Abstract
Hydrogen peroxide (H2O2), an important substance produced by many members of the genus Streptococcus, plays important roles in virulence and antagonism within a microbial community such as oral biofilms. The spxB gene, which encodes pyruvate oxidase, is involved in H2O2 production in many streptococcal species. However, knowledge about its regulation and relation with other genes putatively involved in the same pathway is limited. In this study, three genes – ackA, spxR and tpk – were identified as contributing to H2O2 production in Streptococcus sanguinis by screening mutants for opaque colony appearance. Mutations in all three genes resulted in significant decreases in H2O2 production, with 16–31 % of that of the wild-type. H2O2 production was restored in the complemented strains. Antagonism against Streptococcus mutans by these three S. sanguinis mutants was reduced, both on plates and in liquid cultures, indicating the critical roles of these three genes for conferring the competitive advantage of S. sanguinis. Analysis by qPCR indicated that the expression of spxB was decreased in the ackA and spxR mutants and significantly increased in the tpk mutant.
INTRODUCTION
Hydrogen peroxide (H2O2) is produced by many members of the genus Streptococcus (García-Mendoza et al., 1993; Kreth et al., 2005; Ramos-Montañez et al., 2008) and is important in three aspects. First, H2O2 is reported to correlate with virulence in Streptococcus pneumoniae (Auzat et al., 1999; Ramos-Montañez et al., 2008; Weiser et al., 1994). S. pneumoniae undergoes spontaneous phase variation resulting in opaque and transparent colony forms, and the differences in colony opacity correlate with virulence (Weiser et al., 1994). Recent research indicates that transparent variants are more proficient in colonization, with more production of teichoic acid and H2O2, but with less production of capsule than opaque variants (Ramos-Montañez et al., 2008). Studies also suggest that the H2O2 produced by Streptococcus pyogenes acts as a potential virulence factor by exerting direct damage to host tissues (Ginsburg & Sadovnic, 1998; Ginsburg & Varani, 1993). Second, H2O2 production is related to competition and co-existence within microbial communities such as oral biofilms. Many streptococci are able to produce inhibitory substances such as H2O2 to reduce the growth of co-resident micro-organisms. For example, S. sanguinis can produce H2O2 that will inhibit growth of Staphylococcus aureus (Uehara et al., 2006). S. sanguinis and Streptococcus gordonii demonstrate antagonistic activity against Streptococcus mutans via H2O2 production (Kreth et al., 2005). Other studies also investigated the inhibitory capacity of H2O2 produced by various species of oral streptococci (García-Mendoza et al., 1993; Kreth et al., 2005, 2008). Recently, H2O2 was also shown to contribute to the release of DNA from S. sanguinis and S. gordonii, which appears to support oral biofilm formation and facilitate exchange of genetic material among competent strains (Kreth et al., 2009). Third, H2O2 is a by-product of aerobic metabolism (Jakubovics et al., 2002). Many streptococci produce relatively large amounts of H2O2 during aerobic growth by the action of oxidase enzymes such as pyruvate oxidase and NADH oxidase (Auzat et al., 1999; Tittmann et al., 2005).
S. sanguinis is a member of the human indigenous oral microflora and one of the major microbes colonizing teeth (Kuramitsu et al., 2007; Rosan & Lamont, 2000). It is also one of the most common causative agents of infective endocarditis (Douglas et al., 1993; Mylonakis & Calderwood, 2001; Tleyjeh et al., 2005). On the other hand, S. sanguinis is considered an antagonistic bacterium against S. mutans (Becker et al., 2002; Caufield et al., 2000). Relatively high proportions of S. sanguinis are generally found in dental plaque with lower levels of S. mutans. High levels of S. mutans in the oral cavity correlate with low levels of S. sanguinis (Caufield et al., 2000; Ge et al., 2008b).
In S. pneumoniae, the function of pyruvate oxidase (SpxB) in H2O2 production has been well characterized (Weiser et al., 1994). Pyruvate oxidase plays several critical roles in pneumococcal metabolism and virulence (Ramos-Montañez et al., 2008), including phase variation (Overweg et al., 2000; Pericone et al., 2000, 2002). Though the genetic mechanism behind the phase variation has remained unclear, this morphological change has been successfully used in S. pneumoniae to identify SpxR, a regulator of spxB required for spxB transcription and for full virulence in a murine model of infection (Ramos-Montañez et al., 2008).
Although H2O2 production is involved in important metabolic pathways, potential pathogenic virulence, and interspecies competition, knowledge of its metabolic basis and regulation remains limited in S. sanguinis. The spxB gene is involved in H2O2 production (Weiser et al., 1994; Ramos-Montañez et al., 2008), but its regulation and relation with other genes putatively involved in the same pathway are still unclear. SpxB is a decarboxylase that catalyses the conversion of pyruvate, inorganic phosphate (Pi) and molecular oxygen (O2) to H2O2, carbon dioxide (CO2) and acetyl phosphate, which acts as a high-energy phosphoryl group donor (Ramos-Montañez et al., 2008):
During a study of gene deletion mutants of S. sanguinis SK36, we noticed that some mutants showed opaque colonies, suggesting a possible deficiency in H2O2 production. To identify novel genes relevant to H2O2 production and their relationship with spxB, we screened the single-gene mutant library being constructed in our lab to find H2O2 production-defective mutants. Here, we describe three genes that are involved in pyruvate oxidase-related H2O2 production and antagonism against S. mutans, together with their functional relationship with pyruvate oxidase in S. sanguinis.
METHODS
Bacterial strains and growth media.
The strains used are described in Table 1. S. sanguinis strain SK36 (obtained from Dr Mogens Kilian, Århus University, Denmark) was isolated from human dental plaque (Kilian & Holmgren, 1981). This strain and its derivatives were routinely grown in brain heart infusion broth (BHI; Difco) supplemented with 1.5 % (w/v) agar under microaerobic conditions (7.2 % H2, 7.2 % CO2, 79.6 % N2 and 6 % O2) in an Anoxomat jar (Spiral Biotech) at 37 °C as described previously (Ge et al., 2008a; Paik et al., 2005). S. mutans UA159 or its derivative was also routinely grown in BHI under microaerobic conditions as for S. sanguinis. When needed, medium was supplemented with kanamycin (500 μg ml−1), chloramphenicol (5 μg ml−1) or erythromycin (10 μg ml−1). The mutants were named by using “ssx” to refer the corresponding ssa gene in the NCBI database.
Table 1.
Bacterial strains used in this study
Cm, chloramphenicol; Em, erythromycin; Km, kanamycin.
| Strain | Phenotype or description | Source |
|---|---|---|
| S. sanguinis | ||
| SK36 | Human plaque isolate | Kilian & Holmgren (1981) |
| ssx_0169 | Kmr; Δ0169 : : aphA-3 | This study |
| ssx_0192 | Kmr; ΔackA : : aphA-3 | This study |
| ssx_0192C | Emr; ackA+ : : pSerm | This study |
| ssx_0190 | Kmr; Δssa_0190 : : aphA-3 | This study |
| ssx_0191 | Kmr; Δssa_0191 : : aphA-3 | This study |
| ssx_0193 | Kmr; Δssa_0193 : : aphA-3 | This study |
| ssx_0195 | Kmr; Δssa_0195 : : aphA-3 | This study |
| ssx_0391 | Kmr; ΔspxB : : aphA-3 | This study |
| ssx_0391C | Cmr; spxB+ : : magellan2 | This study |
| ssx_1492 | Kmr; ΔspxR : : aphA-3 | This study |
| ssx_1492C | Cmr; spxR+ : : magellan2 | This study |
| ssx_1494 | Kmr; Δssa_1494 : : aphA-3 | This study |
| ssx_1493 | Kmr; Δssa_1493 : : aphA-3 | This study |
| ssx_1490 | Kmr; Δssa_1490 : : aphA-3 | This study |
| ssx_1489 | Kmr; Δssa_1489 : : aphA-3 | This study |
| ssx_2118 | Kmr; Δtpk : : aphA-3 | This study |
| ssx_2118C | Cmr; tpk+ : : magellan2 | This study |
| ssx_2120 | Kmr; Δssa_2120 : : aphA-3 | This study |
| ssx_2119 | Kmr; Δssa_2119 : : aphA-3 | This study |
| ssx_2117 | Kmr; Δssa_2117 : : aphA-3 | This study |
| ssx_2116 | Kmr; Δssa_2116 : : aphA-3 | This study |
| S. mutans | ||
| UA159 | Wild-type, serotype c | ATCC 700610 |
| smx_42 | Cmr; Δsmu.42 : : magellan2 | This study |
Mutant construction and complementation.
For the construction of precise single gene deletion mutants in S. sanguinis SK36, we developed a PCR-based recombinant method employing linear DNA for deletion construction in vitro (P. Xu and others, unpublished data). Briefly, for each targeted gene, three sets of primers were designed to amplify a linear DNA fragment containing the kanamycin resistance cassette (aphA-3) (Turner et al., 2009) with two flanking arms of DNA upstream and downstream of the targeted gene. The linear recombinant PCR amplicon was directly transformed into S. sanguinis competent cells as described previously (Ge et al., 2008a). A 96-well high-throughput format was used to generate a genome-wide mutant library. The mutants were confirmed by PCR and RT-PCR analyses.
To construct the complemented strain, a DNA fragment containing the targeted gene followed by a selectable marker (either chloramphenicol or erythromycin resistance cassette) (Turner et al., 2009) was integrated via double homologous recombination into the corresponding mutant (Table 1) to replace the kanamycin resistance cassette. Chloramphenicol- or erythromycin-resistant and kanamycin-sensitive transformants were selected and confirmed by PCR analysis.
Screening for H2O2 production-defective mutants by opaque colony morphology.
For opaque colony observation, 5 μl overnight culture of S. sanguinis SK36 or different mutants was spotted on the surface of BHI plates and incubated at 37 °C under microaerobic conditions. For catalase-containing plates, 100 μl catalase from bovine liver (Sigma) was spread on the BHI plate surface (∼880 U cm−2). The plates were air-dried for 10 min in a hood before bacterial inoculation. Bacterial suspension (5 μl) was spotted on the catalase-containing plate and incubated at 37 °C under microaerobic conditions for 2 days. The opaque colonies on the plate were recorded and photographed using a BioDoc-It imaging system. After obtaining the opaque mutants in the assays, the integrity of the mutations was confirmed by using PCR amplification and sequencing.
H2O2 release assays.
H2O2 production was quantified using the Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen) as described by the manufacturer, with minor modifications (Ramos-Montañez et al., 2008). Briefly, 100 μl reaction mixture (50 mM Amplex Red reagent, 0.1 U horseradish peroxidase ml−1 in 0.05 M sodium phosphate buffer, pH 7.4) was dispensed into wells of a 96-well microtitre plate and warmed to 37 °C for 10 min. Exponential cultures of S. sanguinis strains were grown in BHI to OD450 0.15–0.2, centrifuged and then resuspended in fresh BHI to OD450 approximately 0.06. An aliquot of freshly resuspended cells (20 μl) was added in triplicate to the 100 μl pre-warmed reaction mixture. A series of H2O2 concentration standards diluted in BHI together with BHI as the blank control were also included in the plate. The microtitre plate was incubated at 37 °C in a FLUOstar plate reader under aerobic conditions and absorbance was read at 563 nm every 5 min for 20 min. Rates of H2O2 production were calculated and normalized to the OD450 of the cell suspensions. Final values are shown relative to that of the wild-type strain, SK36. Paired t-test was used for statistical analysis.
Competition assays on plates.
To determine the inhibitory effect of S. sanguinis against S. mutans, a previously described protocol (Kreth et al., 2005, 2008) was employed with the following modifications. Briefly, 5 μl of an overnight culture of S. sanguinis SK36 or its derivative in BHI medium was inoculated onto a BHI agar plate. After incubation overnight (16 h), 5 μl of S. mutans UA159 was inoculated next to an S. sanguinis colony. Colonies were inoculated so that they were just touching each other. The plate was incubated again for another night. Growth inhibition was evaluated based on the distance of the inhibition zone from the edges of both colonies.
Competition assays in liquid media.
This was performed as described by Kreth et al. (2008). Cells of S. sanguinis mutants and S. mutans smx_42, a chloramphenicol-resistant derivative of S. mutans, were grown in BHI medium overnight and adjusted to the same OD660 value. S. sanguinis or its mutants (3 μl of each) and S. mutans smx_42 (3 μl) were mixed with 200 μl fresh BHI medium in 96-well microtitre plates in triplicate. The cells were incubated overnight in static culture under microaerobic conditions. Cells were dispersed by vigorous pipetting and serial dilutions were plated on BHI agar plates supplemented with chloramphenicol in triplicate and the c.f.u. was determined.
RNA extraction and qPCR analyses.
Total RNA was prepared from the cells growing in late exponential phase in BHI medium under microaerobic conditions to OD450 0.6–1.0. Cells were lysed after lysozyme treatment and mechanical disruption using FastPrep lysing matrix B (Qbiogene). RNA was isolated by using the RNeasy mini kit (Qiagen). DNA was removed from the RNeasy mini kit column by DNase I treatment. Total RNA was quantified using a NanoDrop ND 1000 spectrophotometer. First-strand cDNA synthesis was performed in a 20 μl reaction mixture containing 100 ng RNA, 0.5 μl random primers (3 μg μl−1), 1.0 μl dNTP mix (10 mM each dNTP), 1.0 μl 100 mM DTT, 1.0 μl RNAout (40 U; Invitrogen) and 0.5 μl SuperScript III reverse transcriptase (200 U μl−1) in first-strand buffer (Invitrogen). Reactions lacking reverse transcriptase were prepared in parallel as controls for possible DNA contamination. First strand cDNA from each reaction was subjected to 80-fold dilutions, and 2 μl of each dilution was used as template for each PCR. Quantitative real-time PCR was performed in reactions containing 5 μl SYBR Green PCR master mix (Applied Biosystems), 1 μl each PCR primer (2 mM) using the ABI 7500 fast real-time PCR system. The housekeeping gene gyrA was used as a normalization control. The data were collected and statistically analysed from triplicates. Serial dilutions of chromosomal DNA from wild-type strain SK36 were used for standard curves.
RESULTS
H2O2 production determines colony morphology in S. sanguinis SK36
During the process of creating a genome-wide single gene deletion mutant library of S. sanguinis SK36, it was found that some mutants showed different colony morphologies. In comparison with the semi-transparent colony of the wild-type strain SK36, certain mutants presented an opaque colony when grown on BHI agar plates under microaerobic conditions. It was reported previously that the colony morphology variation between transparent and opaque colonies in S. pneumoniae is related to H2O2 production and can be detected on tryptic soy agar plates by the addition of catalase (Weiser et al., 1994). Such an opaque colony marker has been used to identify genes involved in H2O2 production in S. pneumoniae (Ramos-Montañez et al., 2008). To investigate the existence of similar morphological variation in S. sanguinis and to establish a condition to screen genes involved in H2O2 production, S. sanguinis SK36 was cultured on BHI plates with and without the addition of excess catalase to hydrolyse peroxides. The colony opacity was compared to examine the effect of catalase. We found that SK36 colonies changed from semi-transparent to opaque after incubation for 2 days when catalase was added (data not shown). This showed that in S. sanguinis, the opaque colony variations were related to H2O2 production. The result also suggested that H2O2 production-defective mutants of S. sanguinis SK36 might be identified by colony morphology in our system. We hypothesized that the mutants with opaque colonies had lower H2O2 production.
Next, to identify the potential genes involved in H2O2 production, we screened over 1000 available single gene deletion mutants for variation in colony morphology to identify potential H2O2-production-defective mutants, as described above. Four mutants showing obvious opaque colonies were identified. The morphological variations of the four mutants were further confirmed by comparison with the wild-type strain SK36 on BHI plates, including a control strain ssx_0169 with kanamycin resistance (data not shown) to determine that the kanamycin resistance gene did not interfere with the phenotypes being investigated. This control strain was selected because it was demonstrated that the ssa_0169 gene did not affect important cellular phenotypes (Turner et al., 2009). The deletion locus of each mutant was confirmed to have the expected structure by PCR analysis and DNA sequencing. One of the mutants (ssx_0391) had a deletion in the spxB gene, whose product, pyruvate oxidase, is known to catalyse the production of H2O2 (Ramos-Montañez et al., 2008). The other three genes identified were ackA, encoding acetate kinase, spxR, encoding a conserved hypothetical protein, and tpk, encoding thiamine pyrophosphokinase (Table 1). The four opaque mutants (including ssx_0391) were further characterized.
Opaque mutants have reduced rates of H2O2 production
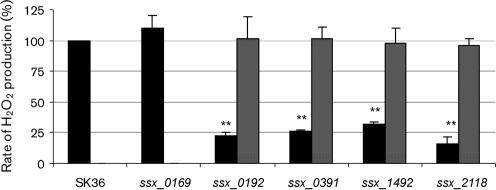
Since H2O2 production is proposed to relate to the opaque morphology, we next quantified H2O2 production in the four mutants identified above, and compared it with that of the wild-type strain SK36. The control strain ssx_0169 was also included in this analysis. All four opaque mutants displayed significantly reduced rates of H2O2 production compared with the semi-transparent parent strain SK36 (Fig. 1). H2O2 production rates of the mutants were only 16–31 % of that of SK36. Similar H2O2 production to the wild-type strain SK36 was found in the kanamycin-resistant control strain (ssx_0169). Though each of the four opaque mutants displayed decreased rates of H2O2 production, none of them lost the capacity of H2O2 production completely, including the spxB mutant. It should be noted that the intact ORF of each mutant was precisely deleted in each of our mutants, so it was impossible that any partial gene function remained. This suggested that the pyruvate oxidase activity might not be the only oxidase activity responsible for H2O2 production.
Fig. 1.
H2O2 production in S. sanguinis strains. H2O2 production normalized to culture densities was determined relative to that produced by the wild-type strain SK36. Data indicate mean±sd from three biological repeats. Statistical significance is indicated (**P<0.01). Black bars, SK36 or a mutant; grey bars, complemented strain.
Next, to ensure that these identified genes function in H2O2 production, we checked the H2O2 production in the relative single gene mutants of their upstream and downstream genes (i.e. ssx_0190, ssx_0191, ssx_0193 and ssx_0195; ssx_1494, ssx_1493, ssx_1490 and ssx_1489; ssx_2120, ssx_2119, ssx_2117 and ssx_2116. The mutant for ssa_1491 is not available because the gene was found to be essential). There was no statistically significant defect in H2O2 production by any of these mutants compared with that of the wild-type. This result supports the hypothesis that the defects in H2O2 production in the identified mutants are not related to the neighbouring genes. To ensure this, we introduced the genes back to the mutants. A chloramphenicol resistance cassette was placed downstream of each gene for selection. After obtaining the complemented strains, their morphology and H2O2 production were examined. The results showed that the morphology of three strains, ssx_0391C, ssx_1492C and ssx_2118C (Table 1), was restored to semi-transparent and the rates of H2O2 production were restored to the wild-type level (Fig. 1). In the first attempt to complement ssx_0192 gene function, we failed to fully restore the phenotype. We then examined the mRNA level of downstream genes ssa_0193, ssa_0195 and ssa_0197 in the mutant ssx_0192, which did not show significant changes compared with that of the wild-type. Given these data, we deduced that there might be some errors with the complemented strain. We therefore performed this complementation again employing an erythromycin resistance cassette (pSerm). The resulting strain ssx_0192C was successfully restored for H2O2 production (Fig. 1). All of the data indicated that the identified genes are involved in H2O2 production.
Opaque mutants demonstrate reduced antagonistic activity against S. mutans UA159 both on plates and in liquid media
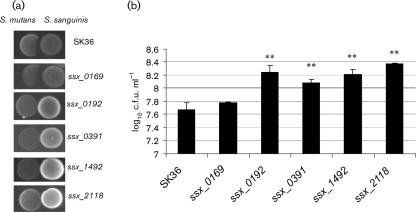
Because the formation of H2O2 in S. sanguinis plays an important role in interspecies interactions within the oral microflora, we performed competition assays to examine whether the four H2O2-defective mutants showed any difference from the parent strain, SK36, in their capacity for antagonism against a primary dental cariogen, S. mutans. We first examined the antagonistic activity on agar plates. S. sanguinis and S. mutans cells were spotted next to one another on agar plates. The inhibition zones of S. sanguinis and the mutants against S. mutans were determined (Fig. 2a). The results showed that all four mutants lost the ability to inhibit S. mutans UA159 on BHI plates under microaerobic conditions (Fig. 2a).
Fig. 2.
Inhibitory effect of S. sanguinis on S. mutans. (a) Inhibition assay on plates. Overnight cultures of different S. sanguinis strains were inoculated on BHI plates, which were incubated for 16 h at 37 °C under microaerobic conditions. S. mutans UA159 was then inoculated next to the pioneer colonizer, and the plates were further incubated overnight and photographed. (b) Inhibition assay in liquid media. Overnight cultures of S. sanguinis SK36 or mutants were adjusted to the same optical density and mixed with the S. mutans UA159 (Cm) in fresh BHI medium. After overnight growth, the cells were serially diluted and plated on BHI plates supplemented with chloramphenicol. The log10 c.f.u. ml−1 values±sd of S. mutans UA159 are shown (data are from triplicate experiments) (**P<0.01 relative to the values obtained for the SK36 mixture).
To further quantify the antagonism of the H2O2-defective mutants against S. mutans, we performed competition assays in liquid culture using mixed species. We first constructed a chloramphenicol-resistant control strain of S. mutans by integrating the chloramphenicol resistance gene magellan2 (Turner et al., 2009) into the S. mutans UA159 chromosome. The smu.42 gene encoding a hypothetical protein (SMU.42) which did not affect its sensitivity to antagonism by S. sanguinis (data not shown) was selected as the target location for integration in the S. mutans genome. Because the S. mutans derivative can be distinguished from S. sanguinis on chloramphenicol selection agar plates, the inhibition effect of S. sanguinis on S. mutans could be determined by bacterial colony numbers on agar plates supplemented with chloramphenicol. We mixed the same amount of each S. sanguinis mutant, as assessed by OD660, with S. mutans and co-cultured the two species mixture. S. mutans cells were counted on BHI plates supplemented with chloramphenicol after 48 h. This indicated that all four mutants were less able to inhibit S. mutans in liquid culture compared with the wild-type strain, SK36, and the control strain ssx_0169 (Fig. 2b).
Transcriptional level of spxB in H2O2 production-defective mutants
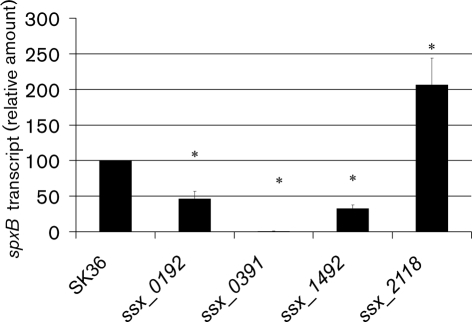
To examine whether spxB expression changes in the mutants with decreased H2O2 production, we determined the transcriptional level of spxB by real-time qPCR (Fig. 3). The results showed that the expression of spxB in ssx_0192 and ssx_1492 decreased significantly compared with SK36. The significant decrease in spxB transcription suggested that the effects of the deleted gene products in ssx_0192 and ssx_1492 on H2O2 production might occur via SpxB. In contrast, the ssx_2118 mutant demonstrated increased expression of spxB, indicating that SSA_2118 affects H2O2 production by a mechanism other than affecting spxB expression (Fig. 3).
Fig. 3.
spxB transcription in S. sanguinis SK36 and mutants. RNA preparation and qPCR were performed as described in Methods. The amount of spxB transcript was normalized to that of gyrA. Data shown are mean±sd from three biological replicates. *Significant difference compared with SK36, P<0.05.
DISCUSSION
In this study, we describe three genes involved in the production of H2O2 and the preliminary study of their effects on spxB expression in S. sanguinis. We also show that all the genes involved in the production of H2O2 identified here were also critical for the antagonism of S. sanguinis against S. mutans.
The three non-spxB mutants identified in this study demonstrated defects in H2O2 production similar to that in the spxB mutant. SSA_0192 is annotated as acetate kinase (Xu et al., 2007), which converts acetyl phosphate, the other product derived from the decarboxylation of pyruvate besides CO2 and H2O2, to acetate. Our results indicated that spxB expression was reduced in the deletion mutant ssx_0192. A possible mechanism of this regulation might be that the gene deletion in ssx_0192 caused acetyl phosphate accumulation, which caused feedback suppression of spxB expression (Wang et al., 1999). We tried to determine the acetyl phosphate concentration using the hydroxamate assay (Gorrell et al., 2005) in ssx_0192 and the wild-type strain to examine this hypothesis. However, the acetyl phosphate concentrations in both strains were too low to give a reliable result. The gene product in ssx_1492 (SSA_1492) showed high identity (76 % identity in amino acid sequence) to SpxR in S. pneumoniae, which was identified as a regulator of spxB (Ramos-Montañez et al., 2008). The conserved domain analysis of SSA_1492 showed that the protein contains a putative helix–turn–helix domain (Ramos-Montañez et al., 2008) located at the amino terminus, followed by a DRTGG-CBS domain, which are hypothesized to bind to DNA and adenosyl compounds (such as AMP and ATP), respectively. Combined with the significantly decreased expression of spxB observed in the ssx_1492 mutant, our data suggest that SSA_1492 might act as a positive regulator of spxB (Kemp, 2004; Ramos-Montañez et al., 2008; Rigali et al., 2002; Scott et al., 2004). It was hypothesized that SpxR in S. pneumoniae regulates spxB transcription in response to energy and metabolic state, and the SpxR regulon includes comparatively few genes (Ramos-Montañez et al., 2008). As some other species of streptococci, including S. mutans, S. pyogenes and Streptococcus agalactiae (group B Streptococcus), lack spxB but contain homologues of SpxR (Ramos-Montañez et al., 2008), presumably the regulatory targets of the SpxR homologues are species-specific. Our study suggests that the role of SpxR in regulating spxB is not confined to S. pneumoniae, because it seems to have the same function in S. sanguinis. The thiamine pyrophosphokinase encoded by ssa_2118 catalyses the transfer of a pyrophosphate moiety from ATP to thiamine and produces thiamine pyrophosphate. Thiamine pyrophosphate has been reported to be an important cofactor for pyruvate oxidase activity (Carlsson & Kujala, 1984; Muller et al., 1994; Tittmann et al., 1998, 2005). The deletion of ssa_2118, therefore, presumably decreases the rate of H2O2 formation by decreasing the activity of pyruvate oxidase (Fig. 1). Our results indicated that SSA_2118 was required for H2O2 production. This is also consistent with the finding that a site-specific mutation of an amino acid in SpxB that is required for thiamine pyrophosphate binding reduces H2O2 production significantly in S. pneumoniae (Ramos-Montañez et al., 2008). At the same time, it is possible that SSA_2118 involves H2O2 production by affecting not only SpxB but also other enzymes that require thiamine pyrophosphate. It was interesting that spxB expression in mutant ssx_2118 showed a significant increase. All three non-spxB mutants exhibited decreased H2O2 production but the expression levels of spxB were distinct. We hypothesize that SSA_0192 and SSA_1492 were required for the normal expression of spxB, while SSA_2118 was required for the thiamine pyrophosphate, which is the cofactor for SpxB. Both SpxB and SSA_2118 were necessary for H2O2 production.
Our study suggests that it is practical to identify H2O2 production-defective mutants in S. sanguinis by their colony morphology variation. In S. pneumoniae, the proposed roles of spxB function and regulation in pneumococcal phase variation have been somewhat contradictory (Ramos-Montañez et al., 2008). Some research suggested that spxB expression level was unlikely to directly determine colony morphology (Overweg et al., 2000) since the spxB mutant still varied in colony morphology, while another study indicated that some opaque variants were later found to be defective in SpxB function (Pericone et al., 2002). It has been suggested recently that SpxB does determine colony morphology and might play a role in phase variation (Belanger et al., 2004). The contradiction may be related to other components contributing to the phenotype, such as a capsule. In S. sanguinis, our results indicated that it was not the expression of spxB that is responsible for the morphological variation, but the H2O2 the strain produces, because all of the four H2O2 production-defective mutants had an opaque appearance, and this appearance was not dependent on the expression of spxB. For example, the expression of spxB in mutant ssx_2118 was significantly increased compared with that in the wild-type strain SK36 (Fig. 3), but the mutant ssx_2118 still presented an opaque morphology, which is presumably due to the H2O2 production deficiency of the mutant.
In S. pneumoniae, a similar screening study was performed by Ramos-Montañez et al. (2008). From screening ∼232 000 colonies, seven spontaneous mutants were identified that showed opaque appearance; six of them were found to produce less H2O2 than the wild-type strain. All were related to spxB and one of the genes, spxR, was found to regulate spxB expression. In our study, in addition to spxB and spxR, we identified two other mutants, ssx_0192 and ssx_2118, that had an opaque appearance and produced less H2O2. It is interesting that these genes were not identified in the S. pneumoniae study, even though we would expect that identical mutants in S. pneumoniae would have the same phenotype. This could be because their spontaneous screen carried out by Ramos-Montañez et al. (2008) was not saturating. It would be interesting to determine whether mutations in the ssa_0192 and ssa_2118 orthologues in S. pneumoniae (spd_1853 and spd_1779) would have also demonstrated this phenotype. If so, they will be potential virulence factors in S. pneumoniae.
The competition between pioneer colonizing oral streptococci in the oral community is of continued interest. Our studies show that the four genes identified are critical for conferring a competition advantage to S. sanguinis. This might contribute to a better understanding of interspecies interactions within oral microbial communities and serve as a foundation on which the molecular mechanisms of H2O2 production and its regulation by oral streptococci could be elucidated.
Acknowledgments
This work was supported by a National Institutes of Health grant (R01DE018138) (P. X). We thank Todd Kitten (VCU School of Dentistry) for critical discussions and suggestions about experiments.
References
- Auzat, I., Chapuy-Regaud, S., Le Bras, G., Dos, S. D., Ogunniyi, A. D., Le, T. I., Garel, J. R., Paton, J. C. & Trombe, M. C. (1999). The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol Microbiol 34, 1018–1028. [DOI] [PubMed] [Google Scholar]
- Becker, M. R., Paster, B. J., Leys, E. J., Moeschberger, M. L., Kenyon, S. G., Galvin, J. L., Boches, S. K., Dewhirst, F. E. & Griffen, A. L. (2002). Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger, A. E., Clague, M. J., Glass, J. I. & LeBlanc, D. J. (2004). Pyruvate oxidase is a determinant of Avery's rough morphology. J Bacteriol 186, 8164–8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, J. & Kujala, U. (1984). Pyruvate oxidase activity dependent on thiamine pyrophosphate, flavin adenine dinucleotide and orthophosphate in Streptococcus sanguis. FEMS Microbiol Lett 25, 53–56. [Google Scholar]
- Caufield, P. W., Dasanayake, A. P., Li, Y., Pan, Y., Hsu, J. & Hardin, J. M. (2000). Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun 68, 4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, C. W., Heath, J., Hampton, K. K. & Preston, F. E. (1993). Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol 39, 179–182. [DOI] [PubMed] [Google Scholar]
- García-Mendoza, A., Liebana, J., Castillo, A. M., de la Higuera, A. & Piedrola, G. (1993). Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol 39, 434–439. [DOI] [PubMed] [Google Scholar]
- Ge, X., Kitten, T., Chen, Z., Lee, S. P., Munro, C. L. & Xu, P. (2008a). Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect Immun 76, 2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y., Caufield, P. W., Fisch, G. S. & Li, Y. (2008b). Streptococcus mutans and Streptococcus sanguinis colonization correlated with caries experience in children. Caries Res 42, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg, I. & Sadovnic, M. (1998). Gamma globulin, Evan's blue, aprotinin A PLA2 inhibitor, tetracycline and antioxidants protect epithelial cells against damage induced by synergism among streptococcal hemolysins, oxidants and proteinases: relation to the prevention of post-streptococcal sequelae and septic shock. FEMS Immunol Med Microbiol 22, 247–256. [DOI] [PubMed] [Google Scholar]
- Ginsburg, I. & Varani, J. (1993). Interaction of viable group A streptococci and hydrogen peroxide in killing of vascular endothelial cells. Free Radic Biol Med 14, 495–500. [DOI] [PubMed] [Google Scholar]
- Gorrell, A., Lawrence, S. H. & Ferry, J. G. (2005). Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. J Biol Chem 280, 10731–10742. [DOI] [PubMed] [Google Scholar]
- Jakubovics, N. S., Smith, A. W. & Jenkinson, H. F. (2002). Oxidative stress tolerance is manganese (Mn2+) regulated in Streptococcus gordonii. Microbiology 148, 3255–3263. [DOI] [PubMed] [Google Scholar]
- Kemp, B. E. (2004). Bateman domains and adenosine derivatives form a binding contract. J Clin Invest 113, 182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, M. & Holmgren, K. (1981). Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect Immun 31, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth, J., Merritt, J., Shi, W. & Qi, F. (2005). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187, 7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth, J., Zhang, Y. & Herzberg, M. C. (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190, 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth, J., Vu, H., Zhang, Y. & Herzberg, M. C. (2009). Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol 191, 6281–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu, H. K., He, X., Lux, R., Anderson, M. H. & Shi, W. (2007). Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev 71, 653–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, Y. A., Schumacher, G., Rudolph, R. & Schulz, G. E. (1994). The refined structures of a stabilized mutant and of wild-type pyruvate oxidase from Lactobacillus plantarum. J Mol Biol 237, 315–335. [DOI] [PubMed] [Google Scholar]
- Mylonakis, E. & Calderwood, S. B. (2001). Infective endocarditis in adults. N Engl J Med 345, 1318–1330. [DOI] [PubMed] [Google Scholar]
- Overweg, K., Pericone, C. D., Verhoef, G. G. C., Weiser, J. N., Meiring, H. D., De Jong, A. P. J. M., De Groot, R. & Hermans, P. W. M. (2000). Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect Immun 68, 4604–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, S., Senty, L., Das, S., Noe, J. C., Munro, C. L. & Kitten, T. (2005). Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect Immun 73, 6064–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone, C. D., Overweg, K., Hermans, P. W. M. & Weiser, J. N. (2000). Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68, 3990–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone, C. D., Bae, D., Shchepetov, M., McCool, T. & Weiser, J. N. (2002). Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J Bacteriol 184, 4392–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Montañez, S., Tsui, H. C., Wayne, K. J., Morris, J. L., Peters, L. E., Zhang, F., Kazmierczak, K. M., Sham, L. T. & Winkler, M. E. (2008). Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol 67, 729–746. [DOI] [PubMed] [Google Scholar]
- Rigali, S., Derouaux, A., Giannotta, F. & Dusart, J. (2002). Subdivision of the helix–turn–helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277, 12507–12515. [DOI] [PubMed] [Google Scholar]
- Rosan, B. & Lamont, R. J. (2000). Dental plaque formation. Microbes Infect 2, 1599–1607. [DOI] [PubMed] [Google Scholar]
- Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G. & Hardie, D. G. (2004). CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest 113, 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittmann, K., Proske, D., Spinka, M., Ghisla, S., Rudolph, R., Hubner, G. & Kern, G. (1998). Activation of thiamin diphosphate and FAD in the phosphate-dependent pyruvate oxidase from Lactobacillus plantarum. J Biol Chem 273, 12929–12934. [DOI] [PubMed] [Google Scholar]
- Tittmann, K., Wille, G., Golbik, R., Weidner, A., Ghisla, S. & Hubner, G. (2005). Radical phosphate transfer mechanism for the thiamin diphosphate- and FAD-dependent pyruvate oxidase from Lactobacillus plantarum. Kinetic coupling of intercofactor electron transfer with phosphate transfer to acetyl-thiamin diphosphate via a transient FAD semiquinone/hydroxyethyl-ThDP radical pair. Biochemistry 44, 13291–13303. [DOI] [PubMed] [Google Scholar]
- Tleyjeh, I. M., Steckelberg, J. M., Murad, H. S., Anavekar, N. S., Ghomrawi, H. M., Mirzoyev, Z., Moustafa, S. E., Hoskin, T. L., Mandrekar, J. N. & other authors (2005). Temporal trends in infective endocarditis: a population-based study in Olmsted County, Minnesota. JAMA 293, 3022–3028. [DOI] [PubMed] [Google Scholar]
- Turner, L. S., Das, S., Kanamoto, T., Munro, C. L. & Kitten, T. (2009). Development of genetic tools for in vivo virulence analysis of Streptococcus sanguinis. Microbiology 155, 2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, Y., Agematsu, K., Kikuchi, K., Matsuzaki, S., Imai, S., Takamoto, M., Sugane, K., Sugiura, T., Konishi, Y. & other authors (2006). Secretory IgA, salivary peroxidase, and catalase-mediated microbicidal activity during hydrogen peroxide catabolism in viridans streptococci: pathogen coaggregation. J Infect Dis 194, 98–107. [DOI] [PubMed] [Google Scholar]
- Wang, H., Tseng, C. P. & Gunsalus, R. P. (1999). The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J Bacteriol 181, 5303–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser, J. N., Austrian, R., Sreenivasan, P. K. & Masure, H. R. (1994). Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62, 2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Alves, J. M., Kitten, T., Brown, A., Chen, Z., Ozaki, L. S., Manque, P., Ge, X., Serrano, M. G. & other authors (2007). Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol 189, 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]