Abstract
Hemiascomycetes, including the pathogen Candida albicans, acquire nitrogen from urea using the urea amidolyase Dur1,2, whereas all other higher fungi use primarily the nickel-containing urease. Urea metabolism via Dur1,2 is important for resistance to innate host immunity in C. albicans infections. To further characterize urea metabolism in C. albicans we examined the function of seven putative urea transporters. Gene disruption established that Dur3, encoded by orf 19.781, is the predominant transporter. [14C]Urea uptake was energy-dependent and decreased approximately sevenfold in a dur3Δ mutant. DUR1,2 and DUR3 expression was strongly induced by urea, whereas the other putative transporter genes were induced less than twofold. Immediate induction of DUR3 by urea was independent of its metabolism via Dur1,2, but further slow induction of DUR3 required the Dur1,2 pathway. We investigated the role of the GATA transcription factors Gat1 and Gln3 in DUR1,2 and DUR3 expression. Urea induction of DUR1,2 was reduced in a gat1Δ mutant, strongly reduced in a gln3Δ mutant, and abolished in a gat1Δ gln3Δ double mutant. In contrast, DUR3 induction by urea was preserved in both single mutants but reduced in the double mutant, suggesting that additional signalling mechanisms regulate DUR3 expression. These results establish Dur3 as the major urea transporter in C. albicans and provide additional insights into the control of urea utilization by this pathogen.
INTRODUCTION
Nitrogen sources play important roles in regulating fungal dimorphism. For example, Ceratocystis ulmi cells grow as yeasts in the presence of proline but as hyphae in the presence of ammonia, arginine and most other nitrogen sources (Kulkarni & Nickerson, 1981). Trigonopsis variabilis cells grow as budding yeasts in the presence of ammonium sulfate and as triangles with methionine (Sentheshanmuganathan & Nickerson, 1962).
Urea is an important nitrogen source for many fungi and is also important for microbial pathogenesis in humans. Urease is a virulence factor for some pathogenic fungi, including Cryptococcus neoformans (Cox et al., 2000a) and Coccidioides immitis (Cole, 1997), and for the bacterial pathogens Helicobacter pylori (Eaton et al., 1991) and Proteus mirabilis (Jones et al., 1990).
The role of urea in the dimorphic human fungal pathogen Candida albicans, however, has been unclear. Although Dastidar and co-workers reported in 1967 that many strains of C. albicans grow well with urea as the sole source of nitrogen (Dastidar et al., 1967), this observation drew little attention based on numerous reports that C. albicans lacks urease (Odds, 1988). This apparent contradiction was recently explained by the discovery that C. albicans uses urea amidolyase to hydrolyse urea (Ghosh et al., 2009). The enzyme urea amidolyase, encoded by DUR1,2, was first characterized in Candida utilis (Roon et al., 1972). Dur1,2 is a multifunctional biotin-dependent enzyme (Roon & Levenberg, 1972) with domains for urea carboxylase and allophanate (urea carboxylate) hydrolase activity (Cooper et al., 1980). Recently, we showed that higher fungi exhibit a dichotomy with regard to urea utilization (Navarathna et al., 2010). All of the hemiascomycetes use urea amidolyase (Dur1,2). Most other higher fungi use urease exclusively, except the Sordariomycetes (Magnaportha, Fusarium and Nectria), which have both urease and urea amidolyase. Still, the vast majority of yeasts/hemiascomycetes preserve the dichotomy by having urea amidolyase but not urease.
Why do Candida and Saccharomyces use an energy-dependent, biotin-containing urea amidolyase system when the same overall reaction could be accomplished by the simpler urease? This question becomes even more germane when we consider that all strains of C. albicans are biotin auxotrophs (Odds, 1988), and that two to four times as much biotin is required for maximum growth of Saccharomyces cerevisiae on urea, allantoic acid or allantoin as sole nitrogen source (Di Carlo et al., 1953). We have suggested that urea amidolyase allows the hemiascomycetes to jettison their sole Ni(II)- and Co(II)-dependent enzyme and thus to require two fewer transition metals (Navarathna et al., 2010). In addition, urea amidolyase may create a urea-dependent signalling pathway, and we have hypothesized that such a pathway might be related to fungal pathogenicity (Ghosh et al., 2009).
While examining the role of arginine-induced germ tube formation in the escape of C. albicans from murine macrophages (Ghosh et al., 2009), we deleted the DUR1,2 gene from the wild-type strain A72. The dur1,2Δ mutant KWN6 was unable to grow on urea as the sole nitrogen source, stimulate germ tube formation in response to l-arginine or urea, or escape from the murine macrophage cell line RAW 264.7. These abilities were restored in the reconstituted strains KWN7 and KWN8 (Ghosh et al., 2009).
Given the divergence in mechanisms for urea degradation, we examined whether a similar dichotomy exists in the mechanisms for urea transport. Here, we define the major urea transporter in C. albicans, and examine the regulation of urea transport and urea utilization by other known nitrogen sources and two transcription factors involved in the induction of genes that are subject to nitrogen catabolite repression (NCR).
METHODS
Strains and growth conditions.
The C. albicans strains used for this study are listed in Table 1. For all experiments, C. albicans cells were grown overnight in 50 ml yeast peptone dextrose (YPD) medium at 30 °C with aeration, as previously described (Navarathna et al., 2005). Cells were harvested by centrifugation at 5000 r.p.m. for 10 min, washed once with 50 ml sterile, non-pyrogenic normal saline (Quality Biological, Inc.) and resuspended in 10 ml saline before quantifying cell numbers using a Petroff–Hausser counting chamber. For gene expression analysis, C. albicans cells were grown for 16 h in 50 ml YPD medium (Kulkarni & Nickerson, 1981), washed three times with PBS, and added to glucose peptone (GP) medium containing 10 mM of the indicated nitrogen sources. Fresh cultures were started at OD600 0.2 using overnight YPD cultures, and grown up to OD600 0.8 (for about 4–5 h) for RNA extraction.
Table 1.
Strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| SC5314 | Wild-type | Gillum et al. (1984) |
| A72 | Wild-type | ATCC* MYA-2430 |
| KWN6 | dur1,2Δ/dur1,2Δ | Ghosh et al. (2009) |
| KWN7 | dur1,2Δ/dur1,2Δ : : DUR1,2 | Ghosh et al. (2009) |
| KWN8 | dur1,2Δ : : DUR1,2/dur1,2Δ : : DUR1,2 | Ghosh et al. (2009) |
| DUR3M4A and -B | dur3Δ/dur3Δ | This study |
| DUR3K2A and -B | dur3Δ/dur3Δ : : DUR3 | This study |
| GAT1M4A | gat1Δ/gat1Δ | Dabas & Morschhäuser (2007) |
| GLN3M4A | gln3Δ/gln3Δ | Dabas & Morschhäuser (2007) |
| Δgln3GAT1M4A | gat1Δ/gat1Δ gln3Δ/gln3Δ | Dabas & Morschhäuser (2007) |
*American Type Culture Collection.
Disruption and complementation of DUR3 (orf 19.781).
A DUR3 deletion cassette was constructed by amplifying the DUR3 upstream and downstream regions from genomic DNA of strain SC5314 with the primer pairs DUR31/DUR32 and DUR33/DUR34, respectively (Table 2), and cloning the SacI/SacII- and XhoI/ApaI-digested PCR products on both sides of the SAT1 flipper cassette of plasmid pSFS1, in which expression of the caFLP gene is controlled by the SAP2 promoter (Reuß et al., 2004). The SacI–ApaI fragment from the resulting plasmid pDUR3M3 was used to sequentially delete the DUR3 alleles of strain SC5314 by the SAT1 flipping strategy, as described previously (Dunkel et al., 2008), resulting in the homozygous dur3Δ mutants DUR3M4A and DUR3M4B. For reintroduction of a functional DUR3 copy, the DUR3 coding region plus 0.3 kb of upstream and 0.5 kb of downstream sequence was amplified with the primers DUR31 and DUR35, and the SacI/SacII-digested PCR product was substituted for the DUR3 upstream fragment in pDUR3M3. The SacI–ApaI fragment from the resulting plasmid pDUR3K1 was used to reintegrate DUR3 at the endogenous locus of the dur3Δ mutants DUR3M4A and DUR3M4B, followed by recycling of the SAT1 flipper cassette, to generate the complemented strains DUR3K2A and DUR3K2B.
Table 2.
Oligonucleotides used in this study
All primers are in 5′→3′ orientation. Introduced restriction sites are underlined.
| Primer | Sequence |
|---|---|
| DUR31 | TTGAGCTCGATAAGACTAAATGACTGCG |
| DUR32 | CACTCCGCGGTAATGGGACATATGAATCAGCC |
| DUR33 | CATTGACGATGCACTCGAGGAGAAGAAAGG |
| DUR34 | GTCAGCATAATCAGGGCCCAATCAGAAGCC |
| DUR35 | TAATCCGCGGTATTCTCATGATTTGCCCC |
| DUR1,2 RT LT | TTTGTTGGTCCATCTGGTGA |
| DUR1,2 RT RT | TCTGGCTTCTTTGGCATCTT |
| DUR RT LT | AGCTTGGCAAAATGAACACC |
| DUR RT RT | GCTGGGGTGACATATCCATC |
| DUR3 RT LT | ACAGTGCTCGAGAAGGTGGT |
| DUR3 RT RT | AAAGCAGCTGAAGCCAATGT |
| DUR4 RT LT | ATCCATGGGCTGGTTATTCA |
| DUR4 RT RT | CACCTGCCTGGTTCAAATCT |
| DUR7 RT LT | GGGTTATTTGGCCAATGTCT |
| DUR7 RT RT | AAAAGGGGACCAATCCAATC |
| NPR2 RT LT | AATGGCGGAAGAATGTCAAG |
| NPR2 RT RT | ACTTCCAGGGTAGGGCAAGT |
| 5915 RT LT | ACGTTGACTCCGATCGAAAC |
| 5915 RT RT | GATTGCAGCACCAGTAGCAA |
| 5017 RT LT | TTTGTCCAGGTGTGATTCCA |
| 5017 RT RT | GTGATATGCCGTCGAAATCC |
| CDC36 RT LT | GAGCGTCCAGTATAAATCCACCAC |
| CDC36 RT RT | TCAAGACGGGCTCCACATTACTAT |
Mouse candidiasis model.
Outbred 6–8-week-old (18–20 g) BALB/c female mice obtained from Charles River Laboratories were randomly allocated to groups of five animals and housed and cared for according to guidelines of the National Cancer Institute (NCI) Animal Care and Use Committee. Each group of mice was inoculated intravenously in the lateral caudal tail vein using a 27-gauge needle with a volume of 0.1 ml containing 106 C. albicans cells (Navarathna et al., 2005, 2007). Clinical signs of illness in each mouse were evaluated three times daily, and moribund mice that displayed sunken eyes, arched back posture, dehydration, ruffled hair or difficulty walking were euthanized immediately by placing them in a closed chamber filled with CO2, and processed for complete necropsy and collection of tissues for histopathological examination.
RNA extraction and gene expression analysis by RT-PCR.
RNA isolation was done using a standard hot phenol procedure (Köhrer & Domdey, 1991). Reverse transcription was conducted using 5 μg total RNA extracted from each sample using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions for oligo-dT priming. Quantitative PCR (qPCR) was conducted as previously described (Navarathna & Roberts, 2010) using ABsolute QPCR SYBR Green Mix (Thermo Scientific), an Opticon I instrument and Opticon I software (Bio-Rad). Samples were analysed by PCR in triplicate and normalized to internal CDC36 mRNA levels. Melting curve analysis was performed to ensure that a single product was produced in each reaction (Navarathna & Roberts, 2010). The qPCR primers used in this study are listed in Table 2, and all products were 75–125 bases in length.
Uptake of [14C]urea.
All C. albicans cells were grown overnight in 50 ml YPD, washed and resuspended in PBS, and used as the inoculum (OD600 0.2) for fresh cultures in glucose phosphate proline (GPP) medium grown at 30 °C for 3–4 h while shaking at 150 r.p.m. Cell numbers were counted and adjusted so that all uptake assays employed an equal number of cells. Assays were done in GP buffer in 14 ml round-bottomed tubes (BD) with or without 5 mM sodium azide, using ∼107 cells ml−1 and 1 μCi (3.7×104 Bq, 1.15 μg ml−1, 19.2 μM) [14C]urea (American Radiolabelled Chemicals). After incubation with orbital shaking at 150 r.p.m. for 5 min, unless otherwise indicated, triplicate 0.5 ml samples were collected. Two 0.2 ml volumes of the cell suspension were sequentially overlaid onto 100 μl 30 % sucrose in 5×49 mm microfuge tubes and centrifuged in a Beckman Microfuge B for 30 s. After removing the upper aqueous layer, the resulting cell pellet containing 4×106 cells was washed by adding 300 μl PBS and recentrifuged. After removing the supernatant fluid, the tips of the microfuge tubes containing the cell pellets were cut off and shaken in scintillation fluid before quantifying the internalized urea using a scintillation counter.
Bioinformatics analysis.
Sequences of fungal proteins were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and the Fungal Genome Initiative (http://www.broad.mit.edu/annotation/fgi/). DUR3 homologues were identified using S. cerevisiae Dur3 (YHL016C) as a query for blastp searches. All blast searches were conducted using default parameters. We examined the distribution of DUR3 in the genomes reported previously (Navarathna et al., 2010).
Statistics.
The probability of survival as a function of time was determined by the Kaplan–Meier method, and significance was determined by the log-rank (Mantel–Cox) test and the Jehan–Breslow–Wilcoxon test using GraphPad Prism software. qRT-PCR data were analysed using the Pfaffl method (Pfaffl, 2001) and were normalized in two steps. CDC36, an abundantly expressed nuclear protein (Collart & Struhl, 1994), was used as the internal control because we found its mRNA to be the most invariant for normalizing C. albicans gene expression analysis (Navarathna & Roberts, 2010; Pendrak et al., 2004a, b, c). First, mean values for the internal CDC36 control were subtracted from the corresponding experimental sample, and the variance was calculated as the square root of the sum of the squares of the standard deviations. Second, each sample group was normalized to C. albicans grown in YPD medium, except in Fig. 2, where GPP was used. The results are presented as fold change using 2−Ct of this normalized Ct value. Statistical differences among groups were analysed with two-way analysis of variance (ANOVA) using GraphPad Prism software.
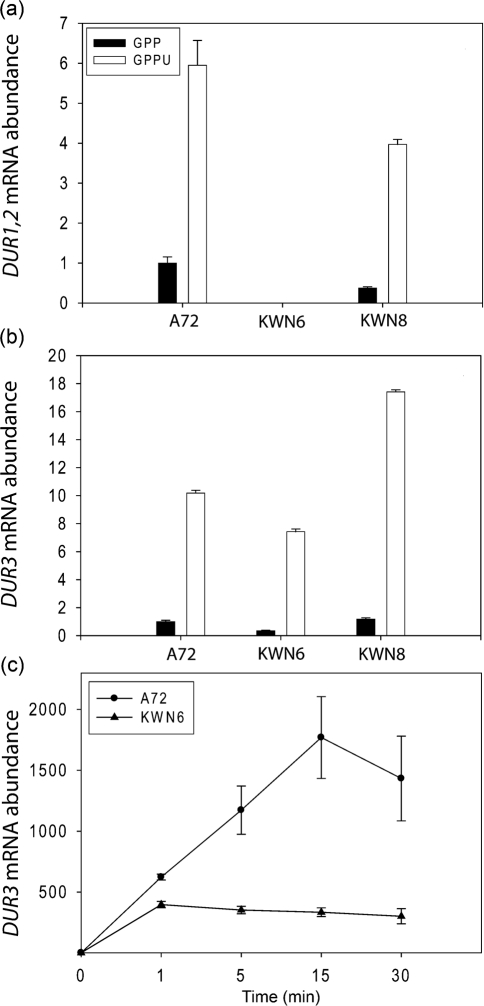
Fig. 2.
Urea induction of DUR1,2 and DUR3 gene expression. mRNA abundance was determined by qPCR for DUR1,2 (a) and DUR3 (b) in C. albicans strains A72 (wild-type), KWN6 (dur1,2Δ/dur1,2Δ) and the reconstituted strain KWN8 (dur1,2Δ : : DUR1,2/dur1,2Δ : : DUR1,2). The cells were grown in defined GPP (filled bars) or GPPU (open bars) medium. (c) Kinetics of induction of DUR3 in the wild-type strain A72 and the dur1,2 knockout strain KWN6 in GPU medium.
RESULTS
Expression of potential urea transporters
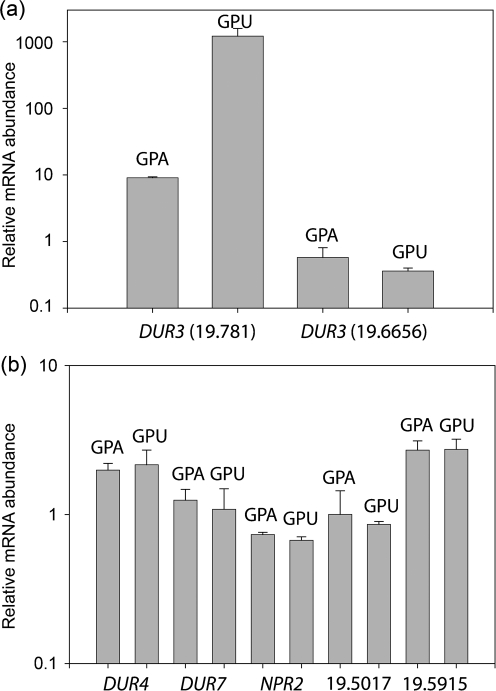
The yeast S. cerevisiae metabolizes urea by means of Dur1,2 and the urea transporters Dur3 and Dur4 (Cooper, 1982). DUR1,2 and DUR3 are inducible by allophanate, whereas DUR4 encodes a constitutive urea-facilitated diffusion transporter (Cooper, 1982). The C. albicans orthologue DUR1,2 was identified by Gene Ontology (GO) annotation. Remarkably, the C. albicans genome contains seven genes encoding putative urea transporters: DUR3 (orf 19.781), DUR3 (orf 19.6656), DUR4 (orf 19.5677), DUR7 (orf 19.7205), NPR2 (orf 19.328), orf 19.5915 and orf 19.5017. The gene name DUR3 has been used for both orf 19.781 and orf 19.6656, but orf 19.781 is the orthologue of DUR3 of S. cerevisiae, and we therefore refer to this gene as DUR3. DUR3, DUR4 and DUR7 are fungal-specific in that they lack human or murine orthologues.
Based on gene expression levels in minimal medium, we separated the putative urea transporters into two categories. DUR3 and orf 19.6656 reproducibly showed high expression, with Ct values less than 22, whereas the others all had Ct values less than 24. Addition of urea instead of ammonia to the growth medium (glucose phosphate urea; GPU) resulted in a 100-fold increase in DUR3 gene expression but no increase for orf 19.6656 relative to the basal expression in YPD (Fig. 1a). None of the other five putative transporter genes showed significant responses to urea relative to that in YPD (Fig. 1b).
Fig. 1.
Effect of urea on the expression of seven putative urea transporters. mRNA abundance was determined by qPCR using C. albicans A72 cells grown in GPA (glucose phosphate ammonia) or GPU medium. Experiments were performed in triplicate; error bars, sem. Genes exhibiting higher expression are shown in (a) and low-expression genes are shown in (b). GPA and GPU expression levels were normalized to those of YPD. Cells grown in GPP medium showed induction of DUR3 similar to that of cells grown in GPA (data not shown).
Urea induction of DUR3 is not dependent upon DUR1,2 expression
The dur1,2Δ mutant KWN6 and the complemented strain KWN8 (Ghosh et al., 2009) were used to determine whether the DUR3 urea response depended on a metabolite downstream of Dur1,2, such as allophanate. Expression of DUR1,2 and DUR3 was increased six- and 10-fold, respectively, for C. albicans A72 grown with urea (glucose phosphate proline urea; GPPU) rather than without (GPP; Fig. 2a, b). Expression and induction of DUR1,2 (Fig. 2a) was lost in the dur1,2Δ mutant, and restored in the fully reconstituted strain KWN8 (Fig. 2a). However, in the dur1,2Δ mutant, DUR3 expression was still increased ∼20-fold by urea (GPPU) relative to the same strain in GPP. Thus, DUR3 basal expression was slightly reduced in the absence of DUR1,2, but its relative induction by urea was preserved.
However, examining the time dependence of DUR3 induction by urea in A72 (wild-type) versus KWN6 (dur1,2Δ) cells revealed a delayed effect of urea metabolism via Dur1,2 on DUR3 induction (Fig. 2c). For this experiment, overnight C. albicans cultures grown in YPD were washed three times before transfer to GPU medium to examine gene expression induced by urea. Within 1 min of addition of urea, mRNA levels increased 400- to 600-fold in both cultures. This immediate induction was largely independent of Dur1,2. However, DUR3 mRNA increased an additional threefold by 15 min in wild-type cells, whereas no further increase was seen in the mutant lacking DUR1,2 (Fig. 2c). Therefore, a metabolite of urea downstream of Dur1,2 may account for a secondary induction of DUR3. Alternatively, upstream feedback inhibition of further DUR3 induction may occur due to urea accumulation in the cell.
We also tested DUR1,2 and DUR3 expression in the presence of 10 mM allantoin. Allantoin, an intermediate of purine degradation that is subsequently converted to urea, increased DUR1,2 mRNA levels 3.8±0.5-fold and DUR3 mRNA levels 9.2±0.5-fold, compared with GPP. Finally, 10 mM parabanic acid (oxaluric acid), a gratuitous inducer of DUR1,2 in S. cerevisiae (Cooper et al., 1980), also induced DUR1,2 mRNA levels 6±0.08-fold and DUR3 mRNA levels 1.8±0.02-fold in C. albicans (data not shown).
Transport of [14C]urea into C. albicans
The functional importance of Dur3 in urea transport was examined by measuring [14C]urea uptake (Table 3). Preliminary experiments established that optimal uptake was attained at 5 min and gradually decreased thereafter due to catabolism (data not shown). By 5 min, urea uptake levels for a dur3Δ mutant were approximately three times lower than those for the wild-type C. albicans. Reconstitution of one allele of DUR3 in the deletion mutant restored urea uptake to wild-type levels (Table 3). This finding is consistent with our observation that the dur3Δ mutant grew very slowly on GPU, with urea as the sole nitrogen source. The remaining urea transport in the dur3Δ mutant (Table 3) was likely achieved by one or more of the other, non-inducible urea transporters (Fig. 1). Urea transport by Dur3 was energy-dependent in that it was reduced four- to fivefold in the presence of 5 mM sodium azide (Table 3).
Table 3.
Urea uptake by wild-type and mutant C. albicans
Values shown are radioactivity internalized in 4–5×106 cells after exposure to [14C]urea for 5 min, and are the mean±sem of triplicate experiments.
| Strain | Treatment | c.p.m. |
|---|---|---|
| SC5314 (wild-type) | GP with [14C]urea | 2707±660 |
| SC5314 (wild-type) | GP with [14C]urea and NaN3 | 517±129 |
| DUR3M4A (dur3Δ) | GP with [14C]urea | 960±280 |
| DUR3K2A (dur3Δ+DUR3) | GP with [14C]urea | 2698±316 |
Distribution of the urea transporter DUR3 in fungi
Because the fungal-specific urea transporter DUR3 resides only 1000 bp upstream of DUR1,2 on chromosome 1 in C. albicans, we expected that its phylogenetic distribution might parallel that of DUR1,2, but it did not. DUR3 orthologues were present as a single copy in all of the 22 ascomycete and basidiomycete genomes searched (Navarathna et al., 2010), except for those of Ustilago maydis and Rhizopus oryzae, where it was absent, and Yarrowia lipolytica, where it was present in two copies. Y. lipolytica also has two copies of DUR1,2. This gene duplication may have occurred relatively recently, at the species level, resulting in co-duplication of DUR1,2 and DUR3. DUR3 orthologues were also not detected in three fungi not included in our previous study, Histoplasma capsulatum, Coccidioides immitis and Paracoccidioides sp.
Regulation of DUR1,2 and DUR3 expression by other nitrogen sources
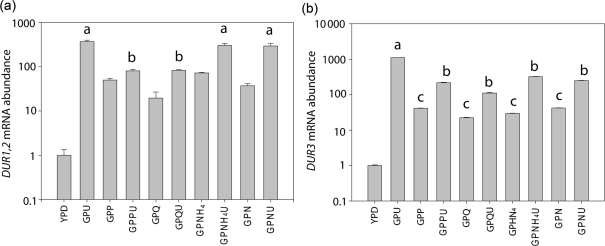
We next explored how urea induction of DUR1,2 and DUR3 relates to NCR regulation of these genes. For this purpose, C. albicans SC5314 cells were grown in defined media containing the well-characterized nitrogen sources proline, glutamine, ammonium sulfate and asparagine, all at 10 mM (Fig. 3). In all cases, expression of DUR1,2 and DUR3 was higher in the GP basal media than in YPD medium. Addition of proline or glutamine at 10 mM in the presence of urea significantly repressed expression relative to urea alone (compare GPPU and GPQU with GPU in Fig. 3). However, addition of 10 mM urea further stimulated expression of DUR1,2 and DUR3 mRNA (compare GPQU, GPNU and GPNH4U with GPQ, GPN and GPNH4, respectively, in Fig. 3). Notably, the levels of gene expression were highest when urea was the sole nitrogen source added to the basal medium. Therefore, the DUR1,2 and DUR3 genes appear to be under limited NCR in that their responsiveness to urea is largely independent of NCR regulation.
Fig. 3.
Effect of nitrogen source on DUR1,2 and DUR3 expression in C. albicans. DUR1,2 (a) and DUR3 (b) mRNA abundance was determined by qPCR for C. albicans SC5314 cells grown with the following additives in defined GP-based media: P, l-proline; U, urea; NH4, ammonia; Q, l-glutamine; N, asparagine; all at 10 mM. Similar results were obtained when A72 wild-type cells were subjected to the same treatments. Experiments were performed in triplicate; error bars, sem. Addition of proline and glutamine significantly reduced DUR1,2 expression (b vs a=P<0.001), and addition of l-proline, glutamine, ammonia and asparagine significantly reduced expression of DUR3 (b vs a=P<0.001 by ANOVA) compared with expression levels in GPU. DUR3 expression was induced by addition of urea, regardless of the nitrogen source tested (c vs b=P<0.001, 0.001, 0.05 and 0.001 for GPP, GPQ, GPNH4 and GPN, respectively, by ANOVA).
Regulation of urea utilization by GATA transcription factors
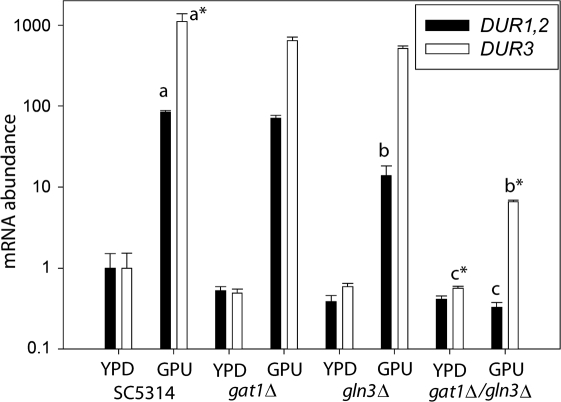
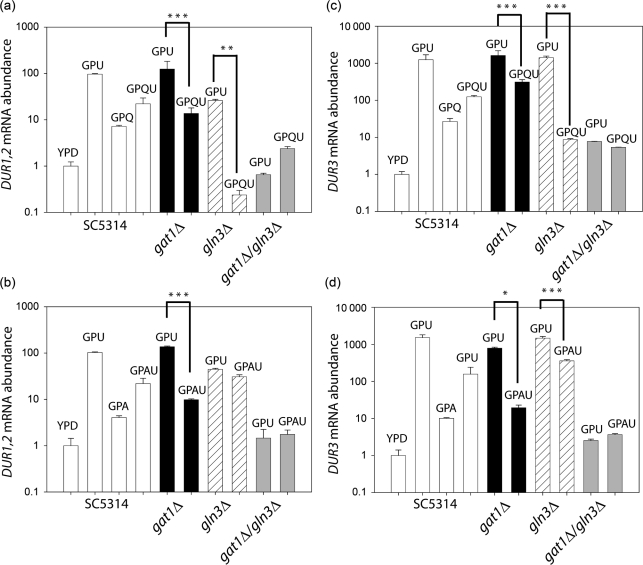
To further examine the contribution of NCR to regulation of these urea utilization and transporter genes, we compared basal (YPD) and induced (GPU) expression of DUR1,2 and DUR3 pairwise in four strains: wild-type C. albicans and mutants lacking one or both of the GATA transcription factors Gat1 and Gln3 (Fig. 4). Induction of DUR1,2 mRNA was not reduced in the gat1Δ mutant, was significantly reduced in the gln3Δ mutant (P<0.001 for gln3Δ vs wild-type), and abolished in the double mutant (P<0.001 for gat1Δ gln3Δ vs gat1Δ, Fig. 4). Interestingly, urea induction of the transporter DUR3 was not affected by deletion of GAT1 or GLN3 individually, but it was significantly reduced in the double mutant (P<0.001 for gat1Δ gln3Δ vs wild-type). However, urea still induced DUR3 expression in the double mutant (P<0.01 for GPU vs YPD), indicating that other transcriptional regulators are involved.
Fig. 4.
Role of GATA transcription factors on expression of DUR1,2 and DUR3. The cells were grown in GPU and YPD. Open bars, DUR3; filled bars, DUR1,2. Lower-case type with an asterisk represents significant changes in DUR3 expression in GPU relative to expression in the wild-type strain in YPD. Lower-case type without an asterisk indicates significant changes in DUR1,2 expression in GPU relative to that of the wild-type strain in YPD medium. Experiments were performed in triplicate; error bars, sem. Compared with the wild-type, the gln3Δ and gln3Δ gat1Δ mutants showed significantly reduced DUR1,2 expression in GPU (P<0.001 for b vs a and c vs a), and DUR1,2 expression was also significantly lower in the gln3Δ gat1Δ double mutant than in the gln3Δ single mutant (P<0.001 for c vs b). In contrast, DUR3 expression was not affected in the gln3Δ and gat1Δ single mutants, although it was significantly reduced (P<0.001 for b* vs a*) in the gln3Δ gat1Δ double mutant compared with the wild-type strain grown in GPU. The strains did not differ in expression when grown in YPD.
NCR regulation of DUR3 involves additional signals
To confirm that, in addition to Gat1 and Gln3, other signalling mechanisms control DUR1,2 and DUR3 expression, we tested DUR1,2 and DUR3 mRNA levels in the gat1Δ, gln3Δ and gat1Δ gln3Δ mutants in the presence of different nitrogen sources. DUR1,2 mRNA levels were high in gat1Δ cells grown in GPU, but addition of glutamine or ammonia significantly reduced its expression (P<0.001, Fig. 5a, b). A similar pattern of DUR1,2 expression was seen in the gln3Δ strain grown with GPU and GPU with glutamine (P<0.002). However, addition of ammonia did not significantly reduce DUR1,2 induction. In the gat1Δ gln3Δ double mutant, urea failed to induce DUR1,2 expression above basal levels.
Fig. 5.
Role of GATA transcription factors in the gene expression of DUR1,2 (a, b) and DUR3 (c, d) with or without other nitrogen sources (10 mM) in the growth media containing urea (10 mM). (a, c) Gene expression was quantified in the presence of glutamine (Q). (b, d) Gene expression was quantified in the presence of ammonia. DUR1,2 expression (a, b) and DUR3 expression (c, d) in the indicated transcription factor mutants was compared with that in the wild-type strain SC5314. Experiments were performed in triplicate; error bars, sem. In the gat1Δ mutant, addition of glutamine and ammonia caused a significant reduction of DUR1,2 expression (P<0.001) compared with the levels in GPU medium. Both the gat1Δ and the gln3Δ mutant exhibited reduced DUR3 expression when glutamine (P<0.001) or ammonia (P<0.05 for gat1Δ and 0.001 for gln3Δ) was added to GPU. *, P<0.05; **, P<0.002; ***, P<0.001.
As shown in Fig. 4, deletion of GAT1 or GLN3 individually did not alter the urea induction of DUR3 expression. Glutamine and ammonia significantly suppressed DUR3 induction in the wild-type as well as in the gat1Δ and gln3Δ single mutants, suggesting that neither GATA transcription factor tested is absolutely required for the induction or repression of this gene (Fig. 5c, d). However, induction of DUR3 by urea was diminished in the double mutant, and addition of an alternative nitrogen source in this mutant did not further reduce its expression.
DUR3 does not contribute to C. albicans virulence in mice
The tail vein injection mouse candidaemia model was used to compare the virulence of the wild-type strain SC5314, the dur3Δ mutant DUR3M4A, and the complemented strain DUR3K2A. No significant differences in the survival of mice infected with the three strains were observed (data not shown).
DISCUSSION
Although urea metabolism in the hemiascomycetes diverges from that of other fungi in using the biotin-containing enzyme Dur1,2 rather than the nickel-containing urease (Navarathna et al., 2010), these fungi have in common a number of putative urea transporters. Among these, we have identified Dur3 as the major urea-inducible transporter in C. albicans. Dur3 is responsible for ∼80 % of the energy-dependent urea transport into C. albicans in basal medium, and presumably accounts for a much higher percentage when induced. The expression of mRNAs for several non-inducible urea transporters in C. albicans (Fig. 1) is consistent with our observation that the dur3Δ mutant exhibited undiminished pathogenicity in a mouse model of disseminated candidiasis. It is also likely that sufficient amounts of other nitrogen sources are available for growth, so urea uptake is not limiting. The fact that the dur3Δ mutant exhibited undiminished pathogenicity is totally consistent with our previous work showing that Dur1,2 is needed for C. albicans to escape from macrophages. In that case, the urea was produced intracellularly from l-arginine. Thus, no import of exogenous urea was needed.
Notably, all other urea transporters are expressed at lower mRNA levels than DUR3. Indeed, some of these putative urea transporters may not be functional or may have other dominant substrates. For instance, Dur3 in S. cerevisiae has recently been shown to be a functional boron transporter (Nozawa et al., 2006).
Although urea can serve as a nitrogen source for C. albicans (Dastidar et al., 1967), the inducible transporter Dur3 and the catabolic enzyme Dur1,2 do not exhibit classical NCR regulation. When rich nitrogen sources such as ammonia and asparagine are available, cells preferentially utilize these sources, while repressing genes that are responsible for breaking down poorer nitrogen sources such as proline, arginine and urea (Cooper, 1982). When cells are starved for nitrogen, these NCR-regulated genes are induced. In S. cerevisiae, urea amidolyase is subject to NCR (Cox et al., 2000b), and we have previously shown in C. albicans that 5–10 mM ammonium salts prevent germ tube formation induced by GlcNAc, arginine or urea, but not that induced by 5 % CO2 (Ghosh et al., 2009). These results were expected, because C. albicans DUR1,2 has at least five (A/T)GATA(A/G) sites upstream of the ORF (Ghosh et al., 2009), and these are the putative binding sites for the GATA transcription factors Gln3 and Gat1, which may mediate gene expression under inducing conditions (Dabas & Morschhäuser, 2007). These sites could mediate both direct and indirect NCR (via ammonium ions) of DUR1,2 expression. However, we show here that urea induction of these genes in GP medium is maintained in the presence of rich and poor nitrogen sources, including proline, glutamine, NH4 and asparagine. The putative GATA sites upstream of DUR3 probably contribute to its induction by urea, although residual induction by urea in the gat1Δ gln3Δ mutant suggests that additional transcription factors mediate DUR3 induction by urea.
In S. cerevisiae, the direct inducer for DUR1,2 is allophanate, also known as urea carboxylate, rather than urea itself (Cooper, 1982). Allophanate is the chemical intermediate for the multifunctional Dur1,2. In C. albicans, allophanate may also be an inducer for DUR1,2, although urea induction of DUR3 must involve a different mechanism. This conclusion is based on the observation that the dur1,2Δ mutant showed the same relative initial DUR3 induction by urea as the wild-type (Fig. 2b). However, following DUR3 expression over time after the introduction of urea revealed that the dur1,2Δ mutant lacked the slow secondary induction of DUR3 expression seen for wild-type cells grown in GPPU (Fig. 2c). Presumably, the longer growth time in GPPU permitted accumulation of another, less efficient inducer (potentially allophanate) and/or the lifting of a partial NCR caused by the metabolism of l-proline.
The expression of DUR1,2 and DUR3 was minimal in the complex medium YPD, suggesting that these genes could be subject to classical NCR. Although the high constitutive expression of both genes in basal medium containing urea was significantly repressed by addition of either good (ammonia) or poor (proline) nitrogen sources (Limjindaporn et al., 2003; Marzluf, 1997), disruption of the NCR transcription factor Gat1 did not prevent induction of DUR1,2 by urea or its repression by either nitrogen source. Similarly, DUR3 induction by urea was refractory to deletion of Gat1 or Gln3, and addition of nitrogen sources significantly repressed DUR3 expression in the presence of urea (Fig. 5c, d). Yet, induction of both genes by urea was dramatically decreased in the gat1Δ gln3Δ double mutant, and this strain was not subject to further repression by ammonia or Gln. These observations indicate that the GATA transcription factors Gat1 and Gln3 play positive roles in the urea induction of these genes, and that another NCR mechanism controls the negative regulation of DUR1,2 and DUR3 expression.
Complexity in the adaptive responses to varying environmental nitrogen sources is not unique to C. albicans. Aspergillus AreA and Neurospora NIT2 play primary roles in derepressing NCR (Caddick et al., 1994). Aspergillus amdS encodes an acetamidase that releases ammonia. This gene is controlled by pathway-specific gene regulation as well as general regulatory proteins (Davis et al., 1993). Gln3 of S. cerevisiae resembles AreA and NIT2 as a positive-acting regulator of transcription (Cooper et al., 1990). Aspergillus nirA encodes a nitrate reductase that requires AreA for induction. A mutation of nirA has been shown to overcome the requirement for AreA, suggesting that both specific and general regulation pathways are required for nitrogen metabolism in Aspergillus (Cove, 1979). These insights from other fungi will guide our future studies of the signalling mechanisms that regulate urea transport in C. albicans.
Although deletion of the inducible urea transporter Dur3 did not decrease virulence in a standard mouse candidaemia model, inducible acquisition of urea may be important for other aspects of C. albicans pathogenesis. Nitrogen metabolism contributes to the pathogenicity of Aspergillus fumigatus (Krappmann & Braus, 2005). Urease is a virulence factor in Cryptococcus neoformans (Cox et al., 2000a) and Coccidiodes immitis (Yu et al., 1997). In addition, urease is a well-established virulence factor in H. pylori. A urease mutant is unable to survive in co-culture with macrophages, whereas the urease in wild-type H. pylori prevents phagosome maturation (Schwartz & Allen, 2006). We know that urea metabolism provides similar protection from clearance by macrophages in C. albicans (Ghosh et al., 2009). Future studies will further examine the role of urea transport and metabolism in pathogenesis and examine the virulence of dur1,2Δ mutants. We will also explore the potential for urea catabolism to contribute to colonization of the kidney, where the organism could be exposed to higher concentrations of urea than in the bloodstream.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research (D. D. R.). Work in J. M.'s laboratory was supported by the Deutsche Forschungsgemeinschaft (DFG; grant MO 846/4). Work in K. W. N.'s laboratory was supported by the University of Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, the John C. and Nettie V. David Memorial Trust Fund, Ann L. Kelsall and the Farnesol and Candida albicans Research Fund.
Abbreviations
NCR, nitrogen catabolite repression
qPCR, quantitative PCR
References
- Caddick, M. X., Peters, D. & Platt, A. (1994). Nitrogen regulation in fungi. Antonie van Leeuwenhoek 65, 169–177. [DOI] [PubMed] [Google Scholar]
- Cole, G. T. (1997). Ammonia production by Coccidiodes immitis and its possible significance to the host–fungus interplay. In Host–Fungus Interplay, pp. 247–263. Edited by Stevens, D. O.. Bethesda, MD. : National Foundation for Infectious Diseases.
- Collart, M. A. & Struhl, K. (1994). NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev 8, 525–537. [DOI] [PubMed] [Google Scholar]
- Cooper, T. G. (1982). Nitrogen metabolism in Saccharomyces cerevisiae. In The Molecular Biology of the Yeast Saccharomyces, pp. 39–99. Edited by Strathern, J. N., Jones, E. W. & Broach, J. R.. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Cooper, T. G., Lam, C. & Turoscy, V. (1980). Structural analysis of the dur loci in S. cerevisiae: two domains of a single multifunctional gene. Genetics 94, 555–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T. G., Ferguson, D., Rai, R. & Bysani, N. (1990). The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol 172, 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove, D. J. (1979). Genetic studies of nitrate assimilation in Aspergillus nidulans. Biol Rev Camb Philos Soc 54, 291–327. [DOI] [PubMed] [Google Scholar]
- Cox, G. M., Mukherjee, J., Cole, G. T., Casadevall, A. & Perfect, J. R. (2000a). Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, K. H., Rai, R., Distler, M., Daugherty, J. R., Coffman, J. A. & Cooper, T. G. (2000b). Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J Biol Chem 275, 17611–17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabas, N. & Morschhäuser, J. (2007). Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot Cell 6, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastidar, S. G., Purandare, N. M. & Desai, S. C. (1967). Growth requriements of Candida species. Indian J Exp Biol 5, 228–232. [PubMed] [Google Scholar]
- Davis, M. A., Kelly, J. M. & Hynes, M. J. (1993). Fungal catabolic gene regulation: molecular genetic analysis of the amdS gene of Aspergillus nidulans. Genetica 90, 133–145. [DOI] [PubMed] [Google Scholar]
- Di Carlo, F. J., Schultz, A. S. & Kent, A. M. (1953). The mechanism of allantoin catabolism by yeast. Arch Biochem Biophys 44, 468–474. [DOI] [PubMed] [Google Scholar]
- Dunkel, N., Blass, J., Rogers, P. D. & Morschhäuser, J. (2008). Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, K. A., Brooks, C. L., Morgan, D. R. & Krakowka, S. (1991). Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun 59, 2470–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum, A. M., Tsay, E. Y. & Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., Navarathna, D. H., Roberts, D. D., Cooper, J. T., Atkin, A. L., Petro, T. M. & Nickerson, K. W. (2009). Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun 77, 1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B. D., Lockatell, C. V., Johnson, D. E., Warren, J. W. & Mobley, H. L. (1990). Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun 58, 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer, K. & Domdey, H. (1991). Preparation of high molecular weight RNA. Methods Enzymol 194, 398–405. [DOI] [PubMed] [Google Scholar]
- Krappmann, S. & Braus, G. H. (2005). Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med Mycol 43 (Suppl. 1), S31–S40. [DOI] [PubMed] [Google Scholar]
- Kulkarni, R. K. & Nickerson, K. W. (1981). Nutritional control of dimorphism in Ceratocystis ulmi. Exp Mycol 5, 148–154. [Google Scholar]
- Limjindaporn, T., Khalaf, R. A. & Fonzi, W. A. (2003). Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol Microbiol 50, 993–1004. [DOI] [PubMed] [Google Scholar]
- Marzluf, G. A. (1997). Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev 61, 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarathna, D. H. & Roberts, D. D. (2010). Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radic Biol Med 49, 1561–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarathna, D. H., Hornby, J. M., Hoerrmann, N., Parkhurst, A. M., Duhamel, G. E. & Nickerson, K. W. (2005). Enhanced pathogenicity of Candida albicans pre-treated with subinhibitory concentrations of fluconazole in a mouse model of disseminated candidiasis. J Antimicrob Chemother 56, 1156–1159. [DOI] [PubMed] [Google Scholar]
- Navarathna, D. H., Nickerson, K. W., Duhamel, G. E., Jerrels, T. R. & Petro, T. M. (2007). Exogenous farnesol interferes with the normal progression of cytokine expression during candidiasis in a mouse model. Infect Immun 75, 4006–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarathna, D. H., Harris, S. D., Roberts, D. D. & Nickerson, K. W. (2010). Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res 10, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, A., Takano, J., Kobayashi, M., von Wiren, N. & Fujiwara, T. (2006). Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett 262, 216–222. [DOI] [PubMed] [Google Scholar]
- Odds, F. C. (1988). Candida and Candidiasis, 2nd edn. London. : Bailliere Tindall.
- Pendrak, M. L., Yan, S. S. & Roberts, D. D. (2004a). Sensing the host environment: recognition of hemoglobin by the pathogenic yeast Candida albicans. Arch Biochem Biophys 426, 148–156. [DOI] [PubMed] [Google Scholar]
- Pendrak, M. L., Yan, S. S. & Roberts, D. D. (2004b). Hemoglobin regulates expression of an activator of mating-type locus α genes in Candida albicans. Eukaryot Cell 3, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendrak, M. L., Chao, M. P., Yan, S. S. & Roberts, D. D. (2004c). Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to α-biliverdin. J Biol Chem 279, 3426–3433. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß, O., Vik, Å., Kolter, R. & Morschhäuser, J. (2004). The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127. [DOI] [PubMed] [Google Scholar]
- Roon, R. J. & Levenberg, B. (1972). Urea amidolyase. I. Properties of the enzyme from Candida utilis. J Biol Chem 247, 4107–4113. [PubMed] [Google Scholar]
- Roon, R. J., Hampshire, J. & Levenberg, B. (1972). Urea amidolyase. The involvement of biotin in urea cleavage. J Biol Chem 247, 7539–7545. [PubMed] [Google Scholar]
- Schwartz, J. T. & Allen, L. A. (2006). Role of urease in megasome formation and Helicobacter pylori survival in macrophages. J Leukoc Biol 79, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentheshanmuganathan, S. & Nickerson, W. J. (1962). Nutritional control of cellular form in Trigonopsis variabilis. J Gen Microbiol 27, 437–449. [DOI] [PubMed] [Google Scholar]
- Yu, J. J., Smithson, S. L., Thomas, P. W., Kirkland, T. N. & Cole, G. T. (1997). Isolation and characterization of the urease gene (URE) from the pathogenic fungus Coccidioides immitis. Gene 198, 387–391. [DOI] [PubMed] [Google Scholar]