Abstract
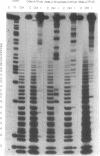
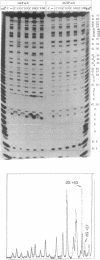
For the first time mosaic nucleic acids composed of 50% RNA and 50% DNA can be obtained as transcripts with T7 RNA polymerase. Two NTPs could be replaced simultaneously in a transcription reaction. This means more than 40 deoxynucleotides were inserted in one transcript. Previously, a maximum of two deoxynucleotides could be incorporated and 2'-O-methyl-NTPs were not substrates at all. We obtained reasonable transcript yields with a maximal level of 99% 2'-O-methyl-NTPs, and the products contained up to 58% 2'-O-methylnucleotides at more than 20 positions. Sequence-specific nucleotide incorporation was monitored by sequence ladders (partial alkali or iodine cleavage). No base misincorporations were detected with 100% dGTP, dCTP and dTTP, and with partial incorporation of dATP alpha S, 2'-O-methyl-GTP alpha S and 2'-O-methyl-CTP alpha S, whereas they were found with dATP, 2'-O-methyl-ATP alpha S and 2'-O-methyl-UTP alpha S. Quantitative data allow predetermined modification levels of partially modified transcripts. Highly modified transcripts can be used for structural and functional studies, in modification interference approaches and for in vitro evolution procedures. Modification interference studies revealed a small number of important phosphate and ribose moieties in RNase P substrates. The conversion of T7 RNA polymerase to a DNA polymerase extends the observation that there is no absolute distinction between RNA and DNA polymerases. Accordingly, an adapted concept of a primordial RNA world is presented.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aurup H., Siebert A., Benseler F., Williams D., Eckstein F. Translation of 2'-modified mRNA in vitro and in vivo. Nucleic Acids Res. 1994 Nov 25;22(23):4963–4968. doi: 10.1093/nar/22.23.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurup H., Williams D. M., Eckstein F. 2'-Fluoro- and 2'-amino-2'-deoxynucleoside 5'-triphosphates as substrates for T7 RNA polymerase. Biochemistry. 1992 Oct 13;31(40):9636–9641. doi: 10.1021/bi00155a016. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Ames B. N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982 Aug 25;257(16):9759–9769. [PubMed] [Google Scholar]
- Burke J. M., Berzal-Herranz A. In vitro selection and evolution of RNA: applications for catalytic RNA, molecular recognition, and drug discovery. FASEB J. 1993 Jan;7(1):106–112. doi: 10.1096/fasebj.7.1.8422956. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Chowrira B. M., Burke J. M. Extensive phosphorothioate substitution yields highly active and nuclease-resistant hairpin ribozymes. Nucleic Acids Res. 1992 Jun 11;20(11):2835–2840. doi: 10.1093/nar/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur R. K., Krupp G. Enzymatic RNA synthesis with deoxynucleoside 5'-O-(1-thiotriphosphates). FEBS Lett. 1993 Jan 2;315(1):56–60. doi: 10.1016/0014-5793(93)81132-j. [DOI] [PubMed] [Google Scholar]
- Gaur R. K., Krupp G. Modification interference approach to detect ribose moieties important for the optimal activity of a ribozyme. Nucleic Acids Res. 1993 Jan 11;21(1):21–26. doi: 10.1093/nar/21.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A. D., Potter B. V., Eperon I. C. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: stereochemistry of transcription by T7 RNA polymerase. Nucleic Acids Res. 1987 May 26;15(10):4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkas J. D. Reverse transcription by Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3834–3838. doi: 10.1073/pnas.70.12.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Roe B. A. Aminoacylation of synthetic DNAs corresponding to Escherichia coli phenylalanine and lysine tRNAs. Science. 1988 Jul 1;241(4861):74–79. doi: 10.1126/science.2455342. [DOI] [PubMed] [Google Scholar]
- Kleineidam R. G., Pitulle C., Sproat B., Krupp G. Efficient cleavage of pre-tRNAs by E. coli RNAse P RNA requires the 2'-hydroxyl of the ribose at the cleavage site. Nucleic Acids Res. 1993 Mar 11;21(5):1097–1101. doi: 10.1093/nar/21.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene. 1988 Dec 10;72(1-2):75–89. doi: 10.1016/0378-1119(88)90129-1. [DOI] [PubMed] [Google Scholar]
- Myers T. W., Gelfand D. H. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry. 1991 Aug 6;30(31):7661–7666. doi: 10.1021/bi00245a001. [DOI] [PubMed] [Google Scholar]
- Paolella G., Sproat B. S., Lamond A. I. Nuclease resistant ribozymes with high catalytic activity. EMBO J. 1992 May;11(5):1913–1919. doi: 10.1002/j.1460-2075.1992.tb05244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992 Jul 20;226(2):399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- Ricchetti M., Buc H. E. coli DNA polymerase I as a reverse transcriptase. EMBO J. 1993 Feb;12(2):387–396. doi: 10.1002/j.1460-2075.1993.tb05670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Feix G. Ribonucleic acid ligase activity of deoxyribonucleic acid ligase from phage T4 infected Escherichia coli. Biochemistry. 1974 Dec 3;13(25):5110–5115. doi: 10.1021/bi00722a009. [DOI] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Sande J. H., Loewen P. C., Khorana H. G. Studies on polynucleotides. 118. A further study of ribonucleotide incorporation into deoxyribonucleic acid chains by deoxyribonucleic acid polymerase I of Escherichia coli. J Biol Chem. 1972 Oct 10;247(19):6140–6148. [PubMed] [Google Scholar]
- Wyatt J. R., Walker G. T. Deoxynucleotide-containing oligoribonucleotide duplexes: stability and susceptibility to RNase V1 and RNase H. Nucleic Acids Res. 1989 Oct 11;17(19):7833–7842. doi: 10.1093/nar/17.19.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H., Usman N., Chartrand P., Cedergren R. Minimum ribonucleotide requirement for catalysis by the RNA hammerhead domain. Biochemistry. 1992 Jun 2;31(21):5005–5009. doi: 10.1021/bi00136a013. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., So A. G., Downey K. M. Mechanisms of error discrimination by Escherichia coli DNA polymerase I. Biochemistry. 1988 Jan 26;27(2):546–553. doi: 10.1021/bi00402a007. [DOI] [PubMed] [Google Scholar]