Abstract
Previous authors have suggested that tumor suppressor expression promotes aging while preventing cancer, but direct experimental support for this cancer-aging hypothesis has been elusive. Here, by using somatic, tissue-specific inactivation of the p16INK4a tumor suppressor in murine T- or B-lymphoid progenitors, we report that ablation of p16INK4a can either rescue aging or promote cancer in a lineage-specific manner. Deletion of p16INK4a in the T lineage ameliorated several aging phenotypes, including thymic involution, decreased production of naive T cells, reduction in homeostatic T-cell proliferation, and attenuation of antigen-specific immune responses. Increased T-cell neoplasia was not observed with somatic p16INK4a inactivation in T cells. In contrast, B lineage–specific ablation of p16INK4a was associated with a markedly increased incidence of systemic, high-grade B-cell neoplasms, which limited studies of the effects of somatic p16INK4a ablation on B-cell aging. Together, these data show that expression of p16INK4a can promote aging and prevent cancer in related lymphoid progeny of a common stem cell.
Introduction
Antagonistic pleiotropy, one of the oldest hypotheses of aging, suggests that certain cellular activities may be of physiologic benefit in youth but may untowardly decrease organismal fitness in later life.1 The cancer-aging hypothesis represents a special case of this model, suggesting that tumor suppressor mechanisms such as cellular senescence can function in an antagonistically pleiotropic manner, ie, preventing malignant transformation in young mammals but promoting aging over a lifespan. Although there are examples of senescence-promoting tumor suppressor proteins such as p16INK4a and p53 contributing to the aging of distinct tissue compartments (reviewed in Campisi2 and Collado et al3), it has not been possible to directly demonstrate a strict trade-off between increased aging and reduced cancer as the result of a cell-autonomous activation of a specific tumor suppressor mechanism within an isolated tissue.
The p16INK4a tumor suppressor, along with 2 other tumor suppressor proteins, p15INK4b and p14ARF (hereafter referred to solely as ARF), originates from the INK4/ARF or CDKN2a/b locus at human chromosome 9p21 (reviewed in Sharpless and DePinho4). Expression of p16INK4a inhibits the cell cycle, promotes cellular senescence, and has been linked to cancer and aging in mammalian systems. Expression of p16INK4a has been shown to markedly increase with age in nearly all mammalian tissues, and caloric restriction, which retards aging in rodents, attenuates this age-induced increase in p16INK4a. Increased expression of p16INK4a is not only associated with aging but appears to play a causal role in some tissues. Elevated expression of p16INK4a with aging has been associated with a decrease in the replicative capacity of hematopoietic stem cells (HSC),5 pancreatic β cells,6,7 and neural stem cells.8 Moreover, germline inactivation of p16INK4a, but not ARF, partially rescues several age-related phenotypes in a progeroid mouse strain.9 Because these experiments have largely relied on germline inactivation or overexpression of p16INK4a, however, the cell autonomous contribution of p16INK4a to aging and its relationship to tumor suppression has not been clearly defined.
In association studies, authors have suggested that altered regulation of p16INK4a expression may contribute to human age-associated phenotypes such as frailty, type 2 diabetes, atherosclerotic disease, cancer susceptibility, and longevity (reviewed in Sharpless and DePinho,4 as well as others10–14). We and others have recently shown that the expression of p16INK4a and other INK4/ARF transcripts is strongly associated with the host genotype of single nucleotide polymorphisms near the INK4/ARF locus,15–17 suggesting the phenotypic impact of these single nucleotide polymorphisms results from altered expression of the INK4/ARF locus.
Some data specifically suggest a role for p16INK4a in aging and tumor suppression in the lymphoid compartment. Transgenic overexpression of p16INK4a in T cells under the control of the Lck promoter arrests thymocyte development at the double-negative (DN) stage.18 Consistent with this result, a role for p16INK4a in peripheral T-cell replicative senescence/hypoproliferation has been proposed19,20 because germline p16INK4a-deficient mice have increased thymocyte and peripheral T-cell numbers.21,22 Of particular importance to the present work, we and others have shown an accumulation of p16INK4a expressing T cells in human peripheral blood with aging.17,23 This expression of p16INK4a appears to be a biomarker of physiologic as opposed to chronologic age, independently correlating with gerontogenic behaviors such as smoking and physical inactivity.17 Although these murine and human data have suggested a role for p16INK4a in lymphoid aging, the cell-autonomous contribution of p16INK4a to immune aging under physiologic conditions has not been previously delineated.
With regard to lymphoid cancers, deletion and silencing of the INK4a/ARF locus are among the most common genetic events in a variety of human lymphoid and plasma cell malignancies, particularly pediatric B- and T-lineage acute lymphoblastic leukemia/lymphoma (ALL).24 Such lesions generally also inactivate p15INK4b and ARF, which clearly play important and specific tumor suppressor roles in hematologic malignancies,25–28 and, therefore, the contribution of p16INK4a to the suppression of these tumor types is not clear. Of particular interest to the present work, increasing expression of p16INK4a and ARF has been associated with an age-dependent decrease in lymphopoiesis as well as transformability of murine B-cell progenitors.29
Although these studies suggest roles for p16INK4a expression in lymphoid tumor suppression and aging, the specific relationship between p16INK4a expression and these phenotypic outcomes is unclear. For example, it is not known whether p16INK4a plays an intrinsic role in the age-related functional decline observed in differentiated lymphocytes. High-level expression of p16INK4a is noted in both lymphocytes and epithelium of the thymus with aging, and functional compromise of either compartment could plausibly lead to immune dysfunction. Moreover, it is not clear whether both the promotion of aging and tumor suppression are affected by p16INK4a in the same tissue or whether there are tissues to which p16INK4a contributes to only one but not both of these phenotypes with aging. To address these issues, we performed lineage-specific deletion of p16INK4a in B- and T-lymphocyte progenitors by using well-characterized, lineage-specific Cre recombinase alleles and a somatically inactivatable p16INK4a-specific allele. This work demonstrates a strong and cell autonomous role for p16INK4a in T-cell aging and B-cell tumor suppression.
Methods
Mice
All mice were bred onto a C57BL/6 genetic background for at least 8 generations. Animals were crossed and aged in positive-pressure–ventilated racking within the University of North Carolina Lineberger Comprehensive Cancer Center Mouse Phase 1 Unit. CD19Cre/+ and Lck-Cre mice were generously provided by Dr Yi Zhang. All experimental procedures were approved by the University of North Carolina, Chapel Hill (UNC-CH) Institutional Animal Care and Use Committee.
Flow cytometry, cell sorting, Western blots, ex vivo activation assays, and nitrophenylacetyl immunization
After labeling with corresponding magnetic microbeads (Miltenyi Biotec), Thy 1.2+ T cells or B220+ B cells were purified from single-cell splenic suspensions generated by the use of the AutoMACS pro separator (Miltenyi Biotec). Cells were then assessed for purity and viability by flow cytometry after CD3 and B220 staining (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All cells were enriched to greater than 90% purity. Multiple-color flow cytometry was performed on a CyAn ADP flow cytometer (Dako Cytomation). Perithymic lymph nodes were carefully dissected away from thymus, and enzymatic thymic digestion was not used to maximize the accuracy of thymocyte counts. Single-cell suspensions from spleen, thymus, and bone marrow were prepared after removal of erythrocytes with ACK buffer. Total cell numbers were then determined by microscopic observation of trypan-blue staining cells by the use of a Neubauer hemocytometer. Antibody information, ex vivo lymphocyte activation assays, nitrophenylacetyl (NP) immunization, germinal center formation, and in vivo 5-bromo-2-deoxyuridine (BrdU) incorporation assays are described in the supplemental text.
Tissue fixation, immunohistochemistry staining, and small animal imaging
Tissues were fixed in 10% neutral-buffered formalin overnight and transferred to 70% ethanol. Histopathology and immunohistochemistry were reviewed by board-certified veterinary (A.B.R.) and human (Y.F.) pathologists. Additional immunohistochemical information and animal imaging and statistical analysis are described in the supplemental text.
Results
Lineage-specific deletion of p16INK4a in lymphocytes
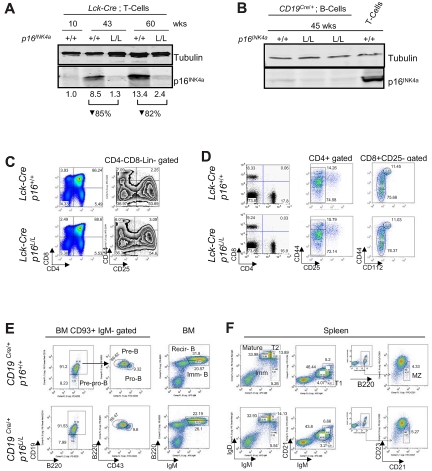
To examine the intrinsic role of p16INK4a in T or B lymphocytes, we crossed a recently described p16INK4a-specific conditional allele30 with Lck-Cre transgenic mice31 to delete p16INK4a in the T lineage or CD19Cre/+ knockin mice32 to delete in the B lineage. The p16INK4a conditional allele has floxed sites flanking exon 1α of the Ink4a/Arf locus, and somatic excision of p16INK4a in this strain does not alter expression of p15INK4b or ARF,30 in contrast to germline p16INK4a inactivation.33,34 Lck-Cre transgenic mice express Cre recombinase in immature thymocytes starting at the DN1-DN3 stage. As in humans,17,23 p16INK4a expression was nearly undetectable in splenic T cells from young adult Lck-Cre p16+/+ mice, but protein expression strongly accumulated with aging in the T-cell fraction. The efficiency of deletion in Lck-Cre p16L/L animals was 82%-85% on the basis of quantification of p16INK4a protein expression in peripheral T cells from 43- to 60-week-old mice (Figure 1A, supplemental Figure 1). In CD19Cre/+ mice, B lineage–restricted Cre expression is seen in early B-cell progenitors of the bone marrow.32
Figure 1.
Lineage-specific deletion of p16INK4a has no effect on lymphocyte development in young mice. (A) Protein expression of p16INK4a in purified mouse T cells from Lck-Cre p16+/+ and Lck-Cre p16L/L mice of indicated ages. Relative quantification of p16INK4a expression after normalization to tubulin is shown. See supplemental Figure 1 for purification scheme. (B) Protein expression of p16INK4a in purified B and T cells from CD19Cre/+ p16L/L and CD19Cre/+ p16+/+ mice of indicated ages. See supplemental Figure 1 for purification scheme. (C) Comparable thymocyte development in young (6-8 weeks old) Lck-Cre p16+/+ mice versus young Lck-Cre p16L/L mice. (Left) Fractions of double-positive, double-negative, and CD4/CD8 single-positive cells. (Right) Fractions of DN1 (CD25−CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44−), and DN4 (CD25−CD44−) after Lin−CD4−CD8− gating. (D) Differentiation of peripheral T cells in young Lck-Cre p16L/L mice versus Lck-Cre p16+/+ mice. (Left) CD4+ or CD8+ SP T-cell percentages in the spleen. (Middle) CD4+ memory (CD25−CD44hi) and naive (CD25−CD44lo/−) T-cell fractions from spleen. (Right) CD8+ memory (CD25−CD44hi CD122+) and naive (CD25−CD44lo/− CD122−) T-cell fractions. (E) Development of B-lineage progenitors in bone marrow (BM) in young CD19Cre/+ p16L/L mice versus CD19Cre/+ p16+/+ mice. (Left) CD93+ immunoglobulin M (IgM)− early B-cell progenitors: pre-pro-B cells, pro-B cells, pre-B cells. (Right) Immature B (Imm-B) cells and mature recirculating B (Recir-B) cells as indicated from mouse bone marrow (BM). (F) Differentiation of splenic B-cell subsets in of young CD19Cre/+ p16L/L mice versus CD19Cre/+ p16+/+ mice. Different B-cell subsets are as shown: mature (IgMloIgDhi), immature (IgMhiIgDlo), transitional 1 (T1, CD21−IgM+), transitional 2 (T2, IgMhiIgDhi), and marginal zone (MZ, CD23−CD21+B220+).
In accord with previous work in humans,17 the expression of p16INK4a in splenic B cells was lower than in contemporaneously harvested T cells from 45-week-old mice (Figure 1B, supplemental Figure 1). Analysis of p16INK4a expression in B cells from CD19Cre/+ p16L/L mice older than 45 weeks was limited by the onset of neoplasia (described in “B cell–specific p16INK4a inactivation accelerates lymphoid tumorigenesis”). Expression of p16INK4a in peripheral B cells from CD19Cre/+ p16L/L mice was reduced compared with B cells from age-matched CD19Cre/+ p16+/+ animals, but low-level expression limited a reliable quantification of the decrease in protein (Figure 1B). These results demonstrate lineage-specific, somatic deletion in p16L/L mice with use of the Lck-Cre and CD19Cre/+ alleles.
Normal lymphocyte development in young mice after deletion of p16INK4a in lymphoid lineage
We next determined whether lineage-specific ablation of p16INK4a has an effect on lymphocyte development and differentiation in young mice. In the present study, T cell–specific deletion of p16INK4a did not affect thymocyte development in young mice (6-8 weeks old, Figure 1C), with comparable numbers of DN1-DN4 thymocytes (right) and single positive thymocyte numbers (left) between Lck-Cre p16+/+ and Lck-Cre p16L/L mice. The peripheral single-positive T cells (Figure 1D left), as well as naive and memory T cells (Figure 1D middle and right) were not significantly altered in young mice. Similarly, B-cell development and differentiation was not affected by p16INK4a deletion in the B lineage, as demonstrated by normal pre-pro-B, pro-B, pre-B, immature B, and mature recirculating B cells in bone marrow (Figure 1E). Likewise, no effect of B-lineage p16INK4a deletion was observed in young mice on peripheral B-cell subsets, including mature B, transitional T1, T2, and marginal zone B cells in spleen (Figure 1F). Consistent with the lack of p16INK4a expression in young animals, these data indicate that p16INK4a does not influence T- and B-lymphocyte development in young adult mice.
Role of p16INK4a in thymic involution and thymocyte development
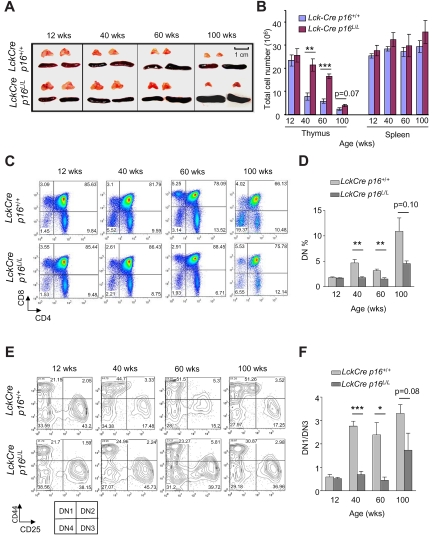
Thymic involution is a familiar age-associated phenotype in humans and mice, and decreasing thymic T-cell output is thought to be a major cause of impaired immunity in human aging. To explore the role of T lineage–specific p16INK4a expression in thymic involution, we examined thymic size in Lck-Cre p16L/L and Lck-Cre p16+/+ mice. Lck-Cre mice express Cre recombinase throughout life in T-cell progenitors. Although no increase in thymic involution has been reported in this strain, this enzyme has produced low levels of DNA damage in other tissues, and therefore its expression may influence T-cell aging. A significant slowing in the rate thymic involution was observed with T-lineage p16INK4a excision, as evidenced by a partial rescue of the decline in thymic size and thymocyte number with aging, particularly in 40- and 60-week-old animals (Figure 2A-B). In contrast, no significant effect on spleen size or splenocyte number was observed in Lck-Cre p16L/L mice.
Figure 2.
T lineage–specific deletion of p16INK4a slows thymic involution and reduces an age-related DN1 to DN3 block. (A) Partial rescue of thymic involution with aging in Lck-Cre p16L/L mice compared with Lck-Cre p16+/+ mice (n = 5 per group). Representative images are shown of thymus and spleen from mice of indicated ages and genotypes. (B) T cell–specific deletion of p16INK4a is associated with a partial but significant rescue of the age-associated decline of total thymocyte number (n = 5 per group). Total numbers of thymocytes and splenocytes from mice of indicated ages are shown. (C) The age-related arrest at the DN thymocyte stage was abolished by T lineage–specific deletion of p16INK4a. (D) Quantification of Lin−CD4−CD8− (DN) thymocyte fractions at indicated ages as shown in panel C; n = 5 mice per group. (E) T lineage–specific deletion of p16INK4a attenuated the age-related DN1-DN3 block. Representative flow analysis of Lin−CD4−CD8− DN1 (CD44+CD25−) and DN3 (CD44−CD25+) fractions in mice of indicated ages and genotypes is shown. (F) The ratio of the absolute number of DN1 to DN3 cells (DN1/DN3) as determined in panel E is shown at indicated ages. See also supplemental Figure 2; n = 5 mice per group. Error bars indicate SEM; *P < .05, **P < .01, ***P < .001.
In accord with previous studies,35 we noted that lineage-negative DN thymocytes accumulate with age (Figure 2C-D) suggesting a partial DN block induced by thymic aging. This age-related block of thymopoiesis was significantly rescued by p16INK4a deletion (Figure 2C-D). Lineage-negative DN cells can in turn be divided into DN1, DN2, DN3, and DN4 cells on the basis of CD44 and CD25 expression, and we further analyzed the role of p16INK4a on DN subfractions during aging. Consistent with work from others,35–37 we observed a significant developmental block from the DN1 to DN3 stage with aging in Lck-Cre p16+/+ mice. This block was significantly reduced in Lck-Cre p16L/L mice (Figure 2E-F; supplemental Figure 2), suggesting this block in part reflects an increased expression of p16INK4a in DN1 cells with aging. In aggregate, these data suggest thymic involution and decreased thymocyte output with aging result from a lymphocyte-specific expression of p16INK4a in DN T-cell progenitors, at least in the setting of persistent Cre expression.
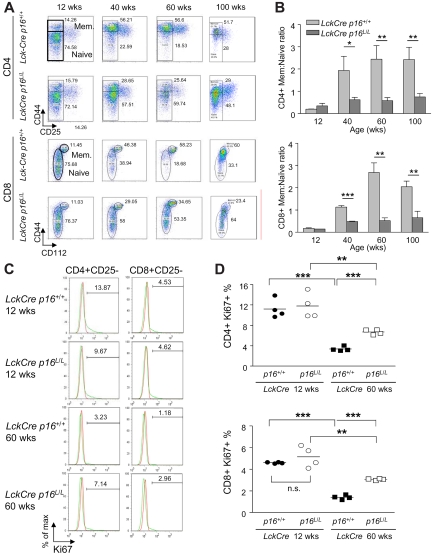
Attenuation of immune aging phenotypes after deletion of p16INK4a in the T lineage
We next determined whether p16INK4a plays an intrinsic role in immune aging phenotypes in addition to thymic involution and thymocyte output. In accord with previous work in both mice and humans,38–40 we found that aging is associated with a decreased production of naive T cells in both the CD4+ and CD8+ compartments (CD4+CD25−CD44− or CD8+CD112−CD44−) and increased memory T cells (CD4+CD25−CD44+ or CD8+CD112−CD44+; Figure 3A), resulting in an increased ratio of memory/naive T cells with aging (Figure 3B). T lineage–specific deletion of p16INK4a, however, was associated with a rescue of naive T-cell production up to 100 weeks of age in both the CD4+ and CD8+ compartments (Figure 3A), with a corresponding maintenance of a “youthful” memory/naive ratio in aged mice (Figure 3A-B).
Figure 3.
Attenuation of peripheral T-cell immune aging phenotypes by T cell–specific deletion of p16INK4a. (A) Representative flow analyses of CD4+ (top) and CD8+ (bottom) splenocytes showing the effect of T cell–specific p16INK4a deletion on age-associated changes in memory and naive T-cell fractions. Memory and naive T cells are identified as described in Figure 1C. (B) Quantification of data from panel A showing the effect of T cell–specific p16INK4a deletion on the ratio of memory versus naive T cells with aging in the CD4+ and CD8+ compartments; n = 5 mice per group. (C) Representative flow analyses showing the effect of aging and T cell–specific p16INK4a deletion on homeostatic proliferation as measured by Ki67 expression in nonactivated (CD25−) CD4+ or CD8+ T cells from mice of indicated ages and genotypes. (D) Quantification of Ki67 expression as shown in panel C, n = 4 mice per group. Error bars indicate SEM; *P < .05, **P < .01, ***P < .001.
Aging is also associated with an increased activation response when cultured ex vivo as measured by induction of CD25 expression after T-cell receptor (TCR) signaling with phorbol 12-myristate 13-acetate plus ionomycin (supplemental Figure 3A-B). This finding likely reflects an increased memory/naive ratio in older mice because memory cells are able to undergo more rapid and sustained activation than naive T cells in response to TCR signaling.41 In accord with the finding of a reduced memory/naive ratio in older Lck-Cre p16L/L animals, we also observed a rescue of this age-associated hyperactivation phenotype by T cell–specific p16INK4a deletion (supplemental Figure 3A-B). Surprisingly, therefore, expression of an antiproliferative protein, p16INK4a, appears to cause an age-associated expansion of more activatable memory T cells late in life, presumably by depressing naive T-cell production. In aggregate, these results demonstrate that, with aging, p16INK4a expression compromises naive T-cell production and thereby indirectly promotes increased memory T-cell expansion.
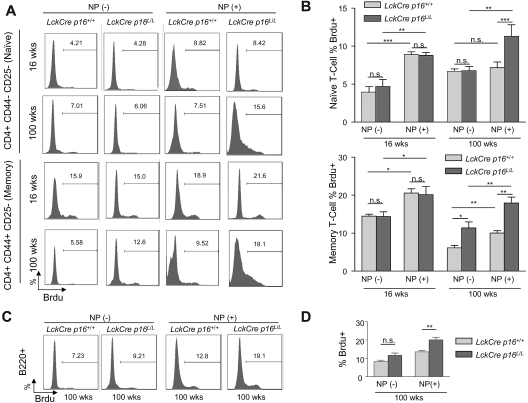
Given that expression of p16INK4a limits cell cycle progression by inhibiting the proliferative cyclin-dependent kinases, we next determined the effect of p16INK4a deletion on homeostatic T-cell proliferation in young and old mice. Expression of a marker of proliferation (Ki67) in nonactivated (CD25−) splenic T cells sharply decreased in both the CD4+ and CD8+ compartments with aging, and this defect was partially rescued after T-cell specific p16INK4a deletion (Figure 3C-D). We further examined homeostatic proliferation of memory and naive T-cell fractions with aging by measuring BrdU incorporation in vivo. Interestingly, although the incorporation of BrdU in nonactivated memory T cells (CD4+CD44+CD25−) decreased with aging, nonactivated naive T cells (CD4+CD44−CD25−) exhibited a modest increase in the incorporation of BrdU with aging (Figure 4A left and Figure 4B). This observation is congruent with previous studies42–44 and suggests that naive T cells in old mice may exhibit increased homeostatic proliferation as compensation for the marked decrease in thymocyte output with aging. T-lineage deletion of p16INK4a almost completely rescued the age-related defect of homeostatic proliferation in memory T cells but did not significantly affect the increased BrdU incorporation of naive T cells in 100-week-old mice (Figure 4A left and 4B). These data suggest that p16INK4a contributes to immune aging by decreasing homeostatic proliferation of memory T cells.
Figure 4.
The effects of p16INK4a deletion on age-related changes in homeostatic or antigen-specific proliferation of memory and naive T cells. (A) Representative flow analyses of in vivo proliferation as measured by BrdU incorporation in memory or naive T cells without NP immunization (homeostatic, −) or with NP immunization (antigen-specific, +) in mice of indicated ages and genotypes. For NP(+) results, BrdU incorporation in memory or naive T cells is determined after reimmunization with nitrophenylacetyl-chicken γ-globulin as described in “Immune function is enhanced in old mice by T-lineage deletion of p16INK4a.” (B) Quantification of BrdU incorporation in memory (bottom) or naive (top) as shown in panel A with and without NP immunization; n = 4-5 mice per group. T cell–specific inactivation of p16INK4a is associated with rescue of an age-associated decline in antigen-specific proliferation in memory and naive T cells and homeostatic proliferation in memory cells. Memory and naive T cells are identified as described in Figure 1C. (C) Representative flow analyses in 100-week-old mice of indicated genotypes showing the effect of T lineage–specific p16INK4a deletion on T-cell helper function as measured by antigen-induced B-cell proliferation (BrdU incorporation). (D) Quantification of antigen-induced B-cell proliferation as measured in panel C; n = 4 mice per group. Error bars indicate SEM; *P < .05, **P < .01, ***P < .001.
Immune function is enhanced in old mice by T-lineage deletion of p16INK4a
The antigen-specific T- and B-cell immune response is compromised in aged mice, with an attendant decrease in malignancy immunosurveillance and pathogen clearance. To investigate whether p16INK4a plays a role in these phenotypes of immune aging, nitrophenylacetyl-chicken γ-globulin, a hapten-protein conjugate, was used for immunization and examination of the antigen-specific immune response. NP immunization induces a T-dependent humoral response that involves both CD4+ T-helper function and B-cell antibody production. Mice were immunized at day 1 and reimmunized at day 28 before primary and memory antigen-specific responses were examined at day 33 by in vivo antigen-specific T- and B-cell proliferation (BrdU incorporation).
In young mice, NP immunization induced significant naive and memory T-cell proliferation, and this proliferative response was not significantly influenced by T-specific p16INK4a deletion (Figure 4A-B). NP immunization in 100-week-old animals, however, induced a very modest proliferative response in memory cells, with no significant response observed in naive T cells. This decreased proliferative response to NP was almost completely rescued in both the memory and naive T-cell fractions from old mice in Lck-Cre p16L/L mice (Figure 4A-B). In addition, in vivo B-cell proliferation after NP reimmunization, which depends on the T-cell helper function, was also significantly increased in aged Lck-Cre p16L/L mice (Figure 4C-D). These data provide evidence that p16INK4a expression plays an important role in the age-related functional decline of the antigen-specific T-cell immune response.
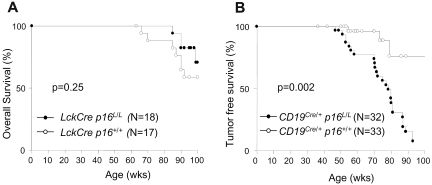
T lineage–specific deletion of p16INK4a is not associated with increased neoplasia
As opposed to studies with B cell–specific inactivation (see “B cell–specific p16INK4a inactivation accelerates lymphoid tumorigenesis”), inactivation of p16INK4a in T cells was not associated with an increase in overall or tumor-associated mortality up to 100 weeks of age (Figure 5A-B). Although a nonsignificant survival advantage was noted in the Lck-Cre p16L/L mice compared with control animals (Lck-Cre p16+/+ and CD19Cre/+ p16+/+), the number of deaths in the T lineage–deleted cohort was too small to assess the effect of the enhanced immune function of Lck-Cre p16L/L mice on nonlymphoid turmorigenesis, autoimmunity, and overall longevity. Also in contrast to B-lineage p16INK4a deletion, inactivation of p16INK4a in T cells from mice up to 100 weeks in age was not associated with the presence of abnormal lymphocyte fractions (see, for example, CD4, CD8, CD25, CD44, and CD112 distributions in Figures 2C and 3A). Likewise, T cell–specific p16INK4a inactivation did not lead to increased T-cell proliferation beyond that observed in young animals (Figures 3C-D and 4A-B). In aggregate, these data indicate a lack of neoplastic proliferation in Lck-Cre p16L/L mice, suggesting that p16INK4a does not contribute to T-lineage tumor suppression in experimentally housed animals up to 100 weeks of age.
Figure 5.
Kaplan-Meier survival curves with p16INK4a deletion in B or T lymphocytes. (A) Overall survival of Lck-Cre p16+/+ and Lck-Cre p16L/L mice. (B) Tumor-free survival of CD19Cre/+ p16+/+ and CD19Cre/+ p16L/L mice.
B lineage–specific deletion of p16INK4a does not affect immune aging in “middle-aged” mice
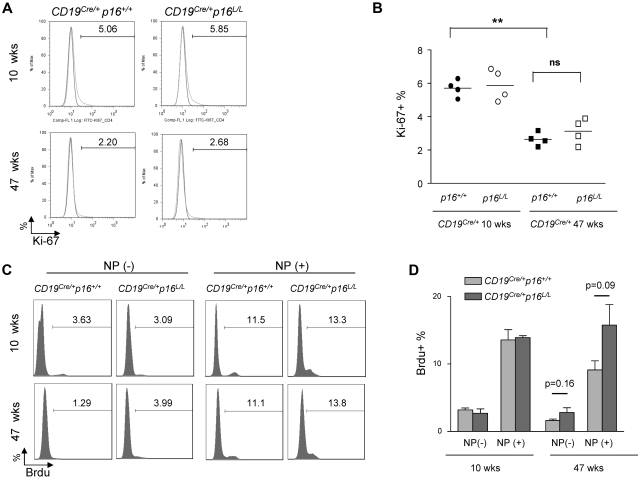
We examined B-cell aging phenotypes in CD19Cre/+ p16+/+ versus CD19Cre/+ p16L/L mice. A variety of assays of B-cell function were tested, including homeostatic proliferation (Figure 6A-D), ex vivo B-cell receptor–dependent B-cell activation (supplemental Figure 4A), and antigen-specific B-cell responses such as proliferative response to reimmunization (Figure 6C-D) and germinal center formation (supplemental Figure 4B-C). In contrast to results seen in T cells of Lck-Cre p16L/L mice, deletion of p16INK4a in B-lineage cells did not significantly rescue any age-related B-cell phenotype in mice analyzed up to 47 weeks (Figure 6A-D, supplemental Figure 4). These data suggest that p16INK4a expression is not a principal cause of B-cell dysfunction in animals up to 47 weeks of age. It is possible that we would have observed an effect of p16INK4a deletion on B-cell aging in older mice, but increased B-cell neoplasia in the CD19Cre/+ p16L/L animals starting at a year of age precluded such an analysis.
Figure 6.
The effect of B lineage–specific p16INK4 deletion on immune aging. (A) Representative flow analysis of B-cell homeostatic proliferation as measured by Ki-67 expression in resting B cells (CD40−) from spleens of mice with indicated ages and genotypes. (B) Quantification of K-i67 expression as shown in panel A. ns indicates not significant. **P < .01. (C) Representative flow analysis showing the effect of B-lineage p16INK4a deletion on antigen-specific B-cell proliferation. Mice were immunized and rechallenged with nitrophenylacetyl (see “NP immunization”) and BrdU incorporation in splenic B cells was measured in mice of indicated ages and genotypes. (D) Quantification of BrdU incorporation as shown in panel C; n = 4 mice per group, P values are as indicated.
B cell–specific p16INK4a inactivation accelerates lymphoid tumorigenesis
In contrast to results in the Lck-Cre cohort, excision of p16INK4a in B-cell progenitors led to a pronounced decrease in overall and tumor-free survival (Figure 5B). Small round cell tumors were first observed in mice younger than a year of age, and nearly 100% of CD19Cre/+ p16L/L mice succumbed to neoplastic disease by 90 weeks of age. A high percentage of CD19Cre/+ p16L/L animals manifested symptoms of central nervous system (CNS) involvement. Histopathologic examination of mice with CNS symptoms showed tumors of the leptomeningeal regions of the spine and/or brain with compression of subjacent CNS parenchyma (supplemental Figure 5A). Despite the meningeal location, immunohistochemistry demonstrated tumor cells to be positive for CD45 leukocyte-common antigen and negative for the neuromeningeal marker S100 (supplemental Figure 5A), establishing a hematopoietic origin. All analyzed tumors focally expressed one or more of the B-lymphocyte markers CD93, CD45R/B220, and/or immunoglobulin (supplemental Figure 5A), consistent with a B-cell origin. Immunohistochemical staining for B-lymphocyte markers typically was most intense in peripheral regions of the tumor, suggesting variable differentiation. These data indicate that somatic p16INK4a deletion causes the highly penetrated development of B-lineage cancers.
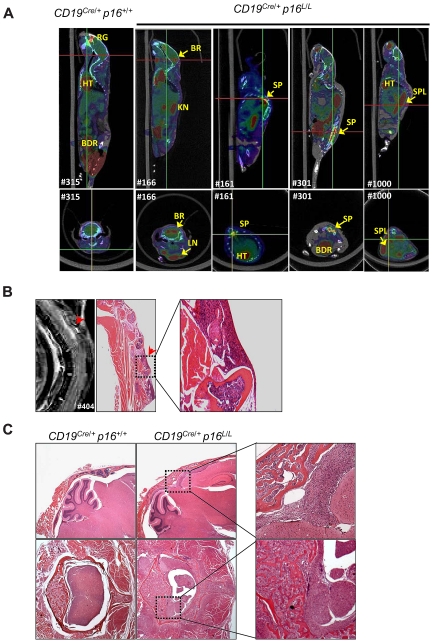
Neoplastic disease in CD19Cre/+ p16L/L mice could be detected before the onset of overt symptoms in some cases by flow cytometric analysis of the peripheral blood (eg, supplemental Figure 5B) or imaging modalities such as positron emission tomography/computed tomography (PET/CT; Figure 7A, supplemental Figure 6) and magnetic resonance imaging (MRI; Figure 7B). Aberrant B-lineage populations were noted in all CD19Cre/+ p16L/L mice (n = 5) analyzed at 52 weeks of age or older but not in littermate CD19Cre/+ p16+/+ animals (supplemental Figure 5B). Radiologic imaging demonstrated tumors involving the spinal cord, cerebellum, lymph nodes, and spleen. Serial PET/CT of a tumor-bearing mouse demonstrated rapid tumor growth during a 4-week period (supplemental Figure 6). Although a high frequency of tumors produced CNS symptoms such as seizures and hind limb paralysis, this appeared to represent direct extension of tumor from vertebral or cranial bone marrow rather than leptomeningeal tropism. In several cases, we were able to note tumor in the bone marrow of skull or spine with contiguous extension multifocally into nearby meninges, brain parenchyma, and spinal cord (Figure 7B-C). In aggregate, the morphologic appearance, immunophenotype, and high-grade systemic nature of these tumors are most reminiscent of systemic human B-cell ALL, a disease with a high frequency of p16INK4a inactivation.24
Figure 7.
B-lineage p16INK4a deletion is associated with high-grade B-cell neoplasms. (A) Representative fused PET/CT images of aged CD19Cre/+ p16+/+ and CD19Cre/+ p16L/L mice identifying tumors with high 18F-FDG uptake in brain (BR), spine (SP), spleen (SPL), and lymph nodes (LN). Background uptakes were observed in heart (HT), bladder (BDR), kidney (KN), and retro-orbital gland (RG). (Top) Sagittal images with red lines indicating level of the associated transverse images (bottom). (B) A T2-weighted MRI of a representative CD19Cre/+ p16L/L mouse indicates a lesion in the spinal cord, which was confirmed by histopathologic analysis to represent lymphoma invading spinal cord from bone marrow. Original magnification ×20. (C) Hematoxylin and eosin–stained images of CNS lymphomas in the brain (top) or spinal cord (bottom) of aged CD19Cre/+ p16L/L mice. Comparison images from littermate CD19Cre/+ p16+/+ mice are shown (left). Original magnification ×40.
Discussion
The present work demonstrates that the p16INK4a cell-cycle inhibitor does not contribute to lymphoid development in young animals, but in old mice, plays a pronounced role in B-cell tumor suppression and T-cell immune aging. These results provide direct experimental support for antagonistic pleiotropy in lymphoid tissues. Although the authors of previous experiments that relied predominantly on germline-deficient mice have suggested roles for tumor suppressor mechanisms in cancer resistance and age promotion in distinct tissues, this cancer-aging trade-off has not been observed within a specific tissue in a cell-autonomous manner. Here, we show that the expression of a single protein is both highly beneficial in mice younger than a year of age (preventing cancer in B-cell progenitors) and detrimental in older mice (promoting immune aging in T cells).
p16INK4a and T-cell aging
We believe the most parsimonious explanation for our data are that p16INK4a expression with aging causes a proliferative compromise in 2 or more distinct T-cell compartments. We observed a pronounced block in thymopoiesis at the DN1-3 stages in aged mice that was largely abrogated by deletion of p16INK4a (Figure 2), suggesting an important role for p16INK4a in regulating cell-cycle traversal of early DN cells during thymocyte development. It is likely that this defect in thymopoiesis in turn contributes to the marked age-associated decline in naive T-cell number, which is almost completely rescued by p16INK4a deletion, even in 2-year-old animals (Figure 3A-B). Of importance, in the oldest (100-week) mice examined, inactivation of p16INK4a only produced a borderline reduction of thymic involution (Figure 2B) and DN1-3 blockade (Figure 2E-F). This observation suggests that DN1-3 blockade may not be the only T-precursor factor that contributes to thymic involution. Other factors such as myeloid-skewing of HSCs and decreased common lymphoid progenitor input to the thymus might exhibit more pronounced effects, particularly in mice at these later ages. Because the Lck-Cre allele does not excise p16INK4a in HSC or common lymphoid progenitor, it is not clear from our experimental approach whether such other influences on thymic involution with aging depend on p16INK4a expression.
We also noted the rescue of age-associated phenotypes in peripheral T cells, notably a rescue of the proliferative decline in antigen-induced proliferation in naive T cells, and of homeostatic and antigen-induced proliferation of memory cells (Figure 4A-B). This T lineage–specific inactivation of p16INK4a was in turn associated with maintenance of a more “youthful” phenotype in functional assays of old mice, including antigen-induced proliferation (Figure 4A-B), T-helper function (Figure 4C-D), and ex vivo activation in response to TCR engagement (supplemental Figure 3). Therefore, our data suggest that physiologic p16INK4a expression is a principal contributor to the observed functional decline of immunity with aging.
It is not clear from our data whether p16INK4a promotes aging in these different T-cell fractions by causing a permanent growth arrest (eg, “cellular senescence”) independently in multiple T-lineage subpopulations, or by contributing to a general “hypoproliferative” state of T-lineage cells, that can be transmitted from progenitors to progeny during T-cell development. We believe this is an important unresolved issue because it has implications for the reversibility of p16INK4a-induced aging in lymphoid tissues.
Although the present study has exclusively focused on murine T-cell aging, we believe existing data also support the possibility that expression of p16INK4a contributes to T-cell aging in humans. The age-dependent exponential increase in p16INK4a expression in circulating T cells in humans is as great or greater than the increase with aging observed in rodents,17,23 and human T cells appear to rely on the same proliferative cyclin-dependent kinases (CDK4 and CDK6) for cellular replication that are inhibited by p16INK4a in both species. In light of these previous findings, the present murine data suggest p16INK4a may play a significant role in human T-cell aging, although specific functional studies in human T cells are needed to test this hypothesis.
p16INK4a and intrinsic versus extrinsic aging
Although p16INK4a expression is typically considered the result of heritable, intrinsic cellular events, we believe our data are also not inconsistent with extrinsic contributions to aging. A variety of extracellular events can rapidly modulate p16INK4a expression in vitro and in vivo including exposure to DNA damaging agents, oxidative stress, suboptimal culture conditions, activators of WNT signaling and various mitogens (reviewed in Sharpless and DePinho4). Moreover, activation of p16INK4a has been reported to mediate “niche” aging signals in muscle stem cells from old mice.45 In the case of lymphoid aging, ample data suggest that an age-related defect in the thymic microenvironment plays a critical role in thymic output with aging.46,47 Therefore, the marked rescue of this phenotype by T-lineage specific p16INK4a excision was perhaps unexpected (Figure 2A-B), although these observations are consistent with a previous report.48 It is worth noting, however, that although the present data establish that expression of p16INK4a within T-cell progenitors contributes to thymic involution with aging, our results do not indicate what feature of aging induces p16INK4a expression in this context. Expression of p16INK4a in early thymic progenitors could reflect a response to an age-associated stress that is cell intrinsic (such as DNA damage) or cell extrinsic (such as degradation of the thymic milieu). We believe an interesting future question in this regard is whether an old thymus can promote aging in adoptively transferred p16INK4a-deficient T-cell progenitors derived from young mice.
p16INK4a and lymphoid cancer
Given the high prevalence of p16INK4a loss in human T-cell progenitor neoplasms such as T-cell ALL, the lack of increased neoplasia in mice without p16INK4a in the T-cell lineage is unexpected. Inactivation of p16INK4a in T-ALL almost always occurs in the setting of somatic deletion, and such lesions invariably target p15INK4b and ARF as well. Therefore, it is possible that p16INK4a plays little if any role in progenitor T-cell tumor suppression. However, the lack of T-cell malignancies in Lck-Cre p16L/L mice may alternatively reflect experimental conditions. For example, chronic immune challenge and viral infection have been associated with lymphoid neoplasia, and loss of p16INK4a expression might be oncogenic in the setting of such T-cell immune challenges not routinely encountered under laboratory conditions. Likewise, although no excess mortality was observed in Lck-Cre p16L/L mice, an increased number and proliferative ability of aged Lck-Cre p16L/L T-cells could predispose to autoimmune sequelae, as observed in p18INK4c null animals.49 Given the lack of spontaneous tumors observed in the Lck-Cre p16L/L mice, it would seem an excellent model to test the effects of enhanced T-cell function on tumorigenesis, autoimmunity and longevity, which are ongoing studies.
Although T cells without p16INK4a do not appear to be prone to tumorigenesis in this model, the opposite is true of B cells lacking p16INK4a. These results are consistent with data from Signer et al demonstrating a prominent and age-associated role of p16INK4a in B-cell progenitor neoplasms.29 It is not clear from our studies whether p16INK4a expression plays a role in B-cell aging. This lack of effect of p16INK4a inactivation on B-cell aging in 47-week-old, “middle-aged” mice could be because of either lower expression of p16INK4a in peripheral B cells compared with T cells (Figure 1B and Liu et al17) or because 47 weeks is not sufficient aging to observe functional B-cell deficits in this mouse model (Figure 6; supplemental Figure 4).
Cancer versus aging
When considered in the context of previous studies, we believe the present work demonstrates that although the p16INK4a tumor suppressor mechanism does exhibit antagonistic pleiotropy at the whole-organism level, this need not be the case at the tissue level. Although the role of p16INK4a on B-cell aging is not clear, recent work suggests that melanocytes represent a clear counter example to the situation observed in T cells. Inactivation of p16INK4a in murine melanocytes is strongly tumor promoting30 but appears to play little if any role in the promotion of melanocyte aging (Inomata et al50 and data not shown). Therefore, we believe these studies, in aggregate, support the existence of cellular compartments in which expression of p16INK4a predominantly modulates: (1) aging but not cancer (eg, T cells), (2) cancer but not aging (eg, melanocytes and possibly B lymphocytes), or (3) both cancer and aging (eg, HSC, neural stem/progenitor cells, pancreatic β cells and possibly B lymphocytes).
In summary, this work directly demonstrates that the p16INK4a tumor suppressor beneficially prevents B-cell neoplasms while untowardly promoting T-cell aging. We believe this work serves as a cautionary tale for those who would seek to ameliorate aging by globally attenuating tumor suppressor function. Tantalizingly, however, this work also provides a first example of a tissue subpopulation, T cells, in which p16INK4a expression appears to play a strong role in the promotion of aging while providing a very limited role in tumor suppression. Likewise, given the known variability in human p16INK4a regulation with respect to aging, these results suggest a genetic basis for heterogeneity of human immune aging phenotypes and lymphoid cancer susceptibility.
Supplementary Material
Acknowledgments
We thank Ken Dorshkind, Alex Seibold, and Christin Burd for their critical reading of the manuscript; Yi Zhang for reagents; and scientists in the University of North Carolina Lineberger Comprehensive Cancer Center Mouse Phase 1 Unit for assistance with animal handling. We thank the UNC-CH Biomedical Research Imaging Center Small Animal Imaging facility for assistance with PET and MRI, the UNC-CH gastrointestinal histology core facility for assistance with immunohistochemistry, and the UNC-CH flow cytometry core facility.
This work was supported by grants from the Burroughs Wellcome Foundation, the American Federation of Aging Research, and the National Institutes of Health (grants CA016086, CA009156, AG024379, F30AG034806, and T32GM008719).
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.L, S.M.J., J.K., and N.E.S. designed and performed research and analyzed data; H.Y., A.B.R., and Y.F. performed additional data analysis; and all authors drafted the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, CB #7295, Departments of Medicine and Genetics, The University of North Carolina School of Medicine, Chapel Hill, NC 27599-7295; e-mail: nes@med.unc.edu.
References
- 1.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 2.Campisi J. Cancer and ageing: rival demons? Nat Rev Cancer. 2003;3(5):339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- 3.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 5.Janzen V, Forkert R, Fleming HE, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy J, Ramsey MR, Ligon KL, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Gu X, Su IH, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molofsky AV, Slutsky SG, Joseph NM, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker DJ, Perez-Terzic C, Jin F, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10(7):825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emanuele E, Fontana JM, Minoretti P, Geroldi D. Preliminary evidence of a genetic association between chromosome 9p21. 3 and human longevity. Rejuvenation Res. 2010;13(1):23–26. doi: 10.1089/rej.2009.0970. [DOI] [PubMed] [Google Scholar]
- 11.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherborne AL, Hosking FJ, Prasad RB, et al. Variation in CDKN2A at 9p21. 3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Sanoff HK, Cho H, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6(4):e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Sanoff HK, Cho H, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagresle C, Gardie B, Eyquem S, et al. Transgenic expression of the p16(INK4a) cyclin-dependent kinase inhibitor leads to enhanced apoptosis and differentiation arrest of CD4−CD8− immature thymocytes. J Immunol. 2002;168(5):2325–2331. doi: 10.4049/jimmunol.168.5.2325. [DOI] [PubMed] [Google Scholar]
- 19.Erickson S, Sangfelt O, Heyman M, Castro J, Einhorn S, Grander D. Involvement of the Ink4 proteins p16 and p15 in T-lymphocyte senescence. Oncogene. 1998;17(5):595–602. doi: 10.1038/sj.onc.1201965. [DOI] [PubMed] [Google Scholar]
- 20.Migliaccio M, Raj K, Menzel O, Rufer N. Mechanisms that limit the in vitro proliferative potential of human CD8+ T lymphocytes. J Immunol. 2005;174(6):3335–3343. doi: 10.4049/jimmunol.174.6.3335. [DOI] [PubMed] [Google Scholar]
- 21.Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413(6851):86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 22.Bianchi T, Rufer N, MacDonald HR, Migliaccio M. The tumor suppressor p16(Ink4a) regulates T lymphocyte survival. Oncogene. 2006;25(29):4110–4115. doi: 10.1038/sj.onc.1209437. [DOI] [PubMed] [Google Scholar]
- 23.Lemster BH, Michel JJ, Montag DT, et al. Induction of CD56 and TCR-independent activation of T cells with aging. J Immunol. 2008;180(3):1979–1990. doi: 10.4049/jimmunol.180.3.1979. [DOI] [PubMed] [Google Scholar]
- 24.Sulong S, Moorman AV, Irving JA, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113(1):100–107. doi: 10.1182/blood-2008-07-166801. [DOI] [PubMed] [Google Scholar]
- 25.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl–induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103(17):6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno T, Yamada Y, Tawara M, et al. Inactivation of p14ARF as a key event for the progression of adult T cell leukemia/lymphoma. Leukemia Res. 2007;31(12):1625–1632. doi: 10.1016/j.leukres.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Bies J, Sramko M, Fares J, et al. Myeloid specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood. 2010;116(6):979–987. doi: 10.1182/blood-2009-08-238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff L, Garin MT, Koller R, et al. Hypermethylation of the Ink4b locus in murine myeloid leukemia and increased susceptibility to leukemia in p15(Ink4b)-deficient mice. Oncogene. 2003;22(58):9265–9274. doi: 10.1038/sj.onc.1207092. [DOI] [PubMed] [Google Scholar]
- 29.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22(22):3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monahan KB, Rozenberg GI, Krishnamurthy J, et al. Somatic p16INK4a loss accelerates melanomagenesis. Oncogene. 2010;29(43):5809–5817. doi: 10.1038/onc.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahama Y, Ohishi K, Tokoro Y, et al. Functional competence of T cells in the absence of glycosylphosphatidylinositol-anchored proteins caused by T cell-specific disruption of the Pig-a gene. Eur J Immunol. 1998;28(7):2159–2166. doi: 10.1002/(SICI)1521-4141(199807)28:07<2159::AID-IMMU2159>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsey MR, Krishnamurthy J, Pei XH, et al. Expression of p16Ink4a compensates for p18Ink4c loss in cyclin-dependent kinase 4/6-dependent tumors and tissues. Cancer Res. 2007;67(10):4732–4741. doi: 10.1158/0008-5472.CAN-06-3437. [DOI] [PubMed] [Google Scholar]
- 34.Krimpenfort P, Ijpenberg A, Song JY, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448(7156):943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 35.Thoman ML. The pattern of T lymphocyte differentiation is altered during thymic involution. Mech Ageing Dev. 1995;82(2–3):155–170. doi: 10.1016/0047-6374(95)01597-s. [DOI] [PubMed] [Google Scholar]
- 36.Aspinall R. Age-associated thymic atrophy in the mouse is due to a deficiency affecting rearrangement of the TCR during intrathymic T cell development. J Immunol. 1997;158(7):3037–3045. [PubMed] [Google Scholar]
- 37.Thoman ML. Early steps in T cell development are affected by aging. Cell Immunol. 1997;178(2):117–123. doi: 10.1006/cimm.1997.1133. [DOI] [PubMed] [Google Scholar]
- 38.Pirruccello SJ, Collins M, Wilson JE, Mcmanus BM. Age-related changes in naive and memory Cd4+ T-cells in healthy-human children. Clin Immunol Immunopathol. 1989;52(2):341–345. doi: 10.1016/0090-1229(89)90185-2. [DOI] [PubMed] [Google Scholar]
- 39.De Paoli P, Battistin S, Santini GF. Age-related changes in human-lymphocyte subsets—progressive reduction of the Cd4 Cd45r (suppressor inducer) population. Clin Immunol Immunopathol. 1988;48(3):290–296. doi: 10.1016/0090-1229(88)90022-0. [DOI] [PubMed] [Google Scholar]
- 40.Ernst DN, Hobbs MV, Torbett BE, et al. Differences in the expression profiles of Cd45rb, Pgp-1, and 3g11 membrane-antigens and in the patterns of lymphokine secretion by splenic-Cd4+ T-cells from young and aged mice. J Immunol. 1990;145(5):1295–1302. [PubMed] [Google Scholar]
- 41.Cho BK, Wang CY, Sugawa S, Eisen HN, Chen JZ. Functional differences between memory and naive CD8 T cells. Proc Natl Acad Sci U S A. 1999;96(6):2976–2981. doi: 10.1073/pnas.96.6.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cicin-Sain L, Messaoudi I, Park B, et al. Dramatic increase in naive T cell turnover is linked to loss of naive T cells from old primates. Proc Natl Acad Sci U S A. 2007;104(50):19960–19965. doi: 10.1073/pnas.0705905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed M, Lanzer KG, Yager EJ, Adams PS, Johnson LL, Blackman MA. Clonal expansions and loss of receptor diversity in the naive CD8 T cell repertoire of aged mice. J Immunol. 2009;182(2):784–792. doi: 10.4049/jimmunol.182.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naylor K, Li GJ, Vallejo AN, et al. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174(11):7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 45.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454(7203):528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aspinall R, Andrew D. Thymic atrophy in the mouse is a soluble problem of the thymic environment. Vaccine. 2000;18(16):1629–1637. doi: 10.1016/s0264-410x(99)00498-3. [DOI] [PubMed] [Google Scholar]
- 47.Mackall CL, Punt JA, Morgan P, Farr AG, Gress RE. Thymic function in young/old chimeras: substantial thymic T cell regenerative capacity despite irreversible age-associated thymic involution. Eur J Immunol. 1998;28(6):1886–1893. doi: 10.1002/(SICI)1521-4141(199806)28:06<1886::AID-IMMU1886>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 48.Min H, Montecino-Rodriguez E, Dorshkind K. Reduction in the developmental potential of intrathymic T cell progenitors with age. J Immunol. 2004;173(1):245–250. doi: 10.4049/jimmunol.173.1.245. [DOI] [PubMed] [Google Scholar]
- 49.Bai F, Pei XH, Pandolfi PP, Xiong Y. p18(Ink4c) and Pten constrain a positive regulatory loop between cell growth and cell cycle control. Mol Cell Biol. 2006;26(12):4564–4576. doi: 10.1128/MCB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inomata K, Aoto T, Binh NT, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137(6):1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.