Summary
The nuclear receptor REV-ERBα is a key negative-feedback regulator of the biological clock. REV-ERBα binds to ROR elements of the Bmal1 (Arntl) promoter and represses Bmal1 transcription. This stabilizing negative loop is important for precise control of the circadian pacemaker. In the present study, we identified a novel synthetic REV-ERBα ligand, which enhances the recruitment of nuclear receptor co-repressor (NCoR) to REV-ERBα. In order to explore REV-ERBα action on resetting responses of the molecular clock, we first established the rhythmic transcription profile and expression level of REV-ERBα in Rat-1 fibroblasts. When applied at different phases of the circadian oscillation to cell models containing stably transfected Bmal1::Luc or Per2::Luc, the REV-ERBα ligand induced phase-dependent bi-directional phase shifts. When the phase changes were plotted against time, a clear phase response curve was revealed, with a significant peak-to-trough amplitude of ca. 5 hours. The phase-resetting effect was also observed when the compound was applied to primary lung fibroblasts and ectopic lung slices from transgenic PER2::Luc mice. Therefore, similar regulation of REV-ERBα function by endogenous ligands, such as heme, is likely to be an important mechanism for clock resetting. In addition, we identify a new means to generate phasic shifts in the clock.
Keywords: Nuclear hormone receptor, Circadian clock, Resetting, Phase response curve
Introduction
In mammals, the master circadian clock is known to reside in the suprachiasmatic nuclei (SCN) of the hypothalamus. Via multiple pathways, output from the SCN synchronizes peripheral oscillators throughout the body. These peripheral clocks can be demonstrated using in vitro culture systems, and operate as independent cell-autonomous oscillators, synchronized to the SCN (Yoo et al., 2004; Hastings et al., 2003).
Circadian transcription is initiated by two bHLH-PAS-domain proteins, CLOCK and BMAL1 (positive limb), which dimerize and activate the transcriptional repressors PERIOD (PER) and CRYPTOCHROME (CRY) through E boxes (negative limb). PER proteins (PER1 and PER2) and CRY proteins (CRY1 and CRY2) accumulate and complex in the cytoplasm, and translocate into the nucleus after a delay of several hours, repressing the activity of constitutively bound CLOCK-BMAL1 complexes (Reppert and Weaver, 2002; Hastings et al., 2003; Lowrey and Takahashi, 2004). Following a further delay, these inhibitory complexes are then degraded through proteasomal degradation mediated by casein kinase Iε/δ (CKIε/δ) (Lee et al., 2001; Gallego and Virshup, 2007) and F-box protein (Godinho et al., 2007; Busino et al., 2007; Siepka et al., 2007), and the de-repression of CLOCK-BMAL1 activity initiates the next circadian cycle of transcription of the genes encoding the PER and CRY proteins.
The ‘orphan’ nuclear receptor REV-ERBα (also known as nuclear receptor subfamily 1, group D, member 1; NR1D1) has been identified as a key component that links the positive and negative limbs of the clock (Preitner et al., 2002). Transcription of Rev-erbα is rhythmical because it is positively regulated (through E boxes) by CLOCK-BMAL1, and negatively regulated by PER-CRY (Hastings et al., 2003; Triqueneaux et al., 2004), by REV-ERBα itself (Adelmant et al., 1996) and post-translationally by GSK3β-phosphorylation-mediated stabilization (Yin et al., 2006). Because REV-ERBα represses Bmal1 transcription, the REV-ERBα oscillations act as an additional stabilizing loop within the clock, and works together with the core loop to maintain the precision of the circadian oscillation.
Nuclear receptors interact with co-modulators in a ligand-regulated manner. The ligand-binding domain of REV-ERBα lacks the typical C-terminal AF2 domain, which has been shown to be important for co-activator binding. As a result, it constitutively represses target genes by recruiting a multimeric co-repressor complex containing nuclear receptor co-repressor (NCoR) and histone deacetylase 3 (HDAC3) (Yin and Lazar, 2005). Recruitment of this repressor complex to REV-ERBα has recently been shown to be enhanced by REV-ERBα binding to heme, intracellular concentrations of which themselves oscillate in a circadian manner (Ceriani et al., 2002; Kaasik and Lee, 2004). Therefore, REV-ERBα is an attractive target for small-molecule manipulation of the circadian clock.
In this study, we reveal the phase-resetting effect of a novel, synthetic REV-ERBα ligand on the molecular oscillators at both the mRNA and protein level. We have modeled our data to define phasic action of this compound on the circadian clock. This compound is the first known pharmacological agent that can reset the circadian clock in a phase-dependent manner and might offer novel approaches to pharmacological treatments of rhythm disorders.
Results
In vitro screening identifies a REV-ERBα–NCoR activator
A novel ligand of REV-ERBα {1,1-dimethylethyl N-[(4-chlorophenyl)methyl]-N-[(5-nitro-2-thienyl)methyl]glycinate} was identified in a REV-ERBα–NCoR fluorescence resonance energy transfer (FRET) assay, which showed an EC50 value of 250 nM (D.P. and J.C., unpublished). In assays of physical binding, the ligand enhanced the recruitment of NCoR peptide to REV-ERBα by up to 70% within 1 hour (Fig. 1A). The compound showed no activity on LRH1, SF1, FXR or RORα using the same FRET assay, and no activity on LXRα or LXRβ in reporter-gene assays.
Fig. 1.
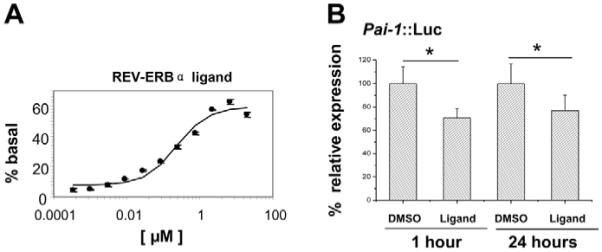
Identification of a ligand that activates recruitment of NCoR to REV-ERBα. (A) Functional EC50 determination of the REV-ERBα ligand. Intensities of the FRET signals following addition of the compound at different concentrations are quantified and normalized to the basal level. Data are shown as mean ± s.e.m. for three independent experiments. (B) Effects of the REV-ERBα ligand on Pai1 transcription. Rat-1 cells were transiently transfected with the Pai1::Luc reporter construct. 48 hours later, cells were treated with the compound or DMSO (as control) for 1 hour or 24 hours. Cell lysates were then prepared and bioluminescence recorded using a luminometer. Four replicates were analyzed for each treatment group, and experiments were performed three times. Results are shown as mean ± s.e.m. *P<0.05, Student’s t-test.
To explore the regulation by this compound of REV-ERBα function in vivo, we generated Pai1-Luc stable transfectants in Rat-1 cells. As expected, these exhibited a circadian oscillation (data not shown). The compound inhibited Pai1 promoter activity by 30% (P<0.05, Student’s t-test) (Fig. 1B). PAI1 is a known target for REV-ERBα, through a well-characterized RORE element in the proximal promoter (Wang et al., 2006). Suppression was still evident following incubation with the ligand for 24 hours (Fig. 1B). At the lower concentrations tested, the compound had little effect on Pai1 transcription (data not shown).
Cell-autonomous oscillations for the three canonical circadian genes in Rat-1 fibroblasts
In order to reveal the true nature of the transcriptional regulation, we flanked our transgenes (Bmal1, Per2 and Rev-erbα) with chicken β-globin HS4 insulator elements. These have long been known to protect target genes from both heterochromatic silencing and activation by nearby enhancer elements (Burgess-Beusse et al., 2002; West et al., 2002). These ‘insulated’ circadian reporters were stably transfected into Rat-1 cells. Using photomultiplier tube (PMT) bioluminescence recording ‘insulated’ mouse Bmal1, Per2-promoter constructs exhibited robust cell-autonomous oscillations. The oscillations of the two promoters were anti-phase to each other when cultures were normalized with respect to synchronization with dexamethasone (Dex; Fig. 2A,B). Rhythms were still detectable 2 weeks after the cells were synchronized, which makes them ideal for the pharmacological-manipulation studies. Oscillations can be reinitiated by changing to fresh recording medium, by Dex treatment (200 nM) or by a combination of both (Fig. 2C).
Fig. 2.
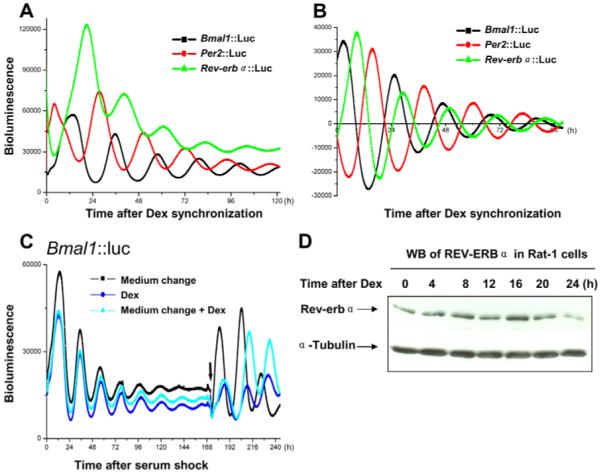
Rhythmic transcription profiles and protein expression of core clock genes in Rat-1 cells. (A) Rat-1 fibroblasts that were stably transfected with different reporter constructs (Bmal1::Luc, Per2::Luc and Rev-erbα::Luc) were synchronized by treatment with 200 nM dexamethasone (Dex) for 1 hour. Cells were then switched to recording medium and bioluminescence (photon counts per minute) was recorded using photon-multiplier tubes. (B) Phase relationships with respect to synchronization were depicted after the data were de-trended by 24 hour moving average. (C) Damped oscillation of Bmal1::Luc Rat-1 cells (synchronized by 50% horse serum) can be re-initiated by medium change, treatment with Dex, or a combination of both. As expected, Dex treatment induced a phase-delay compared with medium-change alone. Arrow shows the time of addition of Dex and/or medium change. (D) Representative western blot (WB) of REV-ERBα in Rat-1 cells synchronized by 200 nM Dex and harvested at 4-hour intervals. The time points indicated are relevant to the time of Dex treatment. This experiment was repeated three times.
Although rhythmic mRNA expression of Rev-erbα has been established in liver, SCN and serum-shocked fibroblasts using quantitative PCR (Q-PCR) or northern blot (Balsalobre et al., 1998; Torra et al., 2000; Yamamoto et al., 2004), transcriptional oscillation using promoter-driven luciferase reporters has not been reported to our knowledge and here we describe for the first time rhythmic transcriptional profiles of the Rev-erbα gene in Rat-1 cells using real-time bioluminescence recording. Cells stably transfected with the 600-bp Rev-erbα-promoter construct demonstrated robust circadian oscillations in culture with high levels of luciferase activity and equivalent amplitude/damping rate compared with Bmal1 and Per2 (Fig. 2A, raw data). Closer examination of the phase relationships (Fig. 2B, de-trended data) revealed the temporal order of peaks of expression of the three canonical circadian genes, which is consistent with previous analysis at the tissue level (Yamamoto et al., 2004), indicating that the organization and temporal regulation of the molecular-clock components are cell-autonomous and can be demonstrated in homogeneous cells.
In order to confirm that the short promoter fragment accurately reflected endogenous events within the molecular clockwork, we undertook western blotting studies to investigate the endogenous REV-ERBα protein levels in Rat-1 cells over circadian cycle. This revealed clearly a circadian expression pattern for REV-ERBα, with a peak expression at the 16-hour time point (Fig. 2D).
Phase-resetting effects of the REV-ERBα ligand on molecular oscillators
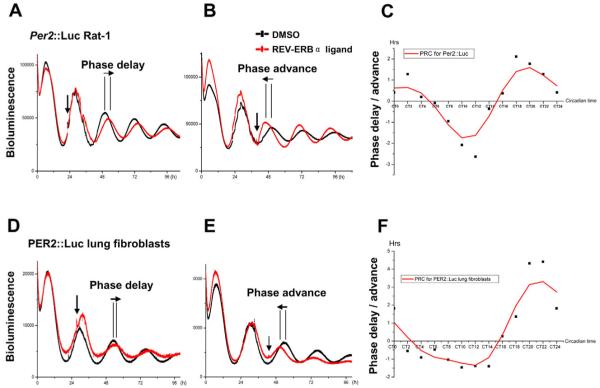
Because Rev-erbα is rhythmically transcribed and western blotting showed rhythmic protein levels in Rat-1 cells, we hypothesized that a ligand for REV-ERBα might have phasic actions on the clock. To explore the action of the REV-ERBα ligand, we applied this compound to the Per2::Luc or Bmal1::Luc cells that had been synchronized and subjected to photon counting. At different phases of the circadian transcriptional oscillation of Per2::Luc rhythms (Fig. 3A,B), the addition of the compound (10 μM) induced phase-dependent bi-directional phase shifts, whereas treatment with solvent alone (DMSO) had no effect. Induction of phase shifts was not associated with alterations in period length after compound treatment (data not shown). When plotted, significant delays and advances were observed, which were dependent on the phase of the cycle in which the drug was applied. Phase delays occurred between circadian time (CT)5 and CT15, whereas phase advances occurred within CT16 to CT4. The resulting phase response curve (PRC) revealed a delay and advance phase of similar duration (ca. 10-12 hours) with no obvious ‘dead-zone’ (Fig. 3C). The maximal peak-to-trough amplitude was around 5 hours. Low concentrations of the compound (100 nM and 1 μM) had little or no phase-resetting effect on rhythms, which is consistent with our observations from the transient transfection assay that these low concentrations of the drug were insufficient to alter REV-ERBα function, as demonstrated on the Pai1 promoter.
Fig. 3.
Phase-resetting effects on the molecular oscillators by the REV-ERBα ligand. Individual cultures of confluent Rat-1 cells stably transfected with Per2::Luc reporter, or primary lung fibroblasts from PER2::Luc mice were synchronized by Dex for 1 hour and monitored using PMT. Cells were then treated with the compound (10 μM, red) at a single phase point for each culture dish. Control cultures were treated with DMSO (black). (A,B,D,E) Representative traces of PMT real-time recording. Compound addition is indicated by vertical arrows and phase changes by horizontal arrows. (C,F) Ligand-induced phase-response curves (PRCs). For the PRC estimates, actual phase changes were plotted against circadian time, with the peak of PER2 expression taken as CT12. Data shown are representative of three independent experiments. Negative values indicate phase delays, whereas positive values refer to phase advances.
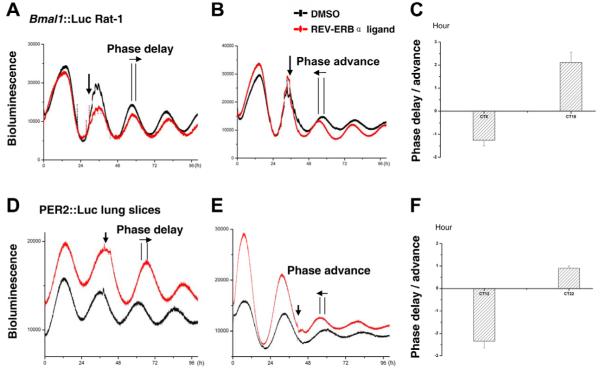
In order to confirm the phase-shifting action of the REV-ERBα ligand on both limbs of the molecular clock, and to explore the mechanism whereby the drug shifted rhythms, we applied the compound to Rat-1 cells that were stably transfected with Bmal1::Luc. Here, we also observed bi-directional phase shifts in the Bmal1 rhythm (Fig. 4A-C), with a phase delay of 1.25±0.24 hours at CT8 and phase advance of 2.11±0.45 hours at CT16 (n=3 for each time point). Notably, there was an acute downregulation of the Bmal1::Luc signal after compound addition, suggesting acute suppression of Bmal1 transcription through enhanced recruitment of NCoR to REV-ERBα bound on ROREs in the Bmal1 promoter.
Fig. 4.
Bi-directional phase shifts induced by the REV-ERBα ligand on Bmal1::Luc Rat-1 cells or PER2::Luc lung slices. Individual cultures of confluent Rat-1 cells stably transfected with Bmal1::Luc reporter or ectopic lung slices from PER2::Luc mice were synchronized and subject to PMT recording. Cells or tissue slices were then treated with the compound (10 μM, red) or DMSO (black) at the indicated circadian time. Representative phase changes (horizontal arrow) after compound treatment (indicated by a vertical arrow) are shown in A and B (for Bmal1::Luc Rat-1), and D and E (for PER2::Luc lung slices). (C,F) Phase changes following the compound treatment are shown as mean ± s.e.m. n=3 for Bmal1::Luc Rat-1 cells, n=5 for the lung slices.
Phase-resetting effects on primary lung fibroblasts with PER2::Luc reporters
The above data demonstrated phase-resetting effects on the transcriptional clock. We next tested the action of this drug on the rhythmic expression of the PER2 protein. Primary lung fibroblasts isolated from PER2::Luc mice, which have a stably integrated luciferase fused to the coding region of the Per2 gene (Yoo et al., 2004), were treated with the drug in an identical manner to the protocol used above. Again, strong phasic resetting was observed (Fig. 3D-F), with a similar PRC to the Per2::Luc Rat-1 cells, suggesting that the compound is capable of resetting the molecular rhythm of PER2 protein expression in primary cells.
Phase-resetting effects on ectopic lung slices with PER2::Luc reporters
In order to establish whether the REV-ERBα ligand resets the molecular clock at the tissue level, we prepared organotypic lung slices from PER2::Luc mice. PMT recording of the circadian PER2 activity in these lung slices revealed robust oscillations over 10 days. Upon treatment with the compound (10 μM) at different circadian phases, bi-directional phase shifts were observed (Fig. 4D-F), with a phase delay of 2.34±0.31 hours at CT12 and phase advance of 0.90±0.11 hours at CT22 (n=5 for each time point). This indicates that the compound is capable of penetrating tissues and acting on the molecular clock in a similar fashion as it does on isolated cells.
Discussion
Over the last decade, the detailed description of the molecular clockwork regulating the circadian clock has elucidated the underlying mechanisms involved in system-wide circadian rhythms in physiology and behavior. In all eukaryotic organisms, the core molecular components conferring rhythmicity are based on conserved transcription factors that feedback to inhibit their own expression and that of other target genes in a periodic manner. This ‘transcriptional-translational’ feedback loop has been dissected by genetic approaches, but has not been accessible to small-molecule approaches, with their attendant advantages for physiological studies. To date, efforts have been directed to targeting key enzymes regulating the phosphorylation and hence stability of clock proteins and, in this regard, inhibitors of casein kinase I are the first known pharmacological drugs that directly target the circadian clock. Such inhibitors lengthen the circadian period in mammalian cells, because they reduce the rate of phosphorylation and hence the decay of PER and other proteins, thereby increasing the length of negative-feedback suppression on the clock (Eide et al., 2005; Reischl et al., 2007).
Within the molecular oscillator, there is a crucial additional feed-forward loop, which stabilizes the molecular clock. In mammals, two members of the retinoid nuclear receptor family, REV-ERBα and RORα, have been identified as a pair with opposing effects. RORα is known to be the key transcriptional activator of the circadian-clock gene Bmal1. This latter gene encodes a protein that dimerizes with CLOCK, to transactivate circadian-clock-controlled genes through E-box sites. Circadian gene products that are regulated in this way are the inhibitory circadian elements PER, CRY and REV-ERBα. PER-CRY dimers feed back to inhibit the transcriptional activity of CLOCK-BMAL1 and, as a result, the Rev-erbα gene is rendered rhythmic (Hastings et al., 2003; Triqueneaux et al., 2004). The REV-ERBα protein competes with RORα for common ROR-elements in the Bmal1 promoter, and acts to recruit a multimeric co-repressor complex containing NCoR and HDAC3 to the gene to repress transcription (Emery and Reppert, 2004; Guillaumond et al., 2005; Yin and Lazar, 2005; Akashi and Takumi, 2005). Consequently, REV-ERBα acts as an additional stabilizing loop within the molecular clockwork, which helps to maintain the precision of the circadian oscillation. Feed-forward loops of this type are wide-spread in nature but, in contrast to PER and CRY, the genetic elements of such loops are not strongly conserved across major phylogenies (i.e. mammals and Drosophila) (Looby and Loudon, 2005).
Previous observations have implicated retinoids as phase-resetting signals for the peripheral clock. For example, retinoic acid can phase-shift the rhythmicity of Per2 mRNA in vivo and in seruminduced smooth-muscle cells in vitro, and this is known to be mediated by the retinoid receptors RXRα and RARα (McNamara et al., 2001). In addition, the synthetic glucocorticoid-receptor agonist Dex has been shown as a potent phase-shifting agent in both cultured cells and peripheral tissues of intact animals (Balsalobre et al., 2000). Intriguingly, glucocorticoids act directly to repress the activity of the human Rev-erbα promoter (Torra et al., 2000).
The role of REV-ERBα on clock function has been explored using gene-deletion approaches. Rev-erbα-knockout mice demonstrate rhythmic locomotor activity with only a slightly shortened period (0.5 hours), but the phase-shifting response to light delivered in the latter half of the night (rising phase of REV-ERBα expression in SCN) is greatly enhanced (Preitner et al., 2002). These studies suggest that REV-ERBα is involved in the resetting responses of the clock to environmental stimuli. More recently, heme was identified as a ligand for REV-ERBα and was shown to regulate REV-ERBα transcriptional activity (Yin et al., 2007; Raghuram et al., 2007), suggesting that REV-ERBα might further couple metabolic activity to the circadian clock, because heme itself has a circadian oscillation in intracellular concentration (Yin et al., 2007). This discovery also suggested that REV-ERBα might be a tractable pharmacological target for manipulation of the clock.
Using a FRET assay, we identified a compound {1,1-dimethylethyl N-[(4-chlorophenyl)methyl]-N-[(5-nitro-2-thienyl)methyl]glycinate} that significantly and specifically enhances the REV-ERBα-NCoR interaction (D.P. and J.C., unpublished). We were able to show the phasic action of this drug on peripheral circadian clocks by real-time assays in reporter cells or tissues. The compound caused a robust and consistent acute suppression of Bmal1, which is compatible with enhanced recruitment of the NCoR repression complex to the promoter-bound REV-ERBα; however, there was no long-term suppression of expression of Bmal1, which presumably reflects the requirement for REV-ERBα protein to mediate the effect of the drug. Indeed, we were able to show a high-amplitude circadian oscillation in REV-ERBα protein in unstimulated cells, and previous evidence shows negative feedback on Rev-erbα expression by REV-ERBα itself (Adelmant et al., 1996). We showed that administration of this pharmacological tool at different phases of the circadian oscillations of Per2-transcription or PER2-protein rhythms, induced bi-directional phase shifts (delays or advances), which revealed a Type-1-like phase-response curve (weak resetting) (Aschoff, 1965). Importantly, the compound did not result in long-term changes in period. This is in contrast to data from studies involving the genetic deletion of Rev-erbα, which showed a small but robust shortening of the circadian period of approximately 0.5 hours. This might reflect the combination of altered potency of REV-ERBα for NCoR recruitment in the continued presence of compound, which might balance increased negative feedback on REV-ERBα protein expression.
Previous studies have employed glucocorticoids to explore the re-setting characteristics of the circadian clock, using Rat-1 cell lines as a model for the peripheral clock. These studies have described robust phasic effects on the re-setting of the underlying oscillator and, in common with our data, reveal that there is no obvious ‘dead zone’ in which cells do not respond to the stimulus (Balsalobre et al., 2000; Izumo et al., 2006). Our data here are the first to describe the impact of the specific targeting of the ROR axis of the circadian clock and we show that this axis is also capable of phasic re-setting. We propose the following model (Fig. 5). Owing to the rhythmic expression level of the REV-ERBα receptor, the efficacy of the ligand will vary across the circadian cycle. As a result, the same concentration of the compound may be predicted to induce different amplitude phase shifts. One prediction of this model is that, because REV-ERBα constitutively suppresses Bmal1 gene transcription, application of a REV-ERBα ligand should enhance repression of Bmal1 expression. The drug-induced decline of Bmal1 expression at the rising phase of Bmal1 rhythm (at phase Y in Fig. 5A and blue in 5B) results in a delayed peak, and hence a phase delay, as seen in Fig. 4A. By contrast, a drug-induced decline of Bmal1 expression at the descending phase of Bmal1 rhythm (at phase X in Fig. 5A and red in 5B) will cause the nadir of the cycle to be reached earlier, hence a phase advance (Fig. 4B). These phase shifts in Bmal1 rhythm will be reflected in altered phasing and amplitude of expression of Per2 mRNA and protein. Collectively, these data suggest that, because the endogenous ligand for REV-ERBα (heme) is known to be produced in a circadian manner in peripheral cells and tissues, altered regulation of this nuclear hormone pathway might play a significant role in setting circadian period.
Fig. 5.
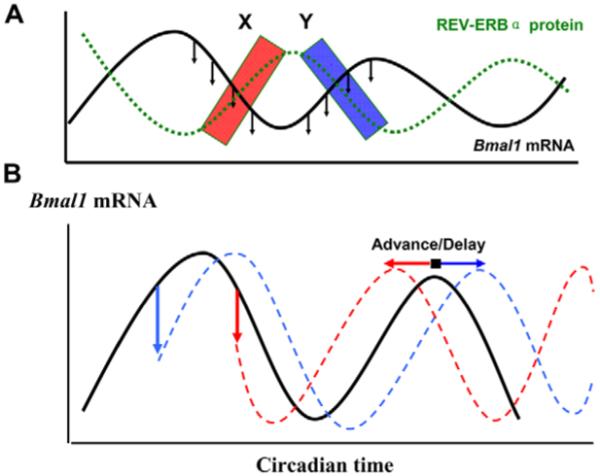
Hypothesis for the mode of action of the ligand. Application of REV-ERBα ligand to the circadian cycle of REV-ERBα protein (A, green) causes the early decline of Bmal1 levels (black). At phase X in A (red box), this leads to an advance in the next Bmal1 cycle (red in B), as demonstrated in Fig. 4B. At phase Y (blue box), a delay results from the same treatment (blue in B), as seen in Fig. 4A.
In summary, we describe the pharmacology of the first orphan retinoid-receptor agonist on circadian timing and demonstrate that this drug acts in a phasic manner on the core molecular clockwork, both in cell lines and primary tissues. Our studies now suggest novel approaches to pharmacological manipulation of circadian rhythms using RORE-acting drugs.
Materials and Methods
Screening for novel REV-ERBα ligands
In order to identify ligands that may enhance or inhibit the REV-ERBα–NCoR peptide interaction, FRET was used for assay development and ligand discovery for REV-ERBα. The FRET signals reported were normalized to the basal level of interaction at which the ligand-binding domain of REV-ERBα interacts with the NCoR peptide in the absence of ligand. On the basis of the abilities of the compounds to facilitate the recruitment of NCoR peptide to REV-ERBα, a normalized response of 0% would be inactive, >0% indicates enhancement of the protein-peptide interaction and <0% (negative values) indicates inhibition of the protein-peptide interaction.
Generation of the reporter constructs
Mouse genomic DNA was prepared from adult mouse ear clips using the GenomicPrep Cells and Tissue DNA Isolation Kit (GE Healthcare). Human genomic DNA was purchased from Sigma. The promoter regions of mouse Bmal1 and Rev-erbα, as well as human PAI1, were PCR amplified from the above genomic DNA and cloned into the pGL4.16[Luc2CP/Hygro] vector (Promega) using NheI-EcoRV (for Rev-erbα or PAI1) or NheI-HindIII (for Bmal1). Our previous studies had indicated that ‘un-insulated’ reporters rapidly become silenced upon stable transfection. Here, we adapted the use of chicken β-globin ‘insulator’ sequences for the protection of target genes. Two tandem copies of core ‘insulator’ sequences (kind gift of Gary Felsenfeld, NIH, Bethesda) were introduced into both sides of the promoter-driven luciferase region to prevent inappropriate gene silencing or enhancing. All the four copies are in the same orientation to ensure optimum protection (West et al., 2002). The mouse Per2 promoter was sub-cloned from a pGL3-Per2 construct (kind gift of Kazuhiro Yagita, Kobe, Japan) into the above ‘insulated’ vector using NheI-EcoRV. Primers used are given here in the order of 5′ to 3′: mBmal1 forward, CGGCGCTAGCGA-GGGATGGGCGAAGAGATG; mBmal1 reverse, CGGCAAGCTTGATCCCGCGG-CGGCGGCG; mRev-erbα forward, CGGCGCTAGCTAGTCACCACTAACCTC-AGGGTG; mRev-erbα reverse, CGGCGATATCGCAACCAGGAAGTAAGTAG-GTGATG; hPai1 forward, CGGCGCTAGCCGGGCAGCTCGAAGAAGTGAAAC; hPai1 reverse, CGGCGATATCAGTTCTCAGAGGTGCCTTGCGATTG.
Transient and stable transfection of Rat-1 cells
Rat-1 fibroblasts were maintained in DMEM (Invitrogen) supplemented with 10% FBS, penicillin/streptomycin and L-glutamine. Cells were cultured at 37°C in a humidified 5% CO2 environment.
For transient-transfection and luciferase assays, cells were seeded into 12-well plates at a density of 5×104 cells/well. When the cells were at 80% confluency, 50 ng/well of Pai1::Luc reporter constructs were delivered with FuGENE 6 reagent (Roche) according to the manufacturer’s instructions. At 48 hour later, cells were treated with the REV-ERBα ligand (concentration range, 100 nM, 1 μM, 10 μM) or DMSO (as control) for 1 hour or 24 hours. Cells were washed twice with ice-cold PBS and lysates were prepared using a luciferase assay system (Promega, UK). The luciferase activity was measured in the presence of luciferin substrate and ATP using a Berthold Mithras 960 luminometer (Berthold Technologies, Redbourn, UK). For statistical analysis, P values were calculated using Student’s t-test.
For stable transfection, Rat-1 cells were cultured in 10-cm dishes at a density of 2×105.
6 μg of each promoter construct (SalI linearized) were mixed with 18 μl of FuGENE 6 and transfected into cells according to the manufacturer’s manual (Roche, Germany). Positive colonies were selected for 3-5 weeks in the presence of Hygromycin (200 ng/ml). Individual colonies were isolated with sterile cloning discs (Sigma, UK) and transferred to 24-well plates until ready for the following experiments.
Lung fibroblast culture from PER2::Luc mice
Primary lung fibroblasts were prepared from PER2::Luc transgenic mice (Yoo et al., 2004), in which endogenous PER2 protein is fused in-frame with luciferase reporter at the C-terminus. This enabled real-time monitoring of the PER2 protein dynamics under bioluminescence recording. Lungs were removed from euthanized adult mice for peripheral fibroblast preparation. Lung tissue was chopped and minced in sterile conditions, and was then washed twice in chilled modified Hank’s Balanced Salt Solution (Sigma). Cells were then dissociated by means of shaking at 37°C for 2 hours in 100 U/ml of Collagenase IA (Sigma) (dissolved in PBS) and 3 mM CaCl2. Once dissociated, cells were filtered through a sterile nylon mesh followed by two washes and centrifugation in chilled Hank’s solution. Pellets were re-suspended in culture medium (DMEM with 4.5 g/l glucose, Glutamax and pyruvate, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin) and plated on a T25 cell-culture flask. Cultures were maintained at 37°C (5% CO2) for 2-3 days until confluent and ready for splitting.
Preparation of lung slices from PER2::Luc mice
PER2::Luc mice were sacrificed using cervical dislocation, and the trachea and diaphragm were exposed. The lung perfusion was adapted from the method by Moreno et al. (Moreno et al., 2007). Briefly, a small incision was made in the upper trachea and fine tubing (OD: 0.96 mm; Harvard Apparatus, Kent) inserted. 1 ml 2% agarose (ultra low gelling temperature agarose, Sigma, in HBSS warmed to 37°C) was perfused through the tubing into the lungs, and the trachea tied off to prevent leakage. The mouse was cooled at 4°C for 15 minutes to allow the agarose to set fully before the lungs were removed on ice. Using a vibraslice (Integraslice 7550 MM; Campden, Loughborough), 275 μm serial sections of lung lobes were cut in 4°C HBSS. Sections were transferred to Glutamax DMEM media (Invitrogen) containing 100 U/ml penicillin and 100 μg/ml streptomycin, and warmed to 37°C in an incubator (5% CO2). Subsequently, the medium was changed four times to remove any traces of agarose. The tissue was left overnight before plating out onto Millicell 30-mm cell-culture plate inserts (Millipore) the next day, prior to synchronization and photon counting.
Bioluminescence real-time recording
Confluent cells or lung slices in 35-mm dishes were synchronized by treatment with 200 nM Dex for 1 hour. The medium was changed to non-phenol-red DMEM supplemented with 0.1 mM Luciferin substrate (Izumo et al., 2003; Yamazaki and Takahashi, 2005). Each 35-mm dish was sealed with vacuum grease and placed in a light-tight and temperature-controlled environment at 37°C. Light emission (bioluminescence) was measured continuously using a Photomultiplier tube (PMT, H6240 MOD1, Hamamatsu Photonics). Data were presented as photon counts per minute. Baseline correction was calculated using a 24-hour moving average.
For the drug treatment, 24 hours after synchronization individual dishes with cells or tissue slices under PMT recording were treated once with the REV-ERBα ligand (concentration range, 100 nM, 1 μM, 10 μM) or DMSO (vehicle control) at a single specific time point. The compound was left continuously with the samples thereafter while the luminescence patterns were recorded for at least 7 days. In total, dishes were treated at 2-hour intervals over a 24-hour cycle. Phase and period were analyzed by RAP software (Okamoto et al., 2005). Changes in the phase of the circadian oscillation were calculated by comparison of treated and control cells and then plotted with reference to circadian time, with the peak of PER2 protein expression taken as CT12.
Western blotting for REV-ERBα
Confluent Rat-1 cells in 35-mm dishes were synchronized with Dex (200 nM) for 1 hour, then replaced with normal culture medium. 24 hours later, cells were harvested at 4-hour intervals. Protein extracts were prepared by lyzing the cells in 400 μl of extraction buffer [0.1 M KCl, 20 mM HEPES (pH 7.5), 5 mM EDTA, 1 mM dithiothreitol, 0.1% Triton X-100, 5% glycerol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM MgCl2, 10 μg/ml of aprotinin, 5 μg/ml of leupeptin, 1 μg/ml of pepstatin A]. Cell lysates were cleared by centrifugation (twice, 10 minutes each, 13,000 g at 4°C). Supernatants were mixed with 2× protein loading buffer and boiled. These were then spun at 13,000 g (1 minute, room temperature). 70 μg of total protein was resolved by electrophoresis through sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and then transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.05% Tween-20 and then incubated with polyclonal anti-REV-ERBα (Cell Signalling) or monoclonal anti-tubulin (Sigma) antibodies. Following secondary-antibody incubation, immunoreactive bands were visualized using enhanced chemiluminescence detection (ECL plus, Amersham Biosciences).
Animal maintenance
All experiments were conducted under the aegis of the 1986 Home Office Animal Procedures Act (UK) and following local ethical review. PER2::Luc transgenic animals were maintained at 20-22°C and maintained on standard rodent breeder or maintenance chow.
Acknowledgments
We thank Kazuhiro Yagita, Osaka, Japan for the kind gift of the Per2 promoter construct; Gary Felsenfeld, NIH, Bethesda for the β-globin insulator construct; Masahiro Ishiura, Japan for the RAP software; Graham Sturton, Imperial College London for technical assistance in lung slice preparation; and the BBSRC (UK) and GSK for financial support.
References
- Adelmant G, Bègue A, Stéhelin D, Laudet V. A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. Proc. Natl. Acad. Sci. USA. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Aschoff J. Response curves in circadian periodicity. In: Aschoff J, editor. Circadian Clocks. Elsevier; Amsterdam: 1965. pp. 95–111. [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-Wide Expression Analysis in Drosophila Reveals Genes Controlling Circadian Behavior. J. Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Reppert SM. A rhythmic Ror. Neuron. 2004;43:443–446. doi: 10.1016/j.neuron.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell. Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by Rev-erb and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: temperature compensation and damping. Proc. Natl. Acad. Sci. USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput. Biol. 2006;2:1248–1261. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Looby P, Loudon AS. Gene duplication and complex circadian clocks in mammals. Trends Genet. 2005;21:46–53. doi: 10.1016/j.tig.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Moreno L, Faro R, Hislop A, Sturton G, Perez-Vizcaino F, Mitchell JA. Endothelium-dependent dilatation of mice pulmonary arteries increases with postnatal maturation. FASEB J. 2007;21:A1164. [Google Scholar]
- Okamoto K, Onai K, Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal. Biochem. 2005;340:193–200. doi: 10.1016/j.ab.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERB controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. β-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J. Biol. Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian Mutant Overtime Reveals F-box Protein FBXL3 Regulation of Cryptochrome and Period Gene Expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erb(alpha) gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 2004;33:585–606. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yin L, Lazar M. The orphan nuclear receptor Rev-erbα regulates circadian expression of Plasminogen Activator Inhibitor Type 1. J. Biol. Chem. 2006;281:33842–33848. doi: 10.1074/jbc.M607873200. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 2004;5:18–27. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Meth. Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erb alpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erb{alpha}, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]