10.1074/jbc.M809544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boguth CA, Singh P, Huang C-c, Tesmer JJ. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29:3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu QM, Cheng ZJ, Gan XQ, Bao GB, Li L, Pei G. The amino terminus with a conserved glutamic acid of G protein-coupled receptor kinases is indispensable for their ability to phosphorylate photoactivated rhodopsin. J. Neurochem. 1999;73:1222–1227. doi: 10.1046/j.1471-4159.1999.0731222.x. [DOI] [PubMed] [Google Scholar]
- 17.Noble B, Kallal LA, Pausch MH, Benovic JL. Development of a yeast bioassay to characterize G protein-coupled receptor kinases. Identification of an NH2-terminal region essential for receptor phosphorylation. J. Biol. Chem. 2003;278:47466–47476. doi: 10.1074/jbc.M308257200. [DOI] [PubMed] [Google Scholar]
- 18.Pao CS, Barker BL, Benovic JL. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009;48:7325–7333. doi: 10.1021/bi900408g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterne-Marr R, Leahey PA, Bresee JE, Dickson HM, Ho W, Ragusa MJ, Donnelly RM, Amie SM, Krywy JA, Brookins-Danz ED, Orakwue SC, Carr MJ, Yoshino-Koh K, Li Q, Tesmer JJ. GRK2 activation by receptors: Role of the kinase large lobe and carboxyl-terminal tail. Biochemistry. 2009;48:4285–4293. doi: 10.1021/bi900151g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J. Biol. Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence data-bases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J. Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wald G, Brown PK. The molar extinction of rhodopsin. J. Gen. Physiol. 1953;37:189–200. doi: 10.1085/jgp.37.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goc A, Angel TE, Jastrzebska B, Wang B, Wintrode PL, Palczewski K. Different properties of the native and reconstituted heterotrimeric G protein transducin. Biochemistry. 2008;47:12409–12419. doi: 10.1021/bi8015444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada T, Le Trong I, Fox BA, Behnke CA, Stenkamp RE, Palczewski K. X-ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J. Struct. Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 27.Fahmy K, Sakmar TP. Regulation of the rhodopsin-transducin interaction by a highly conserved carboxylic acid group. Biochemistry. 1993;32:7229–7236. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 28.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 29.Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J. Biol. Chem. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrens DL, Khorana HG. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 31.Dombkowski AA. Disulfide by Design: A computational method for the rational design of disulfide bonds in proteins. Bioinformatics. 2003;19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 32.Heck M, Hofmann KP. Maximal rate and nucleotide dependence of rhodopsin-catalyzed transducin activation: Initial rate analysis based on a double displacement mechanism. J. Biol. Chem. 2001;276:10000–10009. doi: 10.1074/jbc.M009475200. [DOI] [PubMed] [Google Scholar]
- 33.Fahmy K, Sakmar TP. Light-dependent transducin activation by an ultraviolet-absorbing rhodopsin mutant. Biochemistry. 1993;32:9165–9171. doi: 10.1021/bi00086a023. [DOI] [PubMed] [Google Scholar]
- 34.Schadel SA, Heck M, Maretzki D, Filipek S, Teller DC, Palczewski K, Hofmann KP. Ligand channeling within a G-protein-coupled receptor. The entry and exit of retinals in native opsin. J. Biol. Chem. 2003;278:24896–24903. doi: 10.1074/jbc.M302115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heck M, Schadel SA, Maretzki D, Bartl FJ, Ritter E, Palczewski K, Hofmann KP. Signaling states of rhodopsin. Formation of the storage form, metarhodopsin III, from active metarhodopsin II. J. Biol. Chem. 2003;278:3162–3169. doi: 10.1074/jbc.M209675200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnis S, Hofmann KP. Photoregeneration of bovine rhodopsin from its signaling state. Biochemistry. 1995;34:9333–9340. doi: 10.1021/bi00029a008. [DOI] [PubMed] [Google Scholar]
- 37.Sommer ME, Farrens DL. Arrestin can act as a regulator of rhodopsin photochemistry. Vision Res. 2006;46:4532–4546. doi: 10.1016/j.visres.2006.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baameur F, Morgan DH, Yao H, Tran TM, Hammitt RA, Sabui S, McMurray JS, Lichtarge O, Clark RB. Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in β2-adrenergic receptor and rhodopsin phosphorylation. Mol. Pharmacol. 2010;77:405–415. doi: 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.