Abstract
Poikiloderma with Neutropenia (PN), Clericuzio-Type (OMIM #604173) is characterized by poikiloderma, chronic neutropenia, recurrent sinopulmonary infections, bronchiectasis, and nail dystrophy. First described by Clericuzio in 1991 in 14 patients of Navajo descent, it has since also been described in non-Navajo patients. C16orf57 has recently been identified as a causative gene in PN. The purpose of our study was to describe a spectrum of C16orf57 mutations in a cohort of PN patients including five patients of Athabaskan (Navajo and Apache) ancestry. Eleven patients from eight kindreds were enrolled in an IRB-approved study at Baylor College of Medicine. Five patients were of Athabaskan ancestry. PCR amplification and sequencing of the entire coding region of the C16orf57 gene was performed on genomic DNA. We identified biallelic C16orf57 mutations in all eleven PN patients in our cohort. The seven new deleterious mutations consisted of deletion (2), nonsense (3), and splice site (2) mutations. The patients of Athabaskan ancestry all had a common deletion mutation (c.496delA) which was not found in the six non-Athabaskan patients. Mutations in the C16orf57 gene have been identified thus far in all patients studied with a clinical diagnosis of PN. We have identified seven new mutations in C16orf57 in PN patients. One of these is present in all patients of Athabaskan descent, suggesting that c.496delA represents the PN-causative mutation in this subpopulation.
Keywords: Poikiloderma, Neutropenia, C16orf57, Athabaskan, Navajo, Bronchiectasis, Rothmund-Thomson Syndrome, RECQL4, Mutation
INTRODUCTION
Poikiloderma with Neutropenia (PN), Clericuzio-Type (OMIM #604173) is an autosomal recessive inherited disorder characterized by poikiloderma, pachyonychia (thickened nails), severe neutropenia, recurrent sinopulmonary infections, and chronic lung disease. First described as “immune deficient poikiloderma” by Clericuzio et al. in 1991 in 14 patients of Navajo descent [Clericuzio et al., 1991], it was later referred to as “Navajo poikiloderma” [Erickson, 1999]. This condition is among several genetic disorders enriched in the Southwestern Athabaskan native American populations, which comprises both Navajo and Apache groups [Erickson, 2009]. Subsequently, the disorder was described in patients not of Navajo descent; therefore, Wang and colleagues proposed renaming this disorder “Poikiloderma with Neutropenia” (PN) [Wang et al., 2003a]. In 2005, van Hove et al. described a Turkish family with PN, and he proposed renaming the disorder Clericuzio-type Poikiloderma with Neutropenia [van Hove et al., 2005].
Patients with PN have clinical overlap with Rothmund-Thomson syndrome (RTS) because of the poikilodermatous rash, nail abnormalities, palmar-plantar hyperkeratosis, and growth deficiency. Hematopoietic abnormalities such as aplastic anemia and myelodysplastic syndrome have been described in RTS [Narayan et al., 2001; Rizzari et al., 1996; Knoell et al., 1999]. However, unlike RTS, patients with PN have significant neutropenia that is non-cyclical in pattern and leads to recurrent sinopulmonary infections. Previous reports have shown that the neutropenia responds to growth factors such as Granulocyte Colony Stimulating Factor (GCSF) [van Hove et al, 2005; Arnold et al., 2010]. Bone marrow examinations in some of these patients have demonstrated maturation arrest of the myeloid lineage [van Hove et al., 2005; Mostefai et al., 2008]. Several groups have documented a lack of mutations in the RTS-associated RECLQ4 gene in PN patients [Wang et al., 2003a; van Hove et al., 2005].
In 2010, utilizing linkage analysis and array capture-mediated second-generation sequencing technologies, Volpi et al. identified C16orf57 (NM_024598.2) as a causative gene in PN in a large inbred Italian family [Volpi et al., 2010]. This was confirmed in a Moroccan family [Tanaka et al., 2010] and in another European family [Arnold et al., 2010]. The purpose of the current study was to examine a larger group of PN patients to determine the prevalence and spectrum of C16orf57 mutations in patients with PN including those of Athabaskan descent.
MATERIALS AND METHODS
Patients
Eleven patients from eight kindreds (including three sibling pairs) were enrolled in an Institutional Review Board approved protocol for human subject research at Baylor College of Medicine. All patients or their parents provided signed informed consent to participate in the study. Five patients from four kindreds were of Athabaskan ancestry (Table I). A sibling pair was of Asian Indian descent, and the remaining patients were of Caucasian/European descent. Three kindreds (Families 1, 5 and 6) have been previously described [Wang et al., 2003a]. Patient characteristics are listed in Table I.
Table I.
Poikiloderma with Neutropenia (PN) Patient Information
| Patient # | Family # | Gender | Age at ascertainment | Ethnicity | Clinical Features | C16orf57 Allele 1 Mutation | C16orf57 Allele 2 Mutation |
|---|---|---|---|---|---|---|---|
| 1 | 1 | F | 12 y | 100% Navajo | Neutropenia; poikiloderma*; pachyonychia; recurrent OM; asthma | c.496delA | c.496delA |
| 2 | 1 | F | 15 y | 100% Navajo | Neutropenia; poikiloderma; pachyonychia; recurrent sinopulmonary infections and OM; mastoiditis; asthma | c.496delA | c.496delA |
| 3 | 2 | M | 17 mo | 100% Navajo | Rash started age 5 mo; diagnosed PN age 9 mo due to neutropenia; transient thrombocytopenia; pachyonychia; chronic cough; h/o diarrhea; normal bone marrow studies | c.496delA | c.496delA |
| 4 | 3 | F | 3 y | 100% Apache | Diagnosed PN age 3 y due to rash and neutropenia; recurrent respiratory infections | c.496delA | c.496delA |
| 5 | 4 | F | 5 y | 25% Navajo 75% Caucasian |
Rash age 4 mo; initially diagnosed with RTS age 18 mo; diagnosed with PN age 5 y due to neutropenia; pachyonychia; recurrent OM; sinopulmonary and urinary tract infections; h/o chronic diarrhea; calcinosis cutis | c.496delA | c.489_492del4 |
| 6 | 5 | F | 4 y | Turkish-British | Rash started age 3 mo; diagnosed with PN age 20 mo due to neutropenia and poikiloderma; pachyonychia; hyperkeratosis soles; cellulitis, mastoiditis; h/o mild anemia; bone marrow normal except increased myeloid precursors; received GCSF | c.541C>T/Q181X | c.541C>T/Q181X |
| 7 | 6 | M | 4 y | Scottish | Rash started age 6 mo; neutropenia since birth; transient thrombocytopenia; recurrent sinopulmonary infections; pachyonychia; chronic diarrhea; bone marrow showed delayed maturation of neutrophils | c.489_492del4 | c.693+1G>T |
| 8 | 6 | F | 4 y | Scottish | Rash started age 1 y; neutropenia since birth; recurrent sinopulmonary infections; hyperkeratosis; calcinosis cutis | c.489_492del4 | c.693+1G>T |
| 9 | 7 | F | 7 y | 6.25% Cherokee 93.75% Caucasian |
Rash started age 4 mo; diagnosed with PN age 1 y due to neutropenia; recurrent OM, bronchitis, cellulitis; pachyonychia; autoimmune thyroiditis; h/o chronic diarrhea in infancy; calcinosis cutis; received GCSF; normal bone marrow studies | c.415 C>T/Q139X | c.673 C>T/Q225X |
| 10 | 8 | M | 5 y | Asian Indian (Punjabi) | Diagnosed with PN age 2 y due to rash and neutropenia; recurrent sinopulmonary infections; positive sweat chloride tests | c.266 -1G>A | c.266 -1G>A |
| 11 | 8 | M | 13 mo | Asian Indian (Punjabi) | Starting to develop rash at time of ascertainment at age 13 mo | c.266 -1G>A | c.266 -1G>A |
M Male; F Female; y year; mo month; h/o history of; OM otitis media [Wang et al., 2003a].
All patients had the classic poikiloderma of PN which starts peripherally affecting the limbs first and then extending centrally to the face. This is in contrast to patients with RTS in whom the rash starts on the face and then extends to the limbs [Wang et al., 2003a].
DNA Mutation Analysis
Genomic DNA was isolated from peripheral blood leukocytes or EBV-transformed lymphoblastoid cell lines (LCL) using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). LCLs were cultured in RPMI 1640 medium with L-glutamine (300 mg/L) (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (ATCC, Manassas, VA), in 37°C humidified incubator with 5% CO2. PCR amplification and sequencing of the C16orf57 gene were performed for all eleven patients using previously published primers and conditions [Volpi et al., 2010] and included the entire coding region and conserved splice sites of the C16orf57 gene. PCR amplification was performed using the Advantage-GC complementary DNA PCR kit (Clontech, Palo Alto, CA) on a PTC-200 Peltier Thermal Cycler (Bio- Rad Laboratories, Inc., Hercules, California). PCR conditions consisted of an initial denaturation at 95°C for 3 min, followed by 29 cycles at 94°C for 30 sec, 60°C for 30 sec, 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were purified using Wizard SV Gel and PCR Clean-Up System (Promega, Madison, USA). PCR products were submitted for automated sequencing on an ABI Prism 3730xl automated fluorescent DNA sequencer (Applied Biosystems, Carlsbad, CA) in both forward and reverse directions. Nucleotide sequences were compared to the reference sequences of the wild type C16orf57 gene from the NCBI Entrez Nucleotide database (NT_010498.15, NC_000016.9). Sequence data were analyzed using Sequencher software (version 4.8, Gene Codes Corporation, Ann Arbor, MI).
RT-PCR Analysis of Splice Site Mutations
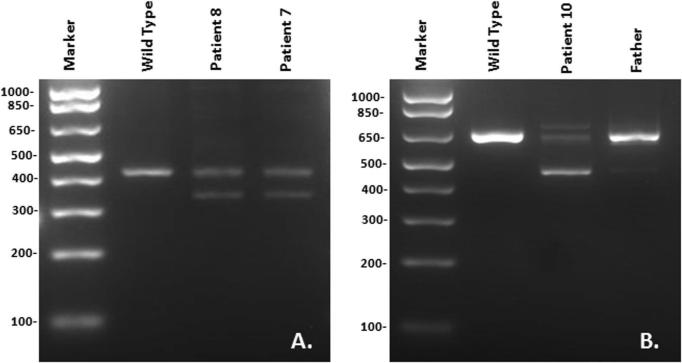
Total RNA was extracted from primary fibroblasts or LCLs using the RNeasy Mini Kit (QIAGEN Inc, Valencia, CA). Fibroblasts were cultured in MEM Alpha medium with L-glutamine (292 mg/L) (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (ATCC, Manassas, VA) in a 37°C humidified incubator with 5% CO2. LCLs were cultured as described above. cDNA was synthesized using oligo(dT) primer and SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). PCR amplification of the C16orf57 gene transcripts was performed for patients, parent and wild type samples using specific primers that spanned the two regions of intronic mutations (primer sequences in Figure 2) and the Advantage-GC complementary DNA PCR kit (Clontech, Palo Alto, CA). PCR conditions consisted of an initial denaturation at 95°C for 3 min, followed by 36 cycles at 94°C for 30 sec, 60°C for 30 sec, 72°C for 1 min, and a final extension at 72°C for 10 min.
FIG. 2.
Reverse-transcriptase polymerase chain reaction (RT-PCR) analysis of splice site mutations in the C160rf57 gene. A: RT-PCR of segment that includes the c.693+1G>T donor splice site mutation and spans exon 3/4 to the 3’ UTR using primers: Forward – CCTCCTTCCACAGATTCTTC. Reverse – GTTCCTCCATCTCAGCCTG. Both siblings (Patients 7 and 8) are heterozygous for this mutation. B: RT-PCR of segment that includes the c.266-1G>A acceptor splice site mutation and spans the 5’ UTR to exon 5 using primers: Forward – CTGCTCTGGTGGTCTTGGAT; Reverse -AGAAGGATCCTGGTAGAAAGTG. The patient is homozygous and his father is heterozygous for this mutation.
RESULTS
Biallelic deleterious mutations in C16orf57 were identified in all PN patients analyzed in our cohort of eleven patients. The seven mutations consisted of deletion (2), nonsense (3), and splice site (2) mutations (Table I). Seven patients were homozygous, and four patients were compound heterozygous for mutations. The five patients of Athabaskan ancestry all shared a deletion mutation (Fig 1) which was not found in the six non-Athabaskan patients. The full Athabaskan patients were homozygous for this mutation while the quarter Navajo patient (Patient #5) was compound heterozygous for this mutation and another deletion mutation. We sequenced the parents of Patient #5 and found that the c.496delA mutation was inherited from the maternal side which was part Navajo. We also sequenced the parents from Families 4-8, and found that the parents were heterozygous for mutations detected in the probands (data not shown), consistent with autosomal recessive inheritance pattern. For the two splice site mutations (c.693+1G>T and c.266-1G>A), to determine whether these intronic changes affected splice products of C16orf57, RT-PCR was performed on the patients’ samples and compared to wild type samples. In both cases, the patients produced bands that differed from wild type controls, suggesting that missplicing of C160rf57 occurred in the patients. For Patient 10, RNA was available from his father, whose splice products were consistent with wild type sequence on one C16orf57 allele and the splice site mutation in the other allele (Fig 2). All C16orf57 mutations in PN patients published to date are summarized in Table II in the order of occurrence in the gene.
FIG. 1.

Chromatograms in forward and reverse directions showing the c.496delA mutation in exon 4 in the homozygous state. Arrow denotes location of the deletion.
TABLE II.
Summary of C16orf57 Mutations in PN Patients
| Mutation | Type | Location | Reference |
|---|---|---|---|
| c.179delC | Deletion/frameshift | Exon 2 | Tanaka et al., 2010 |
| c.243G>A/W81X | Nonsense | Exon 2 | Arnold et al., 2010 |
| c.266-1G>A | Splice site (acceptor) | Intron 2 | This study |
| c.415C>T/Q139X | Nonsense | Exon 3 | This study |
| c.489_492del4 | Deletion/frameshift | Exon 4 | This study |
| c.496delA | Deletion/frameshift | Exon 4 | This study |
| c.502A>G/R168G | Missense | Exon 4 | Volpi et al., 2009 |
| c.504-2A>C | Splice site (acceptor) | Intron 4 | Volpi et al., 2009 |
| c.541C>T/Q181X | Nonsense | Exon 5 | This study |
| c.673C>T/Q225X | Nonsense | Exon 6 | This study |
| *c.683_693+1del12 | Deletion | Exon 6 | Volpi et al., 2009 |
| c.693+1G>T | Splice site (donor) | Intron 6 | This study |
Previously designated c.666_676+1del12 (Volpi et al., 2010)
DISCUSSION
Mutations in the C16orf57 gene have been identified in all patients (n=19) with a clinical diagnosis of PN who have been sequenced thus far, and they include five previously published mutations and seven new mutations from this study. Thus, there is no evidence for genetic heterogeneity in contrast to RTS where only about two-thirds of patients are found to have mutations in the RECQL4 gene [Wang et al., 2003b]. The function of the C16orf57 gene product is currently not known, but it has been suggested to interact directly SMAD4 and indirectly with RECQL4 through SMAD4-mediated signaling [Volpi et al., 2010]. The role of C16orf57 in normal hematopoiesis and its role in pathologic processes such as myelodysplasia are not known. All patients with PN have significant neutropenia leading to recurrent infections, which are predominantly sinopulmonary in location. While the neutropenia is not cyclical in a clockwork fashion, as in the case of cyclic neutropenia, it is variable, as neutrophil counts can range from less than 0.2 × 103/ul to normal levels. The neutropenia has been described in some reports to be responsive to infections as well as to GCSF, both in vivo and in vitro [van Hove et al., 2005; Arnold et al., 2010], although some patients do not seem to mount a response [Mostefai et al., 2008]. Transient thrombocytopenia has been described in several patients, and a few patients have also had transient anemia [van Hove et al., 2005; Mostefai et al., 2008; Arnold et al., 2010], suggesting that this condition may have more global bone marrow involvement rather than a defect isolated to the myeloid lineage. PN patients may be at increased risk for myelodysplasia and leukemic transformation. Thus far there have been reports of three patients with C16orf57 mutations who developed these hematologic disorders. One patient with myelodysplastic syndrome previously diagnosed with RTS was later found to have mutations in C16orf57 [Pianigiani et al., 2001; Volpi et al., 2010]. Two patients recently described by Walne et al. [2010] with C16orf57 mutations developed acute myelogenous leukemia. One had previously been diagnosed with dyskeratosis congenita, while the other had previously been diagnosed with RTS [Porter et al., 1999]. The exact prevalence of hematologic complications in PN is not currently known, but ongoing hematologic monitoring of these patients is warranted. The discovery of the C16orf57 gene defect in PN allows for a definitive diagnostic test for this disorder, provides a basis for genetic counseling and education of medical personnel and families about the infectious risks in these patients, and opens opportunities for basic research into hematopoiesis. Our finding of a uniform gene defect among all patients of Athabaskan descent in our cohort suggests that this mutation is causative in this group and will permit a more targeted and rapid diagnostic approach for PN in this subpopulation.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the patients and families who participated in this research study. They also acknowledge contributions from Moise Levy, MD for clinical input, and Ta-Tara Rideau and JoAnn Milisa for patient coordination support. Portions of this work were supported by National Institutes of Health grants NICHD K08HD42136 (to L.L.W.), NIH-K12 HD41648 (BCM-Child Health Research Center), RR000188-42 (BCM–General Clinical Research Center), NIH-HD024064 (BCM–Intellectual and Developmental Disabilities Research Center, Tissue Culture Core), as well as support from the Doris Duke Charitable Foundation (L.L.W.), Carousel Young Friends of Texas Children's Cancer Center (L.L.W.) and the Holsclaw Family Professorship of Human Genetics and Inherited Diseases (R.P.E.).
REFERENCES
- Arnold AW, Itin PH, Pigors M, Kohlhase J, Bruckner-Tuderman L, Has C. Poikiloderma with neutropenia: a novel C16orf57 mutation and clinical diagnostic criteria. Br J Dermatol. 2010;163:866–869. doi: 10.1111/j.1365-2133.2010.09929.x. [DOI] [PubMed] [Google Scholar]
- Clericuzio, Hoyme HE, Aase JM. Immune deficient poikiloderma: A new genodermatosis. Am.J Hum Genet. 1991;49(Suppl):A661. [Google Scholar]
- Erickson RP. Southwestern Athabaskan (Navajo and Apache) genetic diseases. Genet Med. 1999;1:151–157. doi: 10.1097/00125817-199905000-00007. [DOI] [PubMed] [Google Scholar]
- Erickson RP. Autosomal recessive diseases among the Athabaskans of the southwestern United States: recent advances and implications for the future. Am J Med Genet A. 2009;149A:2602–2611. doi: 10.1002/ajmg.a.33052. [DOI] [PubMed] [Google Scholar]
- Knoell KA, Sidhu-Malik NK, Malik RK. Aplastic anemia in a patient with Rothmund-Thomson syndrome. J Pediatr Hematol Oncol. 1999;21:444–446. doi: 10.1097/00043426-199909000-00021. [DOI] [PubMed] [Google Scholar]
- Mostefai R, Morice-Picard F, Boralevi F, Sautarel M, Lacombe D, Stasia MJ, McGrath J, Taieb A. Poikiloderma with neutropenia, Clericuzio type, in a family from Morocco. Am J Med Genet A. 2008;146A:2762–2769. doi: 10.1002/ajmg.a.32524. [DOI] [PubMed] [Google Scholar]
- Narayan S, Fleming C, Trainer AH, Craig JA. Rothmund-Thomson syndrome with myelodysplasia. Pediatr Dermatol. 2001;18:210–212. doi: 10.1046/j.1525-1470.2001.018003210.x. [DOI] [PubMed] [Google Scholar]
- Pianigiani E, De AG, Andreassi A, Rubegni P, Fimiani M. Rothmund-Thomson syndrome (Thomson-type) and myelodysplasia. Pediatr Dermatol. 2001;18:422–425. doi: 10.1046/j.1525-1470.2001.01971.x. [DOI] [PubMed] [Google Scholar]
- Porter WM, Hardman CM, Abdalla SH, Powles AV. Haematological disease in siblings with Rothmund-Thomson syndrome. Clin Exp Dermatol. 1999;24:452–454. doi: 10.1046/j.1365-2230.1999.00530.x. [DOI] [PubMed] [Google Scholar]
- Rizzari C, Bacchiocchi D, Rovelli A, Biondi A, Cantu'-Rajnoldi A, Uderzo C, Masera G. Myelodysplastic syndrome in a child with Rothmund-Thomson syndrome: a case report. J Pediatr Hematol Oncol. 1996;18:96–97. doi: 10.1097/00043426-199602000-00020. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Morice-Picard F, Lacombe D, Nagy N, Hide M, Taieb A, McGrath J. Identification of a homozygous deletion mutation in C16orf57 in a family with Clericuzio-type poikiloderma with neutropenia. Am J Med Genet A. 2010;152A:1347–1348. doi: 10.1002/ajmg.a.33455. [DOI] [PubMed] [Google Scholar]
- Van Hove JL, Jaeken J, Proesmans M, Boeck KD, Minner K, Matthijs G, Verbeken E, Demunter A, Boogaerts M. Clericuzio type poikiloderma with neutropenia is distinct from Rothmund-Thomson syndrome. Am J Med Genet A. 2005;132A:152–158. doi: 10.1002/ajmg.a.30430. [DOI] [PubMed] [Google Scholar]
- Volpi L, Roversi G, Colombo EA, Leijsten N, Concolino D, Calabria A, Mencarelli MA, Fimiani M, Macciardi F, Pfundt R, Schoenmakers EF, Larizza L. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am J Hum Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Hum Mol Genet. 2010 Sep 17; doi: 10.1093/hmg/ddq371. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Gannavarapu A, Clericuzio CL, Erickson RP, Irvine AD, Plon SE. Absence of RECQL4 mutations in poikiloderma with neutropenia in Navajo and non-Navajo patients. Am J Med Genet A. 2003a;118A:299–301. doi: 10.1002/ajmg.a.10057. [DOI] [PubMed] [Google Scholar]
- Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, Lev D, Rogers M, Zackai EH, Plon SE. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003b;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]