Abstract
The current model of human breast cancer progression proposes a linear multistep process which initiates as flat epithelial atypia (FEA), progresses to atypical ductal hyperplasia (ADH), evolves into DCIS, and culminates in the potentially lethal stage of invasive ductal carcinoma. For several decades a major challenge to human breast cancer research has been the identification of the molecular alterations associated with the different stages of breast cancer progression. Until recently, progress in attaining this goal has been hampered by technical limitations associated with applying advanced molecular technologies to the microscopic preinvasive stages of breast tumorigenesis. Recent advances in comprehensive, high-throughput genetic, transcriptomic and epigenetic technologies in combination with advanced microdissection and ex vivo isolation techniques have provided for a more complete understanding of the complex molecular genetic and molecular biological inter-relationship of the different stages of human breast cancer evolution. Here we review the molecular biological data that suggest breast cancer develops and evolves along two distinct molecular genetic pathways. We also briefly review gene expression and epigenetic data that support the view of the tumor microenvironment as an important coconspirator rather than a passive bystander during human breast tumorigenesis.
Keywords: Flat epithelial atypia, atypical ductal hyperplasia, ductal carcinoma in situ, breast cancer progression, genetics, transcriptomics, epigenetic, tumor environment
Introduction
Breast cancer is the most frequent carcinoma in females, and the second most common cause of cancer related mortality in women. Approximately 54,000 and 207,000 new cases of in situ and invasive breast carcinoma, respectively, are expected to be diagnosed in 2010 in the United States. Within this same period, breast cancer will account for an estimated 39,500 deaths among US women [1]. Over the past two decades, encouraging trends in both breast cancer incidence and mortality have been observed. Overall, breast cancer incidence rates have leveled off since 1990 with a decrease of 3.5 percent per year from 2001 to 2004 [2–4]. Most notably, during this same time period, breast cancer mortality rates have declined 24% with the largest impact among young women and women with estrogen receptor (ER)-positive disease [5, 6]. The decline in breast cancer mortality has been attributed to the combination of early detection with screening programs and the advent of more efficacious adjuvant systemic therapy. Continued advances in our understanding of the molecular biology of breast cancer progression have aided in the discovery of novel pathway-specific targeted therapeutics, and the emergence of such effective therapeutics is currently driving the need for molecular-based, “patient–tailored” treatment planning. Knowledge gained from studying the molecular pathology of human breast cancer progression, and integration and implementation of this knowledge in the clinical setting promises to further reduce breast cancer morbidity and mortality.
Proposed models of human breast cancer progression
The ductal and lobular subtypes constitute the majority of all breast cancers worldwide with the ductal subtype accounting for 40–75% of all diagnosed cases [7, 8]. Epidemiologic and morphologic observations led to the formulation of several linear models of breast cancer initiation, transformation and progression as depicted in Figure 1. For the ductal subtype, two models have been proposed. The first “ductal” model, put forth by Wellings and colleagues, recognizes flat epithelial atypia (FEA), atypical ductal hyperplasia (ADH) and ductal carcinoma in situ (DCIS) as the non-obligate precursors of invasive and metastatic ductal carcinoma [9–13]. The second “ductal” model, supported by epidemiological studies, proposed usual epithelial ductal hyperplasia (UDH) as an intermediate stage of progression between FEA and DCIS [14, 15]. Immunohistochemical [16–18] and recent molecular biological evidence (as described below) strongly suggests that UDH is not a precursor to ADH and that this second model of progression is likely invalid. For the lobular subtype, the progression scheme recognizes atypical lobular hyperplasia (ALH) and lobular carcinoma in situ (LCIS) as the non-obligate precursor lesions to invasive lobular carcinoma.
Figure 1.
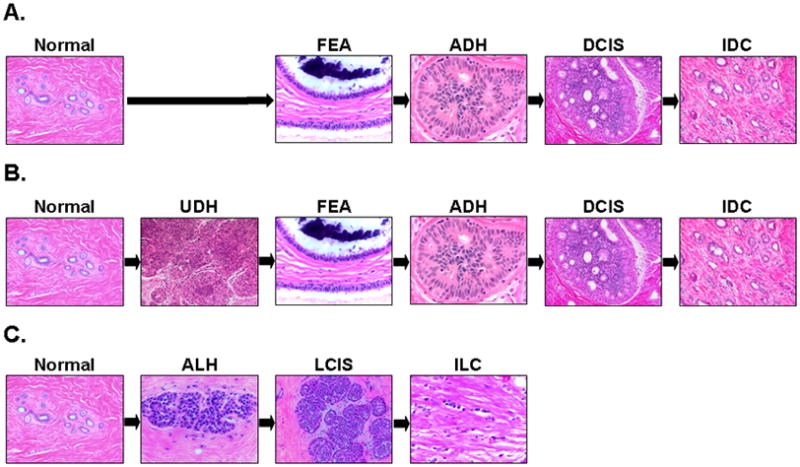
Models of breast cancer progression. (A) The classic model of breast cancer progression of the ductal type proposes neoplastic evolution initiates in normal epithelium (normal), progresses to flat epithelial atypia (FEA), advances to atypical ductal hyperplasia (ADH), evolves to ductal carcinoma in situ (DCIS) and culminates as invasive ductal carcinoma (IDC). Immunohistochemical, genomic and transcriptomic data strongly support the evidence of a continuum from FEA to ADH, DCIS and IDC, indicating FEA as the potential non-obligate precursor of ADH. In contrast, the alternative model of breast ductal cancer progression (B), which was mostly based on epidemiologic and morphologic observations, proposes usual ductal hyperplasia (UDH) instead of FEA as direct precursor to ADH. However recent studies have shown that UDH has a distinct immunohistochemical and molecular profile from FEA and probably represents a biologic dead end. (C) The model of lobular neoplasia proposes a multistep progression from normal epithelium to atypical lobular hyperplasia, lobular carcinoma in situ (LCIS) and invasive lobular carcinoma (ILC).
The cell of origin of breast cancer: the clonal and stem cell hypotheses
The two leading models accounting for breast carcinogenesis are the sporadic clonal evolution model and the cancer stem cell (cSC) model [19–21]. According to the sporadic clonal evolution hypothesis, any breast epithelial cell can be the target of random mutations. The cells with advantageous genetic and epigenetic alterations are selected over time to contribute to tumor progression. The alternative cSC model postulates that only stem and progenitor cells, (representing a small fraction of the tumor cells within the cancer), can initiate and maintain tumor progression. These three hypotheses are not mutually exclusive and it has been suggested that stem cells might undergo clonal evolution providing a dynamic link between the two models [22, 23]. In the remainder of this section, we will provide a concise overview of normal breast stem cells and a review of recent evidence supporting the stem cell hypothesis as it relates to the origin of the different molecular subtypes of human breast cancer.
Normal breast stem cells (nBSC) are long-lived tissue resident cells capable of self-renewal activity and multilineage differentiation that can recapitulate the breast tubulo-lobular architecture that is composed of luminal and myoepithelial cells [24–30]. Normal breast stem cells can be identified and isolated based on their immunophenotypic, and functional features and are generally characterized by low expression of heat stable antigen (CD24), high level expression of the hyaluronic acid receptor (CD44), and expression of aldehyde dehydrogenase (ALDH1) [31–34] Furthermore, nBSCs lack of expression of epithelial cell adhesion molecule (EpCAM), hematopoietic and endothelial markers as well as the estrogen and progesterone receptors. nBSCs mostly reside in ducts in the suprabasal position and are surrounded by proliferating progenitor cells and nBSCs represent a small fraction (estimated at 1:2000 epithelial cells) of mammary gland cells in the human breast [28, 30, 35]. Until recently, maintenance of the BSC population has been poorly understood. Independently, two groups recently provided evidence that progesterone indirectly controls the number and activity of nBSCs through paracrine signaling from the RANK ligand secreted by neighboring PR-positive breast epithelial cells [36, 37]. These findings will likely accelerate our understanding of steroid-hormone control of normal breast development, and provide for a novel mechanism by which steroid hormones can modulate the development of breast cancer.
As normal breast cancer stem cells are long-time tissue residents, it has been proposed that such cells are candidates for accumulating genetic and epigenetic modifications. It has been further proposed that such molecular alterations result in deregulation of normal self-renewal leading to the development of a cancer stem cell (cSC). It is believed that the cSC undergoes asymmetric division maintaining the stem cell population while at the same time differentiating into committed progenitor(s) cells that give rise to the different breast cancer subtypes [21, 38, 39]. A second scenario as it relates to breast cancer development, is one in which the cancer initiating cells are derived from committed progenitor cells that spawn different breast cancer subtypes (Figure 2). In support of the latter scenario, Lim et al. in a series of elegant experiments provide functional and molecular evidence that progenitor cells within the luminal compartment of the human breast represent a cancer initiating population in BRCA1 mutation carriers and possibly other basal subtype breast tumors [40]. Most recently, data from a pre-clinical mouse model of BRCA1 deficiency provide further evidence that sporadic basal-like and BRCA-1 tumors originate from luminal rather than resident normal breast stem cells [41]. Data from this latter study suggest a third possible model of breast cancer tumorigenesis in which the inherent plasticity of stem and progenitor cells during tumorigenesis contributes to the final tumor phenotype beyond that of “cell of orgin” alone [41].
Figure 2.
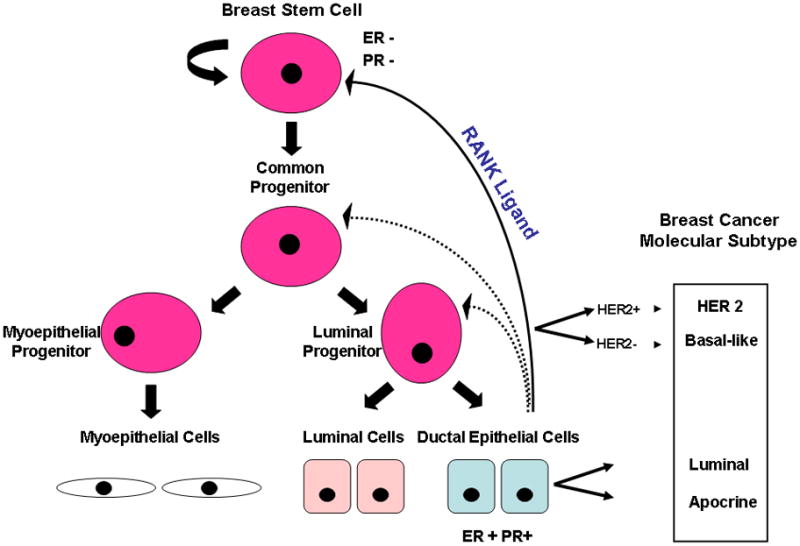
Schematic of the hierarchy of normal breast epithelial development with possible links to the tumor-initiating cells of the different molecular cancer subtypes and to the role of steroid hormones in the control of the mammary stem and progenitor cells. The breast stem cell (BSC) with intrinsic self-renewal potential (curved block arrow) differentiates into a common progenitor that gives rise to committed myoepithelial and luminal progenitors that ultimately differentiate into myoepithelial, luminal and ductal epithelial cells (left hand side). The subpopulations of normal cells show a gene signatures that are similar to that of previously described cancer molecular subtypes (right hand side) providing evidence of a link (bold arrows) between the two. While the luminal progenitor subpopulation is enriched in the HER2 (HER2+) and basal-like cancers (HER2−), the more differentiated ductal epithelial cell subpopulation is enriched in the luminal (A/B) tumors. Mammary stem cells and potentially progenitor cells subpopulations are indirectly profoundly responsive to steroid hormone signaling despite their estrogen and progesterone receptors negative (ER− and PR−) phenotype. During the reproductive cycle and pregnancy the surge of progesterone stimulates the differentiated ductal epithelial cells to release the RANK Ligand that elicits expansion of the stem cell population, via paracrine signaling (blue arrows).
Molecular analysis of the different stages of breast cancer progression
Worldwide, nearly 80% of all diagnosed preinvasive invasive breast cancers are of the ductal subtype. With this in mind, the next two sections of this review will center on a discussion of the genomic and transcriptomic data resulting from the interrogation of the ductal subtype, followed by a discussion of similar molecular data derived from the lobular subtype. A discussion of the less common, special types of breast carcinoma is beyond the scope of this review and a review of the different histological subtypes can be found elsewhere [42].
Invasive breast cancer: genomic and transcriptomic alterations
Over the past decade, the molecular interrogation of invasive human breast cancer at the genomic and transcriptomic (gene expression) levels has been one of the most commonly studied disease entities in all of cancer biology. Data generated from this vast body of work serves as the logical reference point for the subsequent discussion of molecular analysis of the earlier preinvasive stages of breast cancer progression.
Invasive ductal carcinoma (IDC) is characterized by profound heterogeneity at the histomorphologic and clinical levels. As histological grade is closely related to the clinical behavior of tumors, early molecular studies of IDC focused on the relationship of tumor genomic alterations with tumor grade, and such comparisons have contributed significantly to understanding of breast cancer progression [43–45]. Several groups demonstrated that that low-grade IDCs displayed fewer overall chromosomal aberrations as compared with high grade IDCs, and that these quantitative differences are also associated with unique qualitative differences [46–48]. More specifically, low-grade tumors IDCs display frequent recurrent chromosomal loss of 16q and gains of 1q,16p and 8q, whereas high-grade tumors exhibit recurrent losses of 8p,11q,13q, 1p and 18q, recurrent gains of 8q,17q,20q and 16p, and frequent high-level amplifications of 17q12 and 11q13 [47, 48]. Intermediate-grade IDCs display a combination of low-grade and high-grade genomic alterations suggesting that this group of tumors consists of a mixture of “low-grade-like” and “high-grade-like” IDCs [49]. The most important finding emerging from this work as it relates to breast cancer classification and breast cancer progression is the frequent versus infrequent loss of chromosome 16q in low-grade and high-grade IDCs, respectively. This unique pattern of chromosomal loss strongly argues against the epidemiological- and pathological-based hypothesis that most of low-grade IDCs progress to high-grade IDCs through various steps of dedifferentiation [50, 51].
Numerous transcriptional (gene expression) profiling studies have further contributed to not only our understanding of biological and clinical heterogeneity of IDC, but also to our understanding of breast cancer progression. In an effort to understand breast cancer heterogeneity at a molecular level Perou and colleagues performed gene expression profiling that resulted in at least four distinct “intrinsic” subtypes that include two ER positive (luminal A and luminal B) and two ER negative (ERBB2 and basal subtypes) [52–56]. The luminal A and luminal B are the most common subtypes usually representing low to intermediate grade tumors characterized by the expression of genes that are commonly expressed by normal ductal epithelial cells. Luminal A tumors show high expression of ER and PR-related genes, GATA binding protein 3, low expression of proliferation-associated genes, and lack expression of Her-2 [52, 53, 57, 58], while luminal B tumors show decreased expression of ER, overexpression of HER2, and a higher expression of proliferation-related genes [59]; since the original classification by Perou and colleagues, results from several studies suggest that the two luminal subtypes lie on a biological continuum represented by ER expression and proliferation [60–62]. The ERRB2 tumor subtype usually represent high grade tumors, and is characterized by expression of HER2 and other genes on the ERBB2 amplicon such as GRB7, by lack of expression of ER and PR. The basal subtype usually represents high grade tumors displaying necrosis, prominent lymphocytic infiltrate and pushing border. Basal IDCs express cytokeratins and other factors associated with basal/myoepithelial cells and such tumors are most commonly characterized by the lack of expression of ER, PR and HER2 expression. In addition to the luminal, basal and HER2 molecular subtypes, the molecular apocrine subtype has been identified by Farmer et al., and is estimated to represent 8–14% in published microarray cohorts [63]. Molecular apocrine carcinomas are characterized by the presence and absence of the androgen receptor (AR) and the estrogen receptor (ER), respectively, and such tumors represent most ER negative tumors found outside the basal subgroup. Notably, in multiple retrospective-based studies, the gene expression signatures associated with the intrinsic subtypes of IDCs, as well as molecular apocrine carcinoma, were shown to have significant clinical prognostic power [53, 55, 56, 63–65] (reviewed in ref. 65). Until recently, the robustness of the “intrinsic” molecular subtype classification system for human breast cancer samples has been relatively unquestioned. Studies published by several group, including that of Weigelt and colleagues, demonstrate that the molecular breast cancer subtypes (luminal A, luminal B and HER2), with the exception of the basal subtype, are not highly reproducible and that subtle variation in classification methodologies exert a significant effect on classification assignments for individual breast cancer patients [60, 62]. The results of these studies highlight the urgent need for standardization of analytical methods for microarray-based breast cancer classification systems before such stratification schemes are implemented in clinical trials and in clinical practice.
Since the original Perou et al. report [52], multiple transcriptional profiling-based studies have identified other unique gene expression signatures that predict breast cancer outcome in a similar manner to that of the intrinsic subsets [65–69]. Curiously, although these different gene expression signatures consisted of fairly unique gene sets, they all shared the ability to predict breast cancer outcome in a similar manner, suggesting that these signature are tracking with a common biological principle. Data presented from the comparative breast cancer gene expression study by Fan et al. strongly suggested this common biological principle involved molecular pathways centered on tumor grade and cell proliferation [70]. Several subsequent comparative gene expression studies have provided significant evidence in support of this view by demonstrating that simple tumor grade-/proliferation-based gene expression biomarkers possess prognostic equivalence to more complex multigene expression signatures [61, 71–75]. Most notably, these tumor grade-based gene expression studies provide strong evidence that most IDCs can be stratified into either low grade-like (indolent) or high grade-like (aggressive) clinical outcome categories. This recent gene expression data in conjunction with aforementioned genomic data suggest that tumor grade may reflect two divergent evolutionary, biological and clinical behavior pathways.
Regarding invasive lobular carcinomas (ILC) which represent about 10–14% of all invasive breast cancers, conventional cytogenetic and CGH analysis have shown, not surprisingly, that classic ILCs harbor recurrent loss of chromosome 16q, the genetic hallmark of the low-grade ductal carcinomas [76–80]. These data support the concept that classic ILCs share a common evolutionary genetic pathway with low grade IDCs [81]. In a most recent study by Weigelt and colleagues, comparative gene expression analysis of ILCs to grade- and molecular subtype-matched IDCs has revealed that ILCs possess a unique transcriptional profile [82]. This unique ILC gene expression signature is characterized by genes associated cell adhesion, cell-to-cell signaling and actin cytoskeletal signaling, and is consistent with a pattern one might expect in ILCs cells that characteristically display loss of cell-cell cohesion and loss of e-cadherin protein expression. Although several studies have shown that pleomorphic ILCs share genetic changes with both classic ILCs and high-grade IDCs (in the form of loss of 16q, and gain of 1q, and 17q12 amplification, respectively [83, 84]), the studies by Weigelt et al. also demonstrated that classic ILCs and pleomorphic ILCs possess highly similar gene expression patterns (<0.1% of all gene were differentially expressed between the two subtypes) [82]. Thus, consistent with previously observations by Allred et al., this gene expression data support the concept that pleomorphic ILC represents tumor grade progression within a genetic pathway shared by ILC and IDC [85].
Preinvasive stages of breast cancer: genomic and transcriptomic alterations
In the past, our inability to easily and accurately interrogate the preinvasive stages of breast cancer has created a significant impediment to our understanding of breast cancer progression. However, a number of advances in microdissection, ex vivo cellular purification and high-throughput microgenomic technologies have reshaped our view of breast cancer progression. As described in the section below, noteworthy similarities exist between preinvasive and invasive breast cancer.
Morphological, epidemiological and immunohistochemical data support the hypothesis that FEA, ADH and low-grade DCIS represent an evolutionary continuum [11, 86, 87]. Additional support for this hypothesis is provided by several genomic- and transcriptomic-based studies. Similar to that observed for IDC, several comparative genomic-based studies have revealed that DCIS is a genetically advanced process and that distinct patterns of genomic alterations in DCIS are associated with tumor grade. More specifically, using comparative genomic hybridization (CGH), Buerger et al. demonstrated frequent loss of 16q in low-grade DCIS and complex genomic alterations that include 13q loss and high level amplifications of 17q12 and 11q13 in high grade DCIS [88]. Interestingly, CGH analysis of synchronous and metachronous IDC and DCIS, revealed a near identical pattern of genomic alterations supporting a molecular continuum between DCIS and IDC [48, 49, 88]. With regard to ADH and FEA, several loss-of-heterozygosity (LOH)- and CGH-based studies have shed light on the molecular alterations associated with these lesions as they relate to DCIS and IDC. Several LOH-based studies including a comprehensive study performed by O’Connell et al. identified loss of 16q as a hot spot in ADH and revealed that this alteration is most frequently shared with low-grade rather than high grade DCIS [89]. Similarly, Moinfar and colleagues reported a particularly high rate of LOH at chromosome 16q in FEA [90]. Lastly, a recent comprehensive CGH-based study by Simpson and colleagues further strengthened the previous observations by demonstrating that FEA exhibits recurrent chromosomal aberrations (including 16q loss) that have significant overlap with ADH, low grade DCIS and low grade IDC [91]. Notably, this common pattern of genomic alterations is not observed in usual ductal hyperplasia (UDH), a lesion that displays rare and randomly distributed chromosomal alterations that are highly similar to those observed in normal breast tissue [89, 92–94]. Until recently, identification of the precursor lesion of high grade DCIS remained elusive. Immunohistochemical- and CGH-based data by two independent groups support the notion that a subset of microglandular adenosis lesions may represent a precursor to a subset of triple negative, high-grade, DCIS lesions [95, 96]. Together, these studies provide evidence that: 1) DCIS, like IDC, consists of two distinct genetic pathways correlating with tumor grade and that DCIS is direct precursor to IDC, 2) ADH is a precursor to low-grade DCIS, 3) FEA is genetically related to ADH and is likely a precursor to ADH, 4) at the genomic level UDH is most similar to normal breast epithelium and is not a precursor to ADH, and 5) microglandular adenosis may represent a precursor lesion to a subset of high grade DCIS lesions.
Comparative gene expression analysis of the ADH and DCIS further support the concept that low-grade and high-grade DCIS likely arise from two distinct evolutionary pathways and that ADH is the precursor to low-grade DCIS. In one of the earliest and most comprehensive gene expression studies of breast cancer progression, Ma et al. unexpectedly demonstrated that no consistent major transcriptional changes occurred between the preinvasive and invasive stages of breast cancer [97]. Most notably, this study demonstrated that the transition from preinvasive disease to invasive disease is associated with quantitative, rather than qualitative, differences in gene expression, and that this quantative relationship is most prominent in high grade lesions. Furthermore, Ma and colleagues also demonstrated for the first time that unique gene expression signatures are associated with different tumor grades, irrespective of tumor stage. More specifically, ADH and low-grade DCIS (as well as low-grade IDC) share a near identical gene expression profile populated by genes that are associated with the estrogen receptor phenotype, whereas high-grade DCIS (as well as high grade IDC) possesses a uniquely different gene expression profile populated with genes associated mitotic-activity and cell cycle processes. Several breast cancer gene expression profiling studies have confirmed these observations [98–100]. Collectively these studies suggest that breast cancer progression may be more intricate than predicted by the traditional model of molecular progression of activation and inactivation of oncogenes and tumor suppressor genes, respectively, and that progression may be reliant upon such contingencies as quantitative and temporal expression of genes.
As compared with DCIS, few studies have focused on the genomic alterations associated with lobular carcinoma in situ [80, 101–103]. Most notably, akin to that observed for classic ILC, these studies demonstrated a consistent and common loss of chromosome 16q providing evidence that LCIS is a direct precursor to ILC, and that conventional LCIS likely shares a common genetic evolutionary pathway with low-grade DCIS and low-grade IDC. Early CGH-based analysis of ALH and LCIS identified frequent loss of chromosomal material from 16q and 16p in both lesions and observed no statistically significant differences in genomic alterations between ALH and LCIS. More recently, work performed by Mastracci et al. and Morandi et al. demonstrated the common loss of chromosome 16q in ALH, LCIS and ILC supporting an evolutionary link among the three lesions [102, 103]. Lastly, gene expression profiles generated from classic LCIS were remarkably similar to those generated from low-grade IDC. These data are consistent with aforementioned genomic observations and suggest a common evolutionary pathway of progression.
Molecular analysis of the non-epithelial components of the breast cancer microenvironment
Over the past several decades, the major focus of breast cancer research has centered on the breast epithelial cell itself, while the role of the non-neoplastic cells of the tumor microenvironment has been largely unexplored. The non-neoplastic cells of the tumor microenvironment include the myoepithelial and inflammatory cells of the preinvasive stages of breast cancer progression and the fibroblasts, myofibroblasts, adipocytes, inflammatory cells and endothelial cells of the invasive stage of progression. By and large, non-epithelial cells have been considered silent or reactive bystanders. However, emerging in vitro- and in vivo-based data suggest that breast cancer progression results from an orchestrated series of bidirectional signaling between non-epithelial cells and malignant epithelial cell in the tumor microenvironment [104–107].
In an effort to analyze the molecular alterations in cells composing the tumor microenvironment in an unbiased manner, several groups have performed high-throughput genomic and transcriptomic comparative analysis of the different tissue and cellular compartments in the various intramammary stages of breast cancer progression. Using ex vivo purification techniques, Polyak and colleagues characterized the gene expression and genetic profiles of all major cell types in the microenvironment of normal breast tissue and in both in situ and invasive breast cancer [108]. The results of this comprehensive study demonstrated that clonally restricted genetic changes are restricted to the neoplastic epithelial cells and that gene expression changes occur in all cell types during progression. Intriguingly, the comparison of DCIS-associated myoepithelial cells with normal myoepithelial cells yielded the highest number of differentially expressed genes, and a number of such genes encoded for secreted proteins and cell surface receptors that have been implicated in autocrine/paracrine regulatory loops and in invasion and migration processes. Independent studies by Ma et al. combining tissue microdissection with oligonucleotide microarrays tissue microdissection approach reached similar conclusions, as they revealed significant gene expression changes in both the epithelial and non-epithelial compartments of the different stages of breast cancer progression [100]. Notably, unlike the epithelial compartment that demonstrates no or rare gene expression changes at the transition from in situ to invasive carcinoma, the stromal compartment demonstrates significant gene expression changes that include genes encoding for extracellular matrix proteins and matrix metalloproteases [100]. These data in conjunction with previous findings by Hu et al. support the notion that matrix proteases may play an important functional role in driving the DCIS to IDC transition [109].
Significant alterations in gene expression patterns in the non-epithelial compartment of the tumor microenvironment in conjunction with lack of clonally restricted genetic changes in the same cells suggest that epigenetic modifications may be responsible for such gene expression patterns. In fact, a follow up study by Polyak’s group utilizing methylation-specific digital karyotyping identified modified epigenetic patterns not only in tumor epithelial cells but also in tumor-associated stromal cells and in situ-carcinoma-associated myoepithelial cells [110]. Independently, Fiegl and colleagues have confirmed the observation that tumor-associated stroma cells display epigenetic alterations and that the frequency of such epigenetic modifications is greater in HER2 positive than in HER2 negative tumors [111]. Future studies aimed at studying posttranslational modifications of histone proteins of the different stages of breast cancer promise to shed new light on the epigenetic regulatory control of gene expression during breast tumorigenesis.
A Divergent Molecular Model of Human Breast Cancer Progression
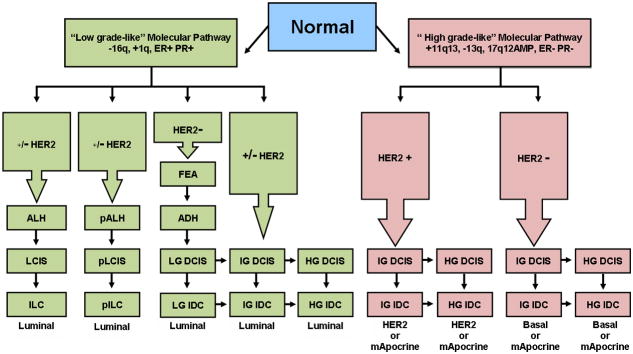
As put forth previously [112, 113], the numerous genetic and transcriptomic analyses of the different phenotypic stages of breast cancer strongly suggest that breast cancer evolves along two divergent molecular pathways of progression (Fig. 3). These two pathways are defined at the genetic level by either the presence or absence of chromosome 1q, 16q, 13q, gain of chromosomal region 11q13 and amplification of the 17q12, and these pathways also strongly correlate with the transcriptomic equivalent of tumor grade/proliferation. The first pathway, designated the “low-grade-like” pathway is characterized by gain of 1q, loss 16q, infrequent amplification of 17q12 and a gene expression signature that is predominantly populated with genes associated with the estrogen receptor phenotype. In general, this pathway consists of neoplastic lesions predominantly of the low to intermediate tumor grade phenotype. However, recent data indicate that a small subset (~9%) of morphologically-defined high grade tumors can evolve from low grade counterparts [85]. Although these data appear inconsistent with a simple two-pathway model, recent array CGH data by Natrajan may provide a unifying corollary [114]. More specifically, Natrajan and colleagues revealed an increased frequency of 16q loss in high grade tumors of the luminal subtype as compared with the basal and HER2 subtypes, suggesting that the progression of a low-grade tumor to a high grade tumor may preferentially occur in breast cancers of the luminal phenotype [114]. The second pathway, designated the “high-grade-like” pathway is most commonly characterized by loss of 13q, gain of chromosomal region 11q13, amplification of 17q12 and by a gene expression signature populated by genes associated with cellular proliferation and cell cycle processes. The high-grade-like pathway consists predominantly of morphologically-defined intermediate and high grade tumors.
Figure 3.
Divergent evolutionary pathways of breast cancer progression. Genomic and transcriptomic data in combination with morphological and immunohistochemical data stratify the majority of breast cancers into a “low-grade-like” molecular pathway and a “high-grade-like” molecular pathway. The low-grade-like pathway (left hand side) is characterized by recurrent chromosomal loss of 16q, gains of 1q, a low-grade-like gene expression signature, and the expression of estrogen and progesterone receptors (ER+ and PR+). The progression (vertical arrows) along this pathway (green rectangles) culminates with the formation of low and intermediate grade invasive ductal, (LG IDC and IG IDC) and invasive lobular carcinomas including both the classic (ILC) and the pleomorphic variant (pILC). The tumors arising from the low grade pathway are classified as luminal consisting of a continuum of gene expression frequently associated with the absence (luminal A) or presence of HER2 expression (luminal B). The vast majority of ILCs and pILCs and their precursors cluster together within the luminal subtype. The high grade-like gene expression molecular pathway (right hand side) is characterized by recurrent gain of 11q13 (+11q13), loss of 13q (13q−), expression of a high-grade-like gene expression signature, amplification of 17q12 (17q12AMP), and lack of estrogen and progesterone receptors expression (ER− and PR−). The progression along this pathway (red rectangles) includes intermediate and high grade ductal carcinomas that are stratified as HER2, or basal-like, depending on the expression/amplification of HER2. The molecular apocrine subtype, characterized by the lack of ER expression and presence of AR expression, arises from the high grade pathway. The model also depicts intra-pathway tumor grade progression (horizontal arrows). Abbreviations: ALH (atypical lobular hyperplasia), LCIS (lobular carcinoma in situ), pALH (pleomorphic atypical lobular hyperplasia), pLCIS (pleomorphic lobular carcinoma in situ), FEA (flat epithelial atypia), ADH (atypical ductal hyperplasia), Basal (basal-like), mApocrine (molecular Apocrine). +/− HER2 in which the “–”sign is bold indicates that the majority of tumors in the pathway lack HER2 overexpression. (This figure is adapted from [112] ).
Although the genomic and transcriptomic data presented in this review support the divergent model of breast cancer progression, our clinical experience indicates that tumors within each pathway are still fairly heterogeneous with respect to clinical outcome suggesting that even this advanced molecular progression scheme is oversimplified. Recent data suggest that intratumoral genetic heterogeneity that is rarely accounted for in diagnosis and treatment of human malignancies may be responsible for the variable clinical behavior of a number of different malignancies [115–117]. With regard to intratumoral genetic heterogeneity in breast cancer, Reis-Filho and colleagues performed genetic analysis of distinct morphological components within a series of metaplastic breast carcinomas [118]. The results of this array-CGH-based study demonstrate that some breast cancers are composed of different “non-modal” clones that possess distinct genetic alterations and that these alterations may underpin morphological heterogeneity within a tumor [118]. In addition to the CGH-based approach by Reis-Filho’s group, two independent groups have recently employed massively parallel sequencing technologies as a means to screen the entire genomes for genetic changes associated with tumor progression [119, 120]. Both groups identified somatic coding mutations in primary breast cancer as compared with patient-matched normal, and each group identified a wide range of mutation frequencies within the primary tumor samples suggesting that considerable genetic heterogeneity exists in the primary tumor [119, 120]. Although most of the original mutations present in the primary tumor are carried forward during progression, additional somatic coding changes were identified in patient-matched metastatic breast cancer samples in both studies [119, 120]. Altogether these studies support the clonal evolution model in which the acquisition of genetic alterations under selective pressure is associated with progression from invasive to metastatic breast cancer. The future application of massively parallel sequencing technologies to the preinvasive stages of breast cancer will assist in assessing intratumoral heterogeneity during the transition from preinvasive to invasive breast cancer, and may assist in identifying early tumor initiating genetic events.
Summary
Over the past decade the integration of numerous genomic and transcriptomic analyses of the various stages of breast cancer has generated multiple novel insights in the complex process of breast cancer progression. First, human breast cancer appears to progress along two distinct molecular genetic pathways that strongly associate with tumor grade. Second, in the epithelial and non-epithelial components of the tumor microenvironment, the greatest molecular alterations (at the gene expression level) occur prior to local invasion. Third, in the epithelial compartment, no major additional gene expression changes occur between the preinvasive and invasive stages of breast cancer. Fourth, the non-epithelial compartment of the tumor micromilieu undergoes dramatic epigenetic and gene expression alterations occur during the transition form preinvasive to invasive disease. Despite these significant advances, we have only begun to scratch the surface of this multifaceted biological process. With the advent of additional novel high-throughput genetic, epigenetic and proteomic technologies, it is anticipated that the next decade of breast cancer research will gain an equally paralleled appreciation for the complexity breast cancer progression. It is with great hope that knowledge gained from such studies will provide for more effective strategies to not only treat, but also prevent breast cancer.
Acknowledgments
We expend special thanks to many collaborators for thoughtful discussions over the years, and to Michelle Forrestall for graphic design assistance. We apologize to all investigators whose research could not be appropriately cited owing to space limitations. D.C. Sgroi is supported in part by NIH RO1-1CA112021-02 (D.C.S), the Department of Defense grant W81XWH-04-1-0606 (D.C. S.), Susan G. Komen Breast Cancer Foundation grants BCTR0402932 (D.C. S.) and the Avon Foundation (D.C.S.).
Footnotes
Author contributions
All authors were involved in writing the paper and had final approval of the submitted and published versions.
Disclosure Statement
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Li CI, Anderson BO, Daling JR, et al. Trends in incidence rates of invasive lobular and ductal breast carcinoma. JAMA. 2003;289:1421–1424. doi: 10.1001/jama.289.11.1421. [DOI] [PubMed] [Google Scholar]
- 3.Li CI, Daling JR, Malone KE. Incidence of invasive breast cancer by hormone receptor status from 1992 to 1998. J Clin Oncol. 2003;21:28–34. doi: 10.1200/JCO.2003.03.088. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 5.Berry DA, Cronin KA, Plevritis SK, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 6.Jatoi I, Chen BE, Anderson WF, Rosenberg, et al. Breast cancer mortality trends in the United States according to estrogen receptor status and age at diagnosis. J Clin Oncol. 2007;25:1683–1690. doi: 10.1200/JCO.2006.09.2106. [DOI] [PubMed] [Google Scholar]
- 7.Tavassoli FADP. World Health Organization Classification of Tumours Pathology and Genetics of Tumours of the Breast and Female Genital Organs. 2003. pp. 9–19. [Google Scholar]
- 8.Rakha EA, Putti TC, Abd El-Rehim DM, et al. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol. 2006;208:495–506. doi: 10.1002/path.1916. [DOI] [PubMed] [Google Scholar]
- 9.Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 10.Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–273. [PubMed] [Google Scholar]
- 11.Oyama T, Maluf H, Koerner F. Atypical cystic lobules: an early stage in the formation of low-grade ductal carcinoma in situ. Virchows Arch. 1999;435:413–421. doi: 10.1007/s004280050419. [DOI] [PubMed] [Google Scholar]
- 12.Oyama T, Iijima K, Takei H, et al. Atypical cystic lobule of the breast: an early stage of low-grade ductal carcinoma in-situ. Breast Cancer. 2000;7:326–331. doi: 10.1007/BF02966399. [DOI] [PubMed] [Google Scholar]
- 13.Lerwill MF. Flat epithelial atypia of the breast. Arch Pathol Lab Med. 2008;132:615–621. doi: 10.5858/2008-132-615-FEAOTB. [DOI] [PubMed] [Google Scholar]
- 14.Page DL, Dupont WD, Rogers LW, et al. Atypical hyperplastic lesions of the female breast. A long-term follow-up study. Cancer. 1985;55:2698–2708. doi: 10.1002/1097-0142(19850601)55:11<2698::aid-cncr2820551127>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 15.Dupont WD, Page DL. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med. 1985;312:146–151. doi: 10.1056/NEJM198501173120303. [DOI] [PubMed] [Google Scholar]
- 16.Kusama R, Fujimori M, Matsuyama I, et al. Clinicopathological characteristics of atypical cystic duct (ACD) of the breast: assessment of ACD as a precancerous lesion. Pathol Int. 2000;50:793–800. doi: 10.1046/j.1440-1827.2000.01121.x. [DOI] [PubMed] [Google Scholar]
- 17.Otterbach F, Bankfalvi A, Bergner S, et al. Cytokeratin 5/6 immunohistochemistry assists the differential diagnosis of atypical proliferations of the breast. Histopathology. 2000;37:232–240. doi: 10.1046/j.1365-2559.2000.00882.x. [DOI] [PubMed] [Google Scholar]
- 18.Boecker W, Moll R, Dervan P, et al. Usual ductal hyperplasia of the breast is a committed stem (progenitor) cell lesion distinct from atypical ductal hyperplasia and ductal carcinoma in situ. J Pathol. 2002;198:458–467. doi: 10.1002/path.1241. [DOI] [PubMed] [Google Scholar]
- 19.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 20.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 22.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 23.Barabe F, Kennedy JA, Hope KJ, et al. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 24.Stingl J, Eaves CJ, Kuusk U, et al. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- 25.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 26.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baik I, Becker PS, DeVito WJ, et al. Stem cells and prenatal origin of breast cancer. Cancer Causes Control. 2004;15:517–530. doi: 10.1023/B:CACO.0000036450.06092.ce. [DOI] [PubMed] [Google Scholar]
- 28.Gudjonsson T, Villadsen R, Nielsen HL, et al. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stingl J, Raouf A, Emerman JT, et al. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- 30.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Ponti D, Zaffaroni N, Capelli C, et al. Breast cancer stem cells: an overview. Eur J Cancer. 2006;42:1219–1224. doi: 10.1016/j.ejca.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pece S, Tosoni D, Confalonieri S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Asselin-Labat ML, Vaillant F, Sheridan JM, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 37.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature. 465:803–807. doi: 10.1038/nature09091. nature09091 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Tu SM, Lin SH, Logothetis CJ. Stem-cell origin of metastasis and heterogeneity in solid tumours. Lancet Oncol. 2002;3:508–513. doi: 10.1016/s1470-2045(02)00820-3. [DOI] [PubMed] [Google Scholar]
- 39.Dontu G, Al-Hajj M, Abdallah WM, et al. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36 (Suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 41.Molyneux G, Geyer FC, Magnay FA, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 44.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]; Histopathology. 2002;41:151–152. discussion 152–153. [PubMed] [Google Scholar]
- 45.Rakha EA, Reis-Filho JS, Baehner F, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tirkkonen M, Tanner M, Karhu R, et al. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 47.Roylance R, Gorman P, Harris W, et al. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res. 1999;59:1433–1436. [PubMed] [Google Scholar]
- 48.Buerger H, Otterbach F, Simon R, et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol. 1999;189:521–526. doi: 10.1002/(SICI)1096-9896(199912)189:4<521::AID-PATH472>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 49.Buerger H, Mommers EC, Littmann R, et al. Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol. 2001;194:165–170. doi: 10.1002/path.875. [DOI] [PubMed] [Google Scholar]
- 50.Duffy SW, Tabar L, Fagerberg G, et al. Breast screening, prognostic factors and survival--results from the Swedish two county study. Br J Cancer. 1991;64:1133–1138. doi: 10.1038/bjc.1991.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajakariar R, Walker RA. Pathological and biological features of mammographically detected invasive breast carcinomas. Br J Cancer. 1995;71:150–154. doi: 10.1038/bjc.1995.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 53.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96–103. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 58.Voduc KD, Cheang MC, Tyldesley S, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 59.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 60.Lusa L, McShane LM, Reid JF, et al. Challenges in projecting clustering results across gene expression-profiling datasets. J Natl Cancer Inst. 2007;99:1715–1723. doi: 10.1093/jnci/djm216. [DOI] [PubMed] [Google Scholar]
- 61.Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weigelt B, Mackay A, A’Hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 63.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 64.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 65.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 66.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 67.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 68.Ma XJ, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 70.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 71.Haibe-Kains B, Desmedt C, Piette F, et al. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 73.Ma XJ, Salunga R, Dahiya S, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 74.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27:3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toussaint J, Sieuwerts AM, Haibe-Kains B, et al. Improvement of the clinical applicability of the Genomic Grade Index through a qRT-PCR test performed on frozen and formalin-fixed paraffin-embedded tissues. BMC Genomics. 2009;10:424. doi: 10.1186/1471-2164-10-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishizaki T, Chew K, Chu L, et al. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer. 1997;74:513–517. doi: 10.1002/(sici)1097-0215(19971021)74:5<513::aid-ijc6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 77.Loveday RL, Greenman J, Simcox DL, et al. Genetic changes in breast cancer detected by comparative genomic hybridisation. Int J Cancer. 2000;86:494–500. doi: 10.1002/(sici)1097-0215(20000515)86:4<494::aid-ijc8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 78.Gunther K, Merkelbach-Bruse S, Amo-Takyi BK, et al. Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J Pathol. 2001;193:40–47. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH745>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 79.Richard F, Pacyna-Gengelbach M, Schluns K, et al. Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer. 2000;89:305–310. [PubMed] [Google Scholar]
- 80.Lakhani SR, Collins N, Sloane JP, et al. Loss of heterozygosity in lobular carcinoma in situ of the breast. Clin Mol Pathol. 1995;48:M74–78. doi: 10.1136/mp.48.2.m74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simpson PT, Reis-Filho JS, Gale T, et al. Molecular evolution of breast cancer. J Pathol. 2005;205:248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 82.Weigelt B, Geyer FC, Natrajan R, et al. The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol. 220:45–57. doi: 10.1002/path.2629. [DOI] [PubMed] [Google Scholar]
- 83.Middleton LP, Palacios DM, Bryant BR, et al. Pleomorphic lobular carcinoma: morphology, immunohistochemistry, and molecular analysis. Am J Surg Pathol. 2000;24:1650–1656. doi: 10.1097/00000478-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 84.Simpson PT, Reis-Filho JS, Lambros MB, et al. Molecular profiling pleomorphic lobular carcinomas of the breast: evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol. 2008;215:231–244. doi: 10.1002/path.2358. [DOI] [PubMed] [Google Scholar]
- 85.Allred DC, Wu Y, Mao S, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 86.Tavassoli FA, Norris HJ. A comparison of the results of long-term follow-up for atypical intraductal hyperplasia and intraductal hyperplasia of the breast. Cancer. 1990;65:518–529. doi: 10.1002/1097-0142(19900201)65:3<518::aid-cncr2820650324>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 87.Page DL, Rogers LW. Combined histologic and cytologic criteria for the diagnosis of mammary atypical ductal hyperplasia. Hum Pathol. 1992;23:1095–1097. doi: 10.1016/0046-8177(92)90026-y. [DOI] [PubMed] [Google Scholar]
- 88.Buerger H, Otterbach F, Simon R, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol. 1999;187:396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 89.O’Connell P, Pekkel V, Fuqua SA, et al. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- 90.Moinfar F, Man YG, Bratthauer GL, et al. Genetic abnormalities in mammary ductal intraepithelial neoplasia-flat type (“clinging ductal carcinoma in situ”): a simulator of normal mammary epithelium. Cancer. 2000;88:2072–2081. [PubMed] [Google Scholar]
- 91.Simpson PT, Gale T, Reis-Filho JS, et al. Columnar cell lesions of the breast: the missing link in breast cancer progression? A morphological and molecular analysis. Am J Surg Pathol. 2005;29:734–746. doi: 10.1097/01.pas.0000157295.93914.3b. [DOI] [PubMed] [Google Scholar]
- 92.Kasami M, Vnencak-Jones CL, Manning S, et al. Loss of heterozygosity and microsatellite instability in breast hyperplasia. No obligate correlation of these genetic alterations with subsequent malignancy. Am J Pathol. 1997;150:1925–1932. [PMC free article] [PubMed] [Google Scholar]
- 93.Boecker W, Buerger H, Schmitz K, et al. Ductal epithelial proliferations of the breast: a biological continuum? Comparative genomic hybridization and high-molecular-weight cytokeratin expression patterns. J Pathol. 2001;195:415–421. doi: 10.1002/path.982. [DOI] [PubMed] [Google Scholar]
- 94.Jones C, Merrett S, Thomas VA, et al. Comparative genomic hybridization analysis of bilateral hyperplasia of usual type of the breast. J Pathol. 2003;199:152–156. doi: 10.1002/path.1280. [DOI] [PubMed] [Google Scholar]
- 95.Geyer FC, Kushner YB, Lambros MB, et al. Microglandular adenosis or microglandular adenoma? A molecular genetic analysis of a case associated with atypia and invasive carcinoma. Histopathology. 2009;55:732–743. doi: 10.1111/j.1365-2559.2009.03432.x. [DOI] [PubMed] [Google Scholar]
- 96.Shin SJ, Simpson PT, Da Silva L, et al. Molecular evidence for progression of microglandular adenosis (MGA) to invasive carcinoma. Am J Surg Pathol. 2009;33:496–504. doi: 10.1097/PAS.0b013e31818af361. [DOI] [PubMed] [Google Scholar]
- 97.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Porter D, Lahti-Domenici J, Keshaviah A, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 99.Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 100.Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu YJ, Osin P, Lakhani SR, et al. Comparative genomic hybridization analysis of lobular carcinoma in situ and atypical lobular hyperplasia and potential roles for gains and losses of genetic material in breast neoplasia. Cancer Res. 1998;58:4721–4727. [PubMed] [Google Scholar]
- 102.Mastracci TL, Shadeo A, Colby SM, et al. Genomic alterations in lobular neoplasia: a microarray comparative genomic hybridization signature for early neoplastic proliferationin the breast. Genes Chromosomes Cancer. 2006;45:1007–1017. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- 103.Morandi L, Marucci G, Foschini MP, et al. Genetic similarities and differences between lobular in situ neoplasia (LN) and invasive lobular carcinoma of the breast. Virchows Arch. 2006;449:14–23. doi: 10.1007/s00428-006-0192-7. [DOI] [PubMed] [Google Scholar]
- 104.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264:169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 105.Price JE, Polyzos A, Zhang RD, et al. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- 106.Mehta RR, Graves JM, Hart GD, et al. Growth and metastasis of human breast carcinomas with Matrigel in athymic mice. Breast Cancer Res Treat. 1993;25:65–71. doi: 10.1007/BF00662402. [DOI] [PubMed] [Google Scholar]
- 107.Sternlicht MD, Kedeshian P, Shao ZM, et al. The human myoepithelial cell is a natural tumor suppressor. Clin Cancer Res. 1997;3:1949–1958. [PubMed] [Google Scholar]
- 108.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 109.Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu M, Yao J, Cai L, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 111.Fiegl H, Millinger S, Goebel G, et al. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res. 2006;66:29–33. doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- 112.Sgroi DC. Preinvasive breast cancer. Annu Rev Pathol. 5:193–221. doi: 10.1146/annurev.pathol.4.110807.092306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, et al. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 57:171–192. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 114.Natrajan R, Lambros MB, Geyer FC, et al. Loss of 16q in high grade breast cancer is associated with estrogen receptor status: Evidence for progression in tumors with a luminal phenotype? Genes Chromosomes Cancer. 2009;48:351–365. doi: 10.1002/gcc.20646. [DOI] [PubMed] [Google Scholar]
- 115.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Godin-Heymann N, Ulkus L, Brannigan BW, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 117.Heinrich MC, Maki RG, Corless CL, et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26:5352–5359. doi: 10.1200/JCO.2007.15.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 119.Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 120.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]