Abstract
DETA-NONOate, a nitric oxide (NO) donor, induced cytostasis in the human breast cancer cells MDA-MB-231, and the cells were arrested in the G1 phase of the cell cycle. This cytostatic effect of the NO donor was associated with the down-regulation of cyclin D1 and hypophosphorylation of the retinoblastoma protein. No changes in the levels of cyclin E or the catalytic partners of these cyclins, CDK2, CDK4, or CDK6, were observed. This NO-induced cytostasis and decrease in cyclin D1 was reversible for up to 48 h of DETA-NONOate (1 mM) treatment. DETA-NONOate (1 mM) produced a steady-state concentration of 0.5 μM of NO over a 24-h period. Synchronized population of the cells exposed to DETA-NONOate remained arrested at the G1 phase of the cell cycle whereas untreated control cells progressed through the cell cycle after serum stimulation. The cells arrested at the G1 phase after exposure to the NO donor had low cyclin D1 levels compared with the control cells. The levels of cyclin E and CDK4, however, were similar to the control cells. The decline in cyclin D1 protein preceded the decrease of its mRNA. This decline of cyclin D1 was due to a decrease in its synthesis induced by the NO donor and not due to an increase in its degradation. We conclude that down-regulation of cyclin D1 protein by DETA-NONOate played an important role in the cytostasis and arrest of these tumor cells in the G1 phase of the cell cycle.
Activated macrophages produce large amounts of nitric oxide (NO), which induces cytostasis and cytotoxicity to tumor cells both in vitro and in vivo (1–8). Macrophages also play a significant role in host resistance to tumors (7–9). Tumor cells cocultured with activated macrophages rapidly develop cytostasis, which precedes the cytotoxic effects (5, 10).
It has been reported that NO-induced early cytostasis begins with a rapid and reversible inhibition of ribonucleotide reductase (RR) (10). However, the possibility of targets other than RR being involved in the NO-induced long-lasting cytostasis was suggested by other investigators (10). Hydroxyurea, a specific inhibitor of RR (10), decreased thymidine incorporation in cultured cells and, after its removal, the thymidine incorporation rapidly returned to control values. The rate of recovery was much faster after withdrawal of hydroxyurea when compared with the rate of recovery observed in cells that initially were exposed either to activated macrophages or to a NO donor and then kept in a NO-free environment. On this basis, it was suggested that targets other than RR may be responsible for producing long-lasting cytostasis in target cells exposed to NO (5, 10, 11). This long-lasting cytostasis, therefore, would be associated with metabolic alterations in target cells, producing long-lasting growth inhibition and requiring a longer time for these cells to resume their normal rate of proliferation.
Because the precise mechanism(s) by which NO induces this long-lasting cytostasis is not completely known, the present work was undertaken to assess this mechanism(s) by using a human breast cancer cell line, MDA-MB-231. This cell line was selected because it represents a highly undifferentiated human breast cancer cell line (12) that does not produce NO (13). This allowed us to assess the effects of exogenous NO without any compounding influence of endogenous NO production. NO has a very short half-life (≈3–7 sec under physiologic conditions), and sustained exposure to NO is required to induce cytostasis. We therefore used DETA-NONOate [2,2′-(hydroxynitrosohydrazono)bis(ethanamine)], a NO donor that has a half-life of approximately 20 h (14), to mimic continuous production of NO as observed with activated macrophages. In this study, we observed that the NO-mediated cytostasis of MDA-MB-231 and the arrest of cells in the G1 phase of the cell cycle were associated with down-regulation of cyclin D1 and this was preceded by hypophosphorylation of the retinoblastoma protein (pRb). This down-regulation of cyclin D1 by NO mainly was due to a decrease in cyclin D1 synthesis.
Materials and Methods
Cell Culture.
MDA-MB-231 was cultured in DMEM supplemented with 6 mM l-glutamine/20 mM Hepes/10 μg/ml human insulin/10% (vol/vol) FBS/100 units/ml penicillin/100 μg/ml streptomycin at 37°C, in 5% CO2.
Reagents.
DMEM low glucose, DMEM high glucose, methionine- and cysteine-free DMEM, streptomycin, and penicillin were purchased from GIBCO/BRL. DETA-NONOate, 1H-[1,2,3]oxadiazolo[4,3-a]quinoxalin-1-one (ODQ), and 2-(4-carboxyphenyl)-4,4,5,5-tetramethyl-1-oxyl-3-oxide (c-PTIO) were from Cayman Chemicals (Ann Arbor, MI); zaprinast, propidium iodide, leupeptin, aprotinin, PMSF, and protein A-Sepharose beads were from Sigma; [3H]thymidine, Tran35S-label, and [35S]cysteine were from ICN; and [α-32P]dCTP and [γ-32P]ATP were from Amersham Pharmacia.
Measurement of Rate of NO Release by DETA-NONOate.
The rate of NO released from 1 mM DETA-NONOate in DMEM medium was assessed over time. Ten milliliters of DMEM containing 5% FBS and 20 mM Hepes was placed in a 100-mm Petri dish and incubated at 37°C. One hundred-microliter samples were taken and injected on Sievers NOA (chemiluminescence) that contained 10 mM NaOH to stop any further liberation of NO from DETA-NONOate. The concentration of NO was determined as pmol/0.1 ml to obtain the actual NO concentration in the medium at any given time (15).
Cytostasis Assay.
Cells were seeded in 6-well plates (7.5 × 105 cells/well). The medium was changed daily, and fresh drug was added. The cells were collected from 0 to 48 h, and viability was determined by trypan blue exclusion method and counted on a hemocytometer.
Cell Cycle Analysis.
Cells suspended in hypotonic DNA staining buffer (0.1% sodium citrate/0.3% Triton X-100/0.01% propidium iodide/0.005% ribonuclease A) were incubated for 15 min at 4°C and subjected to fluorescence-activated cell sorting (FACS) to analyze the percentage of cells in the different phases of the cell cycle (16).
Western Analysis.
Western analysis using an enhanced chemiluminescence detection system (Amersham Pharmacia) was carried out as described (17). The following primary antibodies (1:1,000 dilutions) were used: cyclin D1 (H-295), cyclin E (HE12), CDK4 (C-22), CDK6 (C-21), and CDK2 (D-12) (from Santa Cruz Biotechnology); glyceraldehyde-3-phosphate dehydrogenase (mAb 374) (from Chemicon); and pRb (Ab-1) (from Calbiochem).
Northern Analysis.
Total cellular RNA was extracted by using Trizol Reagent (GIBCO/BRL) according to the manufacturer's instruction. Equal amounts (15 μg/lane) of RNA were resolved by electrophoresis on a 1.2% formaldehyde-agarose gel, blotted onto nylon membrane, and hybridized according to standard protocols. cDNA for cyclin D1 was kindly provided by Andrew Arnold (Harvard Medical School, Cambridge, MA).
Kinase Assay.
Cells (1 × 107) were lysed in the same lysis buffer used in Western analysis. The lysates were immunoprecipitated with CDK4 agarose conjugate (Santa Cruz Biotechnology) for 2 h on ice with gentle shaking (18). The beads were washed and the kinase reaction was initiated by resuspending the beads in 30 μl of kinase buffer and 5 μg of glutathione S-transferase (GST)-Rb fusion protein [plasmid containing GST-Rb was kindly provided by Robert Chiu (University of California, Los Angeles)], which was obtained by transfecting GST-Rb cDNA into Escherichia coli HB 101 and purified from the cells by using the Redi Pack GST purification module. After incubation for 15 min at 30°C and separation on 12% SDS/PAGE, autoradiography was performed.
Metabolic Labeling and Immunoprecipitation to Measure the Synthesis of Cyclin D1.
Control and DETA-NONOate-treated MDA-MB-231 cells at 70–80% confluence were incubated in serum-, methionine-, and cysteine-free DMEM for 30 min. The cells were preincubated for 1 h with 500 μCi of Tran35S-label (1,175 Ci/mmol) and 250 μCi of [35S]cysteine (1,075 Ci/mmol). After labeling, the cells were processed as described (19).
Statistical Analysis.
Each value represents the mean SEM of at least three separate experiments in each group. Comparisons of means were made by using Student's t test for unpaired values. When more than two means were compared, an ANOVA with repeated measurements was used. If a significant F value was found, Scheffé's test for multiple comparisons was used to identify differences among groups. Values were considered significant when P was <0.05.
Results
Rate of NO Release by DETA-NONOate.
The NO released from 1 mM DETA-NONOate rapidly rose to 0.5 μM followed by a relatively constant amount of NO release over a 24-h period. These profiles of DETA-NONOate are similar to macrophages stimulated with IFN-γ and lipopolysaccharide that give steady-state concentrations over 24 h in the media above attached cells (15). This would suggest that NO concentrations of DETA-NONOate at 1 mM in bulk solution is similar to 106 macrophages/ml.
DETA-NONOate Treatment Caused Growth Inhibition in MDA-MB-231 Cells Associated with G1 Arrest and Cyclin D1 Down-Regulation.
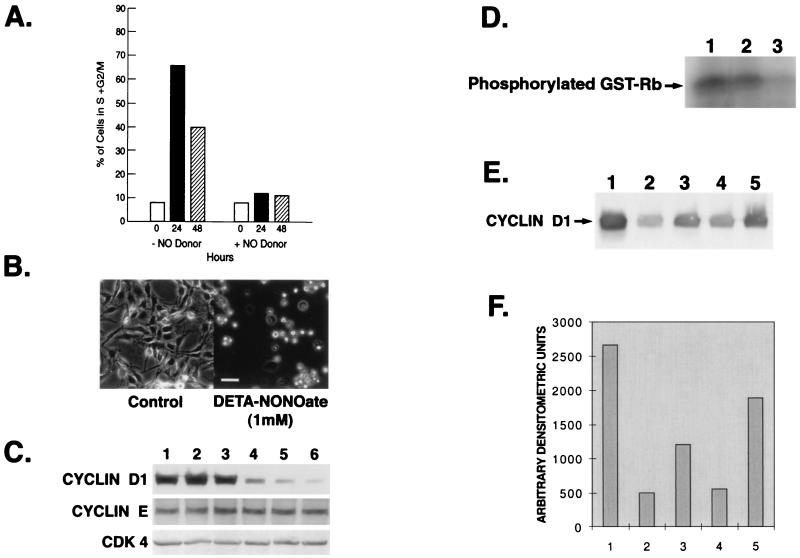
DETA-NONOate at 1 mM led to cytostasis as evidenced by the lack of change in cell number over time (Fig. 1A). Trypan blue exclusion indicated 95% viability of the cells at 48 h after treatment with 1 mM DETA-NONOate. Exposure of cells beyond 48 h induced apoptosis as confirmed by TUNEL assay (data not shown). Because we were interested in the mechanism by which NO induces cytostasis, we confined our studies to the first 48 h of exposure of these cells to DETA-NONOate. FACS analysis revealed that treatment with DETA-NONOate for 48 h resulted in a higher proportion of the cells in G1 (86%) phase of the cell cycle relative to that seen in untreated cells (61%) (Fig. 1B). Removal of DETA-NONOate from the media after exposure of these cells to the NO donor for 36 h led to reversibility of the cytostatic effects, leading to an increase in cell number and decrease of cells in the G1 phase of the cell cycle (data not shown). Western blot analysis was performed to examine the effects of DETA-NONOate on the expression of proteins that participate in regulating G1 progression. DETA-NONOate led to a significant decrease in cyclin D1 protein in a time-dependent manner whereas the levels of cyclin E, CDK2, CDK4, and CDK6 remained unchanged (Fig. 1C). Densitometric scan revealed an approximately 50% decrease in cyclin D1 levels at 24 h, and by 48 h, barely any cyclin D1 was detected by enhanced chemiluminescence (Fig. 1D). However, oxidized DETA-NONOate, which does not release NO, had no effect on the cyclin D1 levels (data not shown). The levels of cyclin D2 and cyclin D3 were negligible in this cell line. Also, the levels of cyclin D1 remained unchanged when the NO donor was added with cPTIO (50 μM), a NO scavenger (20) (data not shown). Hypophosphorylated pRb was found in DETA-NONOate-treated cells, which increased with time (Fig. 1E). The decline in cyclin D1 protein was reversible as removal of DETA-NONOate from the media led to recovery in cyclin D1 levels. Cells exposed to 1 mM DETA-NONOate for 24 h showed a more rapid recovery rate of cyclin D1 protein levels than those exposed to DETA-NONOate for 48 h (Fig. 1F).
Figure 1.
(A) The effect of DETA-NONOate on MDA-MB-231 viable cell count. Cells (7.5 × 105) were treated with 1 mM DETA-NONOate. The medium was changed and fresh drug was added daily. Viability of the cells was assessed every 12 h, up to 48 h, by trypan blue exclusion. The results are representative of four separate experiments (±SE). (B) FACS analysis of 1 mM DETA-NONOate-treated MDA-MB-231 cells at various time points. The cells were prepared for FACS analysis, as described in Materials and Methods, and analyzed in a FACScalibur flow cytometer. The number of cells in each phase of the cell cycle was obtained by modfit software. (C) Effect of DETA-NONOate (1 mM) on cell cycle proteins in MDA-MB-231 cells at different time points. Cells were harvested and prepared for Western blot analysis for cyclin D1, cyclin E, CDK4, CDK6, and CDK2 as described in Materials and Methods. (D) Densitometric scan of Western blot of cyclin D1 from C. Only the arbitrary densitometric units of 0, 24, and 48 h are plotted. (E) Effect of DETA-NONOate on phosphorylation status of pRb in MDA-MB-231 cells at different time points. Western blot analysis shows the phosphorylation status of pRb in the presence of DETA-NONOate. Hypophosphorylated pRb migrates faster than hyperphosphorylated pRb. (F) Western blot analysis showing reversibility of the effect of DETA-NONOate on cyclin D1. MDA-MB-231 cells were treated with DETA-NONOate (1 mM) for either 24 or 48 h. After treatment, the cells were washed and kept in DETA-NONOate-free medium for another 24 h, harvested, and prepared for immunoblotting with cyclin D1 antibody. Lane 1, control cells at 24 h; lane 2, DETA-NONOate treatment for 24 h; lane 3, cells treated with DETA-NONOate for 24 h were washed and allowed to recover for 24 h; lane 4, control cells for 48 h; lane 5, DETA-NONOate treatment for 48 h; lane 6, cells treated with DETA-NONOate for 48 h were washed and allowed to recover for 24 h.
Synchronized Cells Treated with DETA-NONOate Remained Arrested at G1 Phase Associated with Down-Regulation of Cyclin D1.
Because of the possible variations in cyclin D1 levels in unsynchronized cells progressing through different phases of cell cycle, we elected to assess the effects of DETA-NONOate on cyclin D1 levels in synchronized cells. Fig. 2A shows the effect of DETA-NONOate on the cell cycle distribution of MDA-MB-231 cells that were serum-starved (quasi-synchronized) for 48 h and then restimulated to enter the cell cycle by addition of 5% fresh serum. In these experiments, DETA-NONOate was added during the last 12 h of a 48-h serum-starvation period, and the cells remained in the presence of the NO donor after addition of serum. In absence of the NO donor, serum-stimulated cells readily proceeded to enter the S and G2/M phases whereas, in its presence, the cells remained arrested at the G1 phase. Cells stimulated with serum grew exponentially whereas those in presence of the NO donor remained growth-arrested as observed by the characteristic round-shaped morphology (Fig. 2B). Synchronized cells exposed to DETA-NONOate had low to undetectable cyclin D1 levels in contrast to cells without the NO donor, where the level of cyclin D1 was found to be high (Fig. 2C). The levels of cyclin E and CDK4 were the same in cells with or without the NO donor. Cyclin D1 functions after forming a complex with PCNA, p21, and CDK4. We therefore assessed the activity of this complex by a CDK4 kinase assay. The effect of DETA-NONOate on CDK4 kinase activity was examined in these serum-stimulated, synchronized MDA-MB-231 cells by immunoprecipitating cell lysates with anti-CDK4 antibody and assaying the immunocomplex by using GST-Rb as a substrate for the in vitro phosphorylation. CDK4 kinase activity was found to be low in DETA-NONOate-treated cells (Fig. 2D). In additional experiments, we observed that whereas cells grown only in DMEM for 48 h did not show any significant decline in cyclin D1, the addition of DETA-NONOate caused a dramatic decline of the protein (data not shown). To study the recovery of NO donor-mediated decline of cyclin D1 in these cells, the synchronized cells that were exposed to the NO donor for 12 h were washed and allowed to recover in NO-free medium for various time points (Fig. 2E). We observed that there was a 40% and 60% increase in the level of cyclin D1 after 3 and 6 h, respectively, in NO-free medium in these cells, as compared with cells that continued to be exposed to the NO donor (Fig. 2F).
Figure 2.
(A) Effect of DETA-NONOate on the progression of synchronized MDA-MB-231 cells through the cell cycle after stimulation with 5% FBS. At various time points after addition of serum, cells were harvested for DNA analysis by flow cytometry and the percentage of cells in the S + G2/M phase of cell cycle were plotted. (B) Photomicrographs (×50) of synchronized MDA-MB-231 cells stimulated with 5% FBS without (Left) or with DETA-NONOate (Right) for 24 h. (C) Western analysis of cell cycle proteins from synchronized cells stimulated with 5% FBS in the absence or presence of DETA-NONOate. Cells were synchronized in DMEM for 48 h, after which 5% FBS was added to these cells. The cells were harvested at 12, 24, and 48 h (lanes 1, 2, and 3, respectively) after the addition of FBS. Another set of cells were treated with DETA-NONOate for the last 12 of the 48 h of serum starvation. After 48 h of serum starvation, 5% FBS was added to all of the cells and the cells were harvested at 12, 24, and 48 h (lanes 4, 5, and 6, respectively). Cells were harvested and prepared for immunoblotting with cyclin D1, cyclin E, and CDK4 antibodies. (D) Kinase assay as described in Materials and Methods was performed to assess the hypophosphorylated status of pRb. The MDA-MB-231 cells were synchronized and treated with 5% FBS in the presence or absence of DETA-NONOate for different time points. Lane 1, synchronized cells treated with 5% FBS for 12 h; lane 2, DETA-NONOate treatment for 3 h followed by 5% FBS for 12 h; lane 3, DETA-NONOate for 12 h followed by 5% FBS for 12 h. (E) Western analysis showing recovery of cyclin D1. Cells synchronized in DMEM for 48 h followed by 5% FBS stimulation for 12 h. Lane 1, control; lane 2, DETA-NONOate (1 mM) for the last 12 h of the 48-h synchronization period; lane 3, DETA-NONOate for 12 h followed by NO donor-free medium for 3 h; lane 4, DETA-NONOate for 12 h; lane 5, DETA-NONOate for 12 h followed by NO donor-free medium for 6 h. (F) Densitometric scan of cyclin D1 Western blot from E.
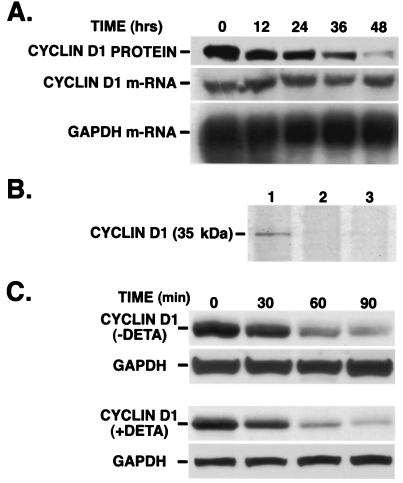
DETA-NONOate Suppressed the Synthesis of Cyclin D1 Without Affecting Its Degradation.
Significant decrease in cyclin D1 protein levels preceded those observed for its mRNA expression (Fig. 3A), indicating the involvement of translational or posttranslational control by NO during cyclin D1 down-regulation. To explore these possibilities, the cells were kept in medium containing Tran35S-label and [35S]cysteine to label newly synthesized proteins. Trichloroacetic acid precipitation of the extract showed that there was a gradual decrease in acid-precipitable counts (data not shown). When equal amounts of protein from labeled cells were immunoprecipitated with cyclin D1 antibody, fractionated on gel, and autoradiographed, we observed that the de novo synthesis of cyclin D1 protein decreased with time in cells treated with the NO donor compared with control cells (Fig. 3B). We next evaluated the effect of the NO donor on the degradation of cyclin D1 protein. Protein synthesis in the cells was stopped by the addition of cycloheximide (100 μg/ml), and cyclin D1 degradation was monitored in control cells with or without DETA-NONOate for 24 h. Western blot analysis (Fig. 3C) followed by densitometric analysis (data not shown) revealed that the half-life of cyclin D1 was not affected by the NO donor. Equal loading of protein in each lane was confirmed by reprobing the stripped membranes with glyceraldehyde-3-phosphate dehydrogenase. The approximate half-life calculated for cyclin D1 was 30 min, which corresponded to other reports (21). Because many of the effects of NO are cGMP-dependent, we tested the possibility of whether cGMP mediated the decline of cyclin D1 by using zaprinast (a phosphodiesterase inhibitor) and ODQ (a guanylate cyclase inhibitor) at concentrations known to inhibit the activities of the respective enzymes (22–24). The effects of 1 mM DETA-NONOate on cyclin D1 protein levels were not changed in the presence of either ODQ or zaprinast, thereby ruling out the role of cGMP in mediating the effects of NO on cyclin D1 protein (data not shown).
Figure 3.
(A) Effects of DETA-NONOate on cyclin D1 protein and mRNA levels. MDA-MB-231 cells were treated with DETA-NONOate for various time periods and prepared for Western and Northern blot analysis as described in Materials and Methods. The membrane for Western blot was immunoblotted with anti-cyclin D1 antibody. The membrane for Northern blot was hybridized with α-32P-labeled cyclin D cDNA. (B) Effect of DETA-NONOate on the synthesis of cyclin D1. The cells were treated with DETA-NONOate for 24 and 36 h. Before the end of the respective time points, the cells were metabolically labeled with Tran35S-label and [35S]cysteine for 2 h as described in Materials and Methods. The cells were lysed, and equal amounts of protein from cell lysates were immunoprecipitated for cyclin D1 and loaded on the gel. Lane 1, control; lane 2, cells treated with DETA-NONOate for 24 h; lane 3, cells treated with DETA-NONOate for 36 h. (C) Effect of DETA-NONOate on the half-life of cyclin D1. Western blot analysis for cyclin D1 was performed in cells treated with or without DETA-NONOate. (Upper) Control cells were exposed to cycloheximide for the specified time points. (Lower) Cells were treated with DETA-NONOate for 24 h and then exposed to cycloheximide for the respective time points, similar to that done with control cells, and prepared for Western blot analysis.
Discussion
The results of our study indicate that NO had a cytostatic effect on the human breast cancer cell line MDA-MB-231, which, after 48 h, induced apoptosis. The primary objective of the present study was to identify targets other than RR in NO-induced cytostasis. We therefore focused our studies on the effects produced by the NO donor on these cells during the first 48 h of exposure. Interference in cell cycle progression has been implicated in arresting cell growth (25). Progression of cells through the G1 phase of the cell cycle is governed mainly by the activity of G1 cyclins, namely, cyclins D and E. Cyclin D is involved in the early- to mid-G1 phase of the cell cycle in association with its catalytic partner CDK4 and CDK6, whereas cyclin E with its catalytic partner, CDK2, is involved mainly in the late-G1 to early-S phase. The cyclin D/CDK complexes phosphorylate pRb in the mid- to late-G1 phase (26–30).
In this study we observed that cytostasis induced by NO was associated with the down-regulation of cyclin D1 protein and that both these effects were reversible after removal of NO from the medium. These experiments indicate cyclin D1 to be a target in the NO-induced, long-lasting cytostasis. The levels of cyclin E, CDK2, CDK4, and CDK6 did not change. The effects of DETA-NONOate were due to the NO released as oxidized DETA-NONOate or DETA-NONOate in the presence of cPTIO was without effect. The arrest of the cells in the G1 phase also was found to be associated with a decline in cyclin D1 levels and hypophosphorylation of pRb. Synchronized cells stimulated with FBS in the presence and absence of DETA-NONOate were used to evaluate the role of cyclin D1 in the progression of these cells from the G1 phase of the cell cycle. Cyclin D1 was found to be down-regulated in synchronized cells stimulated with FBS in the presence of DETA-NONOate, whereas the levels of cyclin E and CDK4 did not undergo significant change. These experiments indicated that cyclin D1 was essential for G1 progression in this cell line. Because hypophosphorylated pRb was found to increase with time in these cells after exposure to the NO donor whereas the CDK4 kinase activity decreased, the arrest of cells in the G1 phase was most likely a result of suppression of the cyclin D1/pRb/CDK pathway.
Down-regulation of cyclin D1 by NO, in contrast to other cell cycle proteins, has important implications because the G1 cyclins, especially the D type cyclins, have been implicated strongly in controlling the progression of cells through the G1 phase of the cell cycle (31, 32). Cyclin D1 has been proven experimentally to be the strongest candidate among the potential oncogenic elements of the cell cycle machinery (33). Cyclin D1 gene is a component of the chromosome 11q13 amplicon involved in the molecular pathogenesis of several common solid tumors, among which breast cancer is clinically the most significant tumor type (34–38). Cyclin D1 is overexpressed in 80% of in situ ductal carcinoma whereas it is low or absent in normal breast tissue. This suggests that its regulation may define a transition from a benign state to carcinoma and that unregulated overexpression of cyclin D1 may be a common early event in mammary carcinomas (31).
Cyclin D1 is up-regulated by various growth factors (39) in breast cancer cells, and we considered the possibility that NO may have inactivated the growth factors in the serum, which subsequently may have led to a decline of cyclin D1 and cell cycle arrest in the G1 phase. This possibility seems unlikely, because when cells were grown in serum-free medium, a similar decline in cyclin D1 still occurred with the NO donor. These results indicate that NO acted on targets inside the cell, which were responsible for down-regulating cyclin D1 levels. We observed that cyclin D1 mRNA levels did not change before the decrease in cyclin D1 protein levels, ruling out the possibility that a decrease in mRNA was the limiting factor. Although there was no effect of DETA-NONOate on the degradation of cyclin D1, because the half-life of the protein was not affected after treatment of the cells with the NO donor, synthesis of cyclin D1 was down-regulated.
Potential mechanisms by which NO may inhibit translation of cyclin D1 need to be investigated. There are reports that NO inhibits protein synthesis (40–42) at the level of translation initiation. Translation initiation depends mainly on the activity of eukaryotic initiation factors (eIFs). Initiation factor eIF2α (43) promotes the expression of cyclins A, E, and D1 (41), whereas eIF4E promotes the expression of cyclin D and ornithine decarboxylase (44, 45). It has been suggested that a NO-induced increase in the phosphorylation of eIF-2α may lead to the activation of an eIF-2α kinase or inhibition of a phosphatase as a likely mechanism for the nonspecific inhibition of protein synthesis (41). This action of NO in inhibiting protein synthesis was found to be guanylate cyclase-dependent (41). However, it seems unlikely that NO decreased protein synthesis in our study by an action of eIF-2α because the decrease in cyclin D1 protein in our study was found to be guanylate cyclase-independent. It is possible that eIF-4E may be very sensitive to the inhibitory action of NO. The expression of this initiation factor is increased consistently in carcinoma of the breast (46, 47) and potentially may explain the increase in cyclin D1 observed in this condition.
NO has been reported to inactivate mitochondrial complexes I and IV (48) and also glyceraldehyde 3-phosphate dehydrogenase (49, 50), both of which are key enzymes responsible for ATP production in the cell. NO also has been reported recently to decrease ATP content of the cells (51), and it would be interesting to investigate whether this could result in the decrease in cyclin D1 synthesis observed in our study. The action of NO in inducing G1 arrest in MDA-MB-231 is independent of p53 because this is mutant in this cell line (52).
We have demonstrated previously that N-hydroxy-l-arginine, an intermediate in the NO biosynthetic pathway, inhibits arginase in MDA-MB-468 cells. This inhibition of arginase led to a decrease in intracellular polyamines and induction of apoptosis in these cells, which was independent of any changes in cyclin D1 or cyclin E levels (17). However, MDA-MB-231 cells do not express arginase (17), and NO in this cell line inhibited cyclin D1 translation, which initially led to cytostasis for 48 h followed by apoptosis. In conclusion, our studies indicate the existence of another novel mechanism by which NO, the end product of NO synthase-catalyzed reaction, can induce long-lasting cytostasis. This effect of NO in inhibiting synthesis of cyclin D1 may be relevant only in cell lines that overexpress this protein.
Acknowledgments
We acknowledge the constructive suggestions of Dr. Salvador Moncada during the preparation of this manuscript. We also thank Janis Cuevas and Svetlana Arutyunova for their technical assistance. This work was supported in part by Palomba Weingarten, The Allegra Charach Cancer Research Fund, and U.S. Public Health Service Grant CA-78357 (to G.C.).
Abbreviations
- RR
ribonucleotide reductase
- DETA-NONOate
2,2′-(hydroxynitrosohydrazono)bis(ethanamine)
- pRb
retinoblastoma protein
- FACS
fluorescence-activated cell sorting
- GST
glutathione S-transferase
References
- 1.Bartlett D, Church D F, Bounds P L, Koppenol W H. Free Radical Biol Med. 1995;18:85–92. doi: 10.1016/0891-5849(94)e0133-4. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 3.MacMicking J, Xie Q W, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 4.Hibbs JB, Jr, Taintor R R, Vavrin Z. Science. 1987;235:473–476. doi: 10.1126/science.2432665. [DOI] [PubMed] [Google Scholar]
- 5.Stuehr D J, Nathan C F. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farias-Eisner R, Chaudhuri G, Aeberhard E, Fukuto J M. J Biol Chem. 1996;271:6144–6151. doi: 10.1074/jbc.271.11.6144. [DOI] [PubMed] [Google Scholar]
- 7.Farias-Eisner R, Sherman M P, Aeberhard E, Chaudhuri G. Proc Natl Acad Sci USA. 1994;91:9407–9411. doi: 10.1073/pnas.91.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yim C Y, Bastian N R, Smith J C, Hibbs JB, Jr, Samlowski W E. Cancer Res. 1993;53:5507–5511. [PubMed] [Google Scholar]
- 9.Thomsen L L, Miles D W, Happerfield L, Bobrow L G, Knowles R G, Moncada S. Br J Cancer. 1995;71:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepoivre M, Flaman J M, Bobae P, Lemaire G, Henry Y. J Biol Chem. 1994;269:21891–21897. [PubMed] [Google Scholar]
- 11.Kwon N S, Stuehr D J, Nathan C F. J Exp Med. 1991;174:761–767. doi: 10.1084/jem.174.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedl A, Jordan V C. Eur J Cancer. 1994;30A:1559–1564. doi: 10.1016/0959-8049(94)00293-e. [DOI] [PubMed] [Google Scholar]
- 13.Zeillinger R, Tantscher E, Schneeberger C, Tschugguel W, Eder S, Sliutz G, Huber J C. Breast Cancer Res Treat. 1996;40:205–207. doi: 10.1007/BF01806216. [DOI] [PubMed] [Google Scholar]
- 14.Mooradian D L, Hutsell T C, Keefer L K. J Cardiovasc Pharmacol. 1995;25:674–678. [PubMed] [Google Scholar]
- 15.Espey M G, Miranda K M, Pluta R M, Wink D A. J Biol Chem. 2000;275:11341–11347. doi: 10.1074/jbc.275.15.11341. [DOI] [PubMed] [Google Scholar]
- 16.Krishan A. J Cell Biol. 1975;66:88–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R, Pervin S, Karimi A, Cederbaum S, Chaudhuri G. Cancer Res. 2000;60:3305–3312. [PubMed] [Google Scholar]
- 18.Musgrove E A, Hamilton J A, Lee S L, Sweeny J E, Watts K W, Sutherland R L. Mol Cell Biol. 1993;13:3577–3587. doi: 10.1128/mcb.13.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kujubu D A, Reddy S T, Fletcher B S, Herschman H R. J Biol Chem. 1993;268:5425–5430. [PubMed] [Google Scholar]
- 20.Ishida A, Sasaguri T, Kosaka C, Nojima H, Ogata J. J Biol Chem. 1997;272:10050–10057. doi: 10.1074/jbc.272.15.10050. [DOI] [PubMed] [Google Scholar]
- 21.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 22.Brunner F, Schmidt K, Nielsen E B, Mayer B. J Pharmacol Exp Ther. 1996;277:48–53. [PubMed] [Google Scholar]
- 23.Cellek S, Kasakov L, Moncada S. Br J Pharmacol. 1996;118:137–140. doi: 10.1111/j.1476-5381.1996.tb15376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams P D, Kaelin WG., Jr Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 25.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 26.Lam E W, La Thangue N B. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 27.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 28.Slansky J E, Farnham P J. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 29.Scherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 30.Hebeiss K, Kilbinger H. Br J Pharmacol. 1996;118:2073–2078. doi: 10.1111/j.1476-5381.1996.tb15646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantl V, Smith R, Brookes S, Dickson C, Peters G. Cancer Surv. 1993;18:77–94. [PubMed] [Google Scholar]
- 32.Lammie G A, Fantl V, Smith R, Schuring E, Brookes S, Michalides R, Dickson C, Arnold A, Peters G. Oncogene. 1991;6:439–444. [PubMed] [Google Scholar]
- 33.Michalides R, van Veelen N, Hart A, Loftus B, Wientjens E, Balm A. Cancer Res. 1995;55:975–978. [PubMed] [Google Scholar]
- 34.Peters G, Fantl V, Smith R, Brookes S, Dickson C. Breast Cancer Res Treat. 1995;33:125–135. doi: 10.1007/BF00682720. [DOI] [PubMed] [Google Scholar]
- 35.Schuuring E, Verhoeven E, van Tinteren H, Peterse J L, Nunnink B, Thunnissen F B, Devilee P, Cornelisse C J, van de Vijver M J, Mooi W J, et al. Cancer Res. 1992;52:5229–5234. [PubMed] [Google Scholar]
- 36.van Diest P J, Michalides R J, Jannink L, van der Valk P, Peterse H L, de Jong J S, Meijer C J, Baak J P. Am J Pathol. 1997;150:705–711. [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstat-Saslow D, Merino M J, Manrow R E, Lawrence J A, Bluth R F, Wittenbel K D, Simpson J F, Page D L, Steeg P S. Nat Med. 1995;1:1257–1260. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 38.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 39.Musgrove A, Sarcevic B, Sutherland R L. J Cell Biochem. 1996;60:363–378. doi: 10.1002/(SICI)1097-4644(19960301)60:3%3C363::AID-JCB8%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 40.Curran R D, Ferrari F K, Kispert P H, Stadler J, Stuehr D J, Simmons R L, Billiar T R. FASEB J. 1991;5:2085–2092. doi: 10.1096/fasebj.5.7.1707021. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y M, Son K, Hong S J, Green A, Chen J J, Tzeng E, Hierholzer C, Billiar T R. Mol Med. 1998;4:179–190. [PMC free article] [PubMed] [Google Scholar]
- 42.Kolpakov V, Gordon D, Kulik T J. Circ Res. 1995;76:305–309. doi: 10.1161/01.res.76.2.305. [DOI] [PubMed] [Google Scholar]
- 43.Aktas H, Fluckiger R, Acosta J A, Savage J M, Palakurthi S S, Halperin J A. Proc Natl Acad Sci USA. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenwald I B, Lazaris-Karatzas A, Sonenberg N, Schmidt E V. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, DeBenedetti A. Int J Cancer. 1995;64:27–31. doi: 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- 47.Nathan C A, Carter P, Liu L, Li B D, Abreo F, Tudor A, Zimmer S G, DeBenedetti A. Oncogene. 1997;15:1087–1094. doi: 10.1038/sj.onc.1201272. [DOI] [PubMed] [Google Scholar]
- 48.Clementi E, Brown G C, Feelisch M, Moncada S. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimmeler S, Lottspeich F, Brune B. J Biol Chem. 1992;267:16771–16774. [PubMed] [Google Scholar]
- 50.Kots A Y, Skurat A V, Sergienko E A, Bulgarina T V, Severin E S. FEBS Lett. 1992;300:9–12. doi: 10.1016/0014-5793(92)80153-8. [DOI] [PubMed] [Google Scholar]
- 51.Lowenstein C J, Snyder S H. Cell. 1992;70:705–707. doi: 10.1016/0092-8674(92)90301-r. [DOI] [PubMed] [Google Scholar]
- 52.Jones C B, Clements M K, Redkar A, Daoud S S. Int J Oncol. 2000;17:1043–1051. doi: 10.3892/ijo.17.5.1043. [DOI] [PubMed] [Google Scholar]