Abstract
Gene transfer using adeno-associated virus (AAV) vectors has great potential for treating human disease. Recently, questions have arisen about the safety of AAV vectors, specifically, whether integration of vector DNA in transduced cell genomes promotes tumor formation. This study addresses these questions with high-dose liver-directed AAV-mediated gene transfer in the adult mouse as a model (80 AAV-injected mice and 52 controls). After 18 months of follow-up, AAV-injected mice did not show a significantly higher rate of hepatocellular carcinoma compared with controls. Tumors in mice treated with AAV vectors did not have significantly different amounts of vector DNA compared with adjacent normal tissue. A novel high-throughput method for identifying AAV vector integration sites was developed and used to clone 1029 integrants. Integration patterns in tumor tissue and adjacent normal tissue were similar to each other, showing preferences for active genes, cytosine-phosphate-guanosine islands, and guanosine/cysteine-rich regions. Gene expression data showed that genes near integration sites did not show significant changes in expression patterns compared with genes more distal to integration sites. No integration events were identified as causing increased oncogene expression. Thus, we did not find evidence that AAV vectors cause insertional activation of oncogenes and subsequent tumor formation.
Introduction
Liver-directed gene transfer using adeno-associated virus (AAV) vectors has the potential to serve as therapy for several inherited hematologic diseases. One such disease is the bleeding disorder hemophilia B, caused by a deficiency in coagulation factor IX (FIX). Currently, there are 2 clinical trials for hemophilia B that use liver-directed AAV-mediated gene transfer of the F9 gene (www.clinicaltrials.gov; identifiers NCT00515710 and NCT00979238). One of these trials reported transient efficacious circulating FIX levels (∼ 10%) with the use of the vector AAV2-hFIX16.1
Although AAV vectors are predominantly nonintegrating, with most of the transgene expression from stable episomes,2 it has been shown through direct sequencing that integration can occur.3,4 When integration takes place, there is a preference for integrating in regions where DNA breaks occur. These can be regions of endonuclease cleavage,5 active transcription,6–8 cytosine-phosphate-guanosine (CpG) islands,7,8 and palindromes.9 All of these studies describing AAV vector genome integration identified vector integration sites through plasmid rescue of vectors containing bacterial origins of replication (ori).4 Amplification of these plasmids in bacterial culture allows for sequencing of the integration junction between vector and host genome. Because of the bacterial selection involved in this method, bias may occur against recovering integrants whose size or sequence negatively affect bacterial growth, resulting in incomplete mapping of the full spectrum of integrants.
Vector genome integration has been associated with adverse events; integrating γ-retroviral vectors were implicated in the clonal expansion of transduced cells in 3 clinical studies, 2 for X-linked severe combined immunodeficiency10,11 and the other for chronic granulomatous disease.12,13 Although AAV vectors integrate at a much lower frequency than retroviral vectors, low-level AAV vector integration in transduced cells may still be a concern. A compelling argument supporting low genotoxic risk of AAV vectors comes from long-term follow-up of liver-directed AAV-mediated gene transfer in canine and murine models. Of 77 dogs receiving AAV vector at doses up to 3.4 × 1012 vector genomes(vg)/kg and followed for ≤ 10 years, none developed liver tumors as assayed by ultrasound, computed tomographic (CT) scan, and magnetic resonance imaging (MRI)14,15 (K.A.H., V.R.A., and Timothy C. Nichols, unpublished data, October 15, 2010). Similarly, > 300 mice receiving AAV vectors with a therapeutic transgene at doses up to 4 × 1013 vg/kg and followed ≤ 14 months have not shown a difference in tumor incidence compared with control mice.16,17
However, a study by one group reported an increase in tumor incidence that was attributed to AAV vectors.18 These investigators reported that administration of an AAV serotype 2 (AAV2) vector encoding β-glucuronidase in neonatal mice resulted in a significant increase in incidence of hepatocellular carcinoma (HCC), a tumor commonly found in the mouse strain used, compared with control mice. This increase was only detected in mice surviving > 13 months of age. Subsequent work showed a fraction of these tumors contained integrated vector DNA within a 6-kb window on chromosome 12, near the Rian and Mirg genes that encode regulatory RNAs of unknown function.19 This locus was termed the “AAV-HCC locus.” The investigators also showed increased expression of the genes near the AAV-HCC locus in the tumors. It has been argued that the results of this study are consistent with a classical model of malignant transformation after insertional oncogene activation by gene transfer vectors.19,20 Because the different gene expression pattern and developmental state in neonatal mice compared with adult mice may affect AAV vector integration, questions have been raised as to how relevant the observations of Donsante et al18,19 are to AAV-mediated gene transfer in general.
We thus sought to assess whether integration of AAV vectors into the host genome causes activation of oncogenes, leading to an increased incidence of HCC in adult mice. We performed liver-directed gene transfer with the single-stranded AAV2-hFIX16 vector,1 which contains the liver specific apoprotein E enhancer and human α-1-antitrypsin promoter controlling expression of a human FIX mini-gene. AAV2-hFIX16 vector was generated by transient transfection of human embryonic kidney 293 cells and purified with the use of a manufacturing process comparable to that used for preparation of clinical grade vectors. The goals of the study were to assess HCC incidence and to investigate whether integrated AAV vector DNA played a role in oncogenesis by identifying vector integration sites and examining expression of genes nearby. After following injected mice for 18 months, the HCC incidence of mice receiving AAV2-hFIX16 was not significantly different from control mice. To determine whether vector integration played a role in HCC formation, we developed a novel high-throughput method for identifying AAV vector integration sites, using linker-mediated polymerase chain reaction (LM-PCR) and 454 pyrosequencing, that identified 1029 unique AAV vector integrations. Analysis of the distribution of these sites within tumor tissue and adjacent normal tissue, as well as expression analysis of genes near integration sites, provided no evidence supporting the idea that integrated AAV2-hFIX16 contributed to HCC oncogenesis.
Methods
Vectors
AAV2-hFIX16 vector was prepared by current Good Manufacturing Practices–comparable processes (ie, performed at the same scale and using the same steps used for current Good Manufacturing Practices–grade vectors prepared for human clinical studies). Vector generation was by helper virus–free transient transfection of human embryonic kidney 293 cells grown in roller bottles21,22; the 11 277-bp vector plasmid containing the FIX expression cassette, flanked by AAV2 inverted terminal repeats (ITRs), contains a liver-specific human SERPINA1 (alpha1-antitrypsin) promoter, the apoprotein E enhancer and hepatic control region, coupled to the human F9 cDNA interrupted by a 1.4-kb fragment of intron 1, as previously described.23–25 The vector was purified by combined column chromatography with the use of Poros 50HS and cesium chloride density centrifugation as previously described26,27 with minor modifications. The final purified vector was formulated in 10mM sodium phosphate, 180mM sodium chloride, 0.1% pluronic F68, pH 7.3, and sterile filtered. Titering was performed by quantitative PCR (qPCR) with the use of linearized plasmid standards.28
Mice
Male C57BL/6 mice were obtained from Charles River Laboratories and housed in the Children's Hospital of Philadelphia Laboratory Animal Facility. Portal vein injections were performed as previously described.29 Plasma was collected by retro-orbital bleed and was quantified with a previously described enzyme-linked immunosorbent assay assay.30 Livers of mice that died before18 months of follow-up were preserved in 10% formalin for the duration of the study and embedded in paraffin for histopathologic analysis. At 18 months after injection mice were killed, livers were removed by dissection and examined for abnormalities by “breadloaf” sectioning at 3-mm intervals. Raised nodular lesions or discolored areas (suspected tumors) or both of each liver were collected in 20-mg sections and snap-frozen in liquid nitrogen. A 20-mg section of normal liver adjacent to the tumor was collected and similarly snap-frozen. If no liver abnormalities were observed, a 20-mg section of the median lobe was collected and frozen. Liver tissue for histopathologic evaluation was collected, fixed overnight in 4% paraformaldehyde, and embedded in paraffin. Tissue sections were stained with hematoxylin and eosin and examined for primary neoplasia by a veterinary pathologist specializing in rodent histopathology who was blinded to treatment status. Tumors were classified with a standard classification system.31 All procedures performed in this study were approved by the Children's Hospital of Philadelphia Institutional Animal Care and Use Committee.
Vector genome copy quantification
Frozen liver tissue was homogenized in lysis buffer and DNA isolated with the use of the DNEasy kit (QIAGEN). AAV2-hFIX16–specific sequence was amplified from 60 ng of genomic DNA with the use of primers hFIX-Gen1For (5′-ACCAGCAGTGCCATTTCCA-3′) and hFIX-Gen1Rev (5′-GAATTGACCTGGTTTGGCATCT-3′) in a model epGradient S Real-Time PCR machine (Eppendorf) with SYBR Green PCR master mix (Applied Biosystems). AAV2-hFIX16 copy number was calculated by comparing amplification signal with a standard curve created by spiking known amounts of linearized AAV2-hFIX16 production plasmid into 60 ng of liver genomic DNA from a saline-injected mouse.
Integration site cloning
Genomic DNA (1 μg) was digested overnight with MseI (New England Biolabs), and 1 μg of genomic DNA was digested overnight with CviQ1 (New England Biolabs) for 16 hours at 37°C. Digested DNA was purified with the use of a PCR purification kit (QIAGEN), and a previously described double-stranded adapter32 was ligated to digested DNA ends with the use of T4 DNA Ligase (New England Biolabs) for 16 hours at 16°C. Integration junctions were PCR amplified with the use of an adapter primer (5′-GTAATACGACTCACTATAGGGC-3′) and either a 5′ end vector primer (5′-AGGTCAGCAGGCAGGGAGGG-3′) or a 3′ end vector primer (5′-CAGCAAGGGGGAGGATTGGG-3′). PCR products were then diluted 1 in 200 in TE buffer, and integration junctions were re-amplified with the use of a second adapter primer (5′-GCCTCCCTCGCGCCATCAGnnnnnnnnAGGGCTCCGCTTAAGGGAC-3′, where nnnnnnnn is a sample-specific barcode) and either a second 5′ end vector primer (5′-GCCTTGCCAGCCCGCTCAGnnnnnnnnCTGAGGGGTTGGAAGGGGGC-3′, where nnnnnnnn is a sample-specific barcode) or a second 3′ end vector primer (5′-GCCTTGCCAGCCCGCTCAGnnnnnnnnAGGCATGCTGGGGATGCGGT-3′, where nnnnnnnn is a sample-specific barcode). Amplified integration junctions were sequenced with the use of a Genome Sequencer FLX pyrosequencer (Roche/454).
Integration site analysis
Pyrosequencing reads were first decoded with DNA barcodes included in the second round of PCR and then trimmed to remove ITR and adapter sequences.33 The resulting collection of sequences was mapped to the AAV2-hFIX16 genome to filter out any sequences where vector integrated into itself and to identify boundaries of AAV vector sequence versus genomic sequence. Any sequences beginning or ending with AAV counterpart were aligned against mouse genome with the use of BLAST-like alignment tool (University of California Santa Cruz; mm8, > 98% match score). Only sequences that showed unique best alignments, which began within the first 3 base pairs, were used in downstream analyses. All integration sites were defined by ≥ 1 sequence reads as comparison of integration preferences showed no obvious difference between sites defined by a single read and sites defined by multiple reads. For any integrants with identical sequence reads appearing in > 1 sample (n = 25), the integrant was assigned to the sample containing the highest clone count of sequence reads and discarded from all others as sample cross-contamination. Any integrants (n = 7) having the same number of sequence reads (clone count = 1 for all) in different samples were discarded as PCR crossover events. For comparison, matched random control sets were generated computationally by randomly choosing 3 genomic sites lying the same distance from MseI or CviQI sites as each of the integration sites. This method for generating matched random controls accounts for restriction site biases in the recovery of integration sites on the basis of their proximity to restriction sites and allows for more accurate statistical analysis.32,34 For a description of Genomic Features, please see the “Genomic Features Heatmap Guide” link (http://microb230.med.upenn.edu/protocols.html).
Expression analysis
Frozen liver tissue was homogenized in lysis buffer, and RNA was isolated with the use of the RNeasy kit (QIAGEN). cDNA was synthesized and hybridized to a Mouse Gene 1.0ST chip (Affymetrix). Fold difference in expression was obtained by dividing the expression signal observed in tumor tissue by the expression signal observed in adjacent normal tissue. Data series is available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE23680).
Integration junction copy quantification
Integration junction–specific sequences for the M24-Ntrk1 integrant and the M50-Ntrk1 integrant were amplified from 200 ng of genomic DNA with the use of primers 24Ntrk1 For (5′-GAGCCCAGAAACTCCTGTGT-3′), 24Ntrk1 Rev (5′-TGCCTCTCACTTGGATTTGA-3′), 50Ntrk1 For (5′-ACTCCTGTGTGCCTCTGAGC-3′), and 50Ntrk1 Rev (5′-CTTCCAACTTGGGCTGACAT-3′) and were detected with the use of 24Ntrk1 Probe (5′-FAM-AGGAACCCCTAGTAGATCTCAATATCC-TAMRA-3′) and 50Ntrk1 Probe (5′-FAM-CCGCCCGACGCTGGAATT-TAMRA-3′), respectively, in a model epGradient S Real-Time PCR machine (Eppendorf) with Taqman Universal PCR master mix (Applied Biosystems). Integration junction copy number was calculated by comparing amplification signals to standard curves created by spiking known amounts of linearized plasmid containing synthesized M24-Ntrk1 integration junction or M50-Ntrk1 integration junction (Genscript) into 200 ng of liver genomic DNA from a saline-injected mouse.
Statistics
Fisher exact test, Student t test, Mann-Whitney U test, and statistical power calculation were performed when appropriate. Statistical methods used for comparisons to matched random controls in Figure 2B are described in Berry et al35 and Brady et al.36 Differences were considered significant when P < .05.
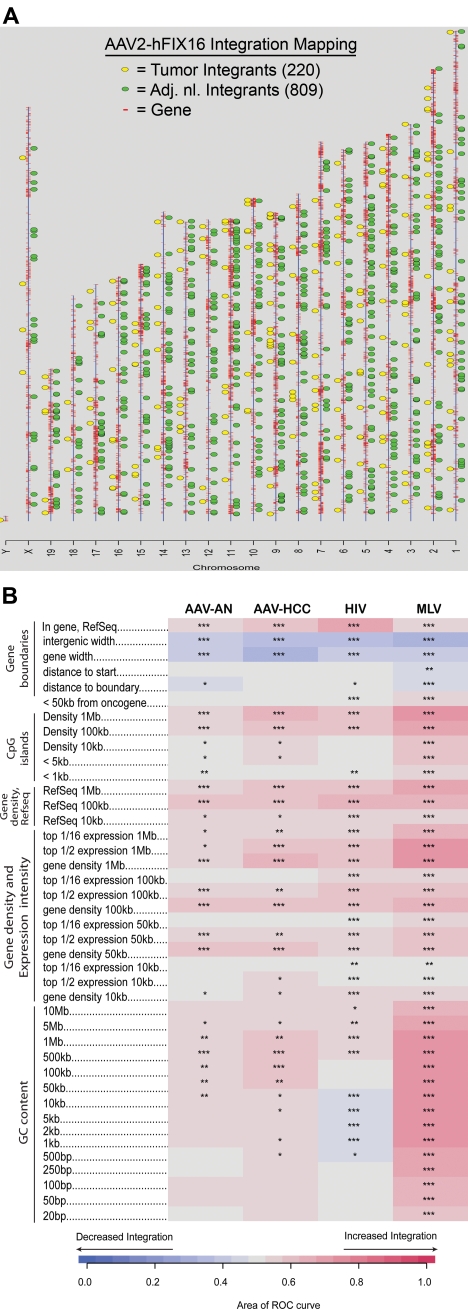
Figure 2.
Vector integration site distribution and preferences in normal and tumor tissues. (A) Ideogram of integration patterns from hepatocellular carcinoma and adjacent normal datasets across mouse genome. (B) Genomic heatmap of integration frequency relative to genomic features. Integration site dataset names are shown above the columns. Genomic features analyzed are shown to the left of the corresponding row of heatmap. The heatmap compares each experimental dataset to the matched random controls relative to frequency of the indicated genomic feature. A colored receiver operating characteristic (ROC) area scale is shown along the bottom of the panel with increasing shades of blue indicating negative correlation relative to the genomic feature and increasing shades of red indicating positive correlation relative to the comparison set. Comparisons to genomic features were carried out as previously described.35,52 Asterisks summarize the statistical significance of departures from random (*P < .05; **P < .01; ***P < .001).
Results
Prospective study of HCC risk
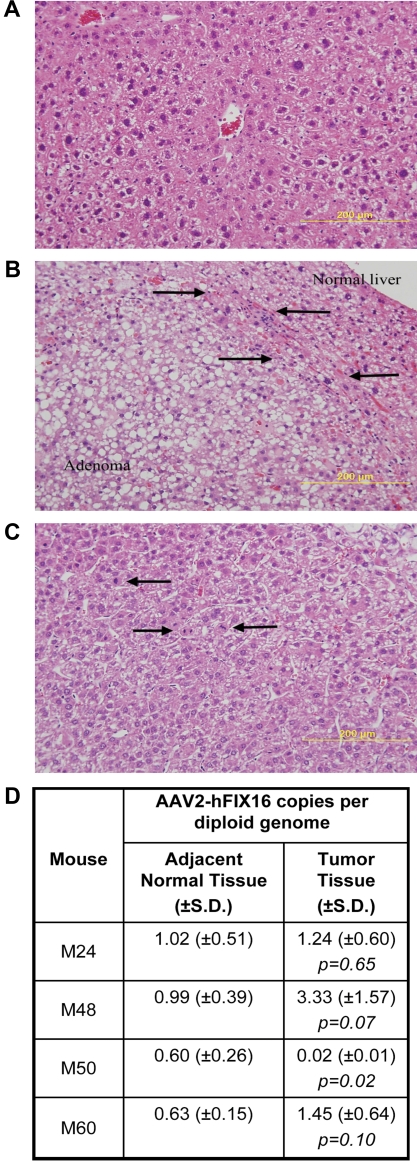
To determine whether liver-directed gene transfer with the use of AAV2-hFIX16 increases HCC risk, we performed a longitudinal study to compare HCC incidence in mice receiving either AAV vector or control treatments. We performed portal vein injections on 120 wild-type male C57BL/6J mice at 2-3 months of age. We injected 60 mice with 5 × 1012 vg/kg AAV2-hFIX16 (dose A) and 20 mice with 1 × 1014 vg/kg AAV2-hFIX16 (dose B). As controls, we injected 20 mice with 1 × 1014 vector capsids/kg AAV2 empty capsid and 20 mice with saline excipient. We also followed an additional 12 mice that did not receive portal vein injections for the same duration of time. The size of this study (80 mice receiving AAV, 52 controls) had 85% power to detect a difference between a 25% HCC rate in AAV-injected mice and 5% in controls. Seven of the dose A recipients, 4 of the dose B recipients, 2 of the empty capsid recipients, and 1 of the saline recipients did not survive the portal vein injection procedure. We followed the remaining mice for 18 months, preserving liver samples for histopathologic analysis from all mice that died before the end of the study. At 18 months we measured circulating hFIX levels before necropsy to confirm stable gene transfer (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We performed necropsy followed by complete liver histopathology to determine the incidence of HCC in the experimental groups. Tumors were characterized as HCCs when packets or sheets of atypical hepatocytes were observed.31 Mitotic figures were also common in HCCs (Figure 1C). HCCs were distinguished from adenomas because adenomas were characterized by well differentiated hepatocytes (with some variation in cell morphology) that had well-defined borders and often caused compression of the adjacent parenchyma31 (Figure 1B). The reported lifetime (median survival, 22-27 months) rate of spontaneous HCC formation in male C57BL/6J mice is 8.8% (www.informatics.jax.org/mtbwi/index.do). One of 49 mice in the 3 control groups developed HCC and 4 of 69 mice in the 2 AAV groups developed HCC (mice M24, M48, M50, M60). The overall HCC rates were not significantly different between groups: 5.8% for AAV-treated mice and 2.0% in control mice (P = .31, one-tailed Fisher exact test; Table 1).
Figure 1.
HCC tissue differs histopathologically from normal and adenomatous liver tissue but has no difference in vector genome copy number. Hematoxylin and eosin–stained sections for histopathologic diagnosis of (A) normal liver; (B) hepatic adenoma, arrows denote zone of compression between adenoma and normal liver; and (C) hepatocellular carcinoma, arrows indicate mitotic figures. Images were captured with the use of a Zeiss Axiophot microscope (Carl Zeiss Imaging, Inc) with a 20×, 0.40 aperture EC PLAN NEOFLUAR objective lens at room temperature. Images were acquired with the use of an Olympus DP70 (Olympus America Inc) camera and DP Manager Version 1.21.107 software, with subsequent image cropping performed with Adobe Photoshop. (D) AAV2-hFIX16 vector genome copy number in tumor and normal liver was quantified by qPCR, performing 3 independent measurements on total DNA isolated from tumor and normal liver tissue. P values from 2-tailed Student t test between 3 independent measurements of adjacent normal liver tissue and tumor tissue.
Table 1.
HCC incidence in AAV-injected and control mice
| Treatment group | AAV2-hFIX16 dose | HCC rate, n/N (%) |
|---|---|---|
| Uninjected | N/A | 1/12 (8.3) |
| Saline | N/A | 0/19 (0) |
| Empty capsid | 1 × 1014 vc/kg | 0/18 (0) |
| Combined controls | 1/49 (2.0) | |
| AAV dose A | 5 × 1012 vg/kg | 2/53 (3.8) |
| AAV dose B | 1 × 1014 vg/kg | 2/16 (12.5) |
| Combined AAV* | 4/69 (5.8) |
All mice were of the C57BL/6J strain, were 2-3 months old at the time of injection, and were followed for 18 months. The injection route was through the portal vein for those mice receiving injections. Bold indicates combined values.
vc indicates vector capsids; vg, vector genomes; and N/A, not applicable.
P value from 1-tailed Fisher exact test compared HCC rates of combined controls and combined AAV (P = .31).
Comparison of AAV2-hFIX16 copy number in tumor and adjacent normal tissue
To determine whether there was a relationship between amount of vector DNA in tissues and tumorigenesis, we measured AAV2-hFIX16 vector genome copy number by qPCR in HCC tumor tissue and adjacent normal tissue from the 4 AAV-treated mice that developed HCC. Three independent measurements on total DNA isolated from tumor tissue and adjacent normal tissue showed no significant difference in AAV2-hFIX16 copy number in mice M24, M48, and M60 (P > .05 for each) and a significantly lower copy number in tumor tissue compared with adjacent normal tissue in mouse M50 (P = .02) (Figure 1D)
Profiling of AAV2-hFIX16 vector integration sites
To investigate whether insertional activation of oncogenes occurred in the 4 AAV-treated mice with HCC, we cloned integrated AAV2-hFIX16 vector genomes to determine vector insertion sites within the mouse genome. Because AAV2-hFIX16 is intended for use as a therapeutic gene transfer vector, no bacterial origin of replication is present in its DNA sequence. Thus, we used LM-PCR32,37–40 to clone integrated AAV vector genomes from tumor tissue and adjacent normal tissue of mice M24, M48, M50, and M60. To determine genomic insertion sites of the AAV vector integrants, we used a high-throughput pyrosequencing method,41 which previously identified large numbers of integration sites from retroviral and lentiviral vectors.32,40,42,43
As a quality control filter, only sequence reads that contained both AAV2-hFIX16 vector DNA and mouse genomic DNA were analyzed. This process resulted in the identification of 1029 unique AAV vector integrants, 809 from adjacent normal tissue and 220 from tumor tissue. Given that the cloning procedure began with 2 μg of genomic DNA per sample and given the DNA mass of a diploid genome, we calculated a recovery frequency of one integrated AAV vector per 1661 diploid genome equivalents (dge) in normal liver. This is equal to 0.06% of diploid genomes containing a single integrated vector, although the efficiency of recovery is unknown. No AAV vector integrations were identified within 1 Mb of the AAV-HCC locus19 (supplemental Table 1). Integration junctions between vector and chromosomal DNA were characterized by frequent ITR deletions and microhomologies. These characteristics have previously been observed in smaller scale AAV vector integration site studies.7,8
We first examined the distribution of integration sites in normal tissue and determined the likelihood that integrants were located near annotated genomic features. In normal liver tissue there was a preference for integrating near genes (P < .001), CpG islands (P < .05), and guanosine/cysteine-rich (G-C rich) regions (P < .05) (Figure 2B). We then compared the integration site profile of normal tissue with tumor tissue, as well as of previously published integration profiles for HIV- and murine leukemia virus (MLV)–based vectors.35 In comparing AAV sites in tumor tissue with normal tissue, we found that the strength of associations with annotated genomic features were generally similar over many types of comparisons. A few measures did show detectable differences. We found that integration sites in tumor tissue were more likely to be located within 1 Mb of CpG islands (P < .05), 1 Mb of expressed genes (P < .05), and within Refseq genes (P < .05) than integration sites in normal tissue (Figure 2B). No attempt was made to correct the statistical tests for multiple comparisons, and the importance of these differences is unclear. No significant difference in likelihood to integrate near oncogenes was seen between tumor and adjacent normal tissue.
Compared with HIV-based vectors, AAV vector integrants were more likely to be located near G-C rich regions (P < .05) and less likely to be located near genes and CpG islands (P < .05) (Figure 2B). Compared with MLV-based vectors, AAV vector integrants were less likely to be located near G-C rich regions (P < .001), CpG islands (P < .001), and genes (P < .05) (Figure 2B). AAV vector integrants were also less likely than MLV-vectors to be located near oncogene 5′ ends (P < .05), a consequence of the general tendency for MLV-vectors to integrate near gene 5′ ends (Figure 2B).
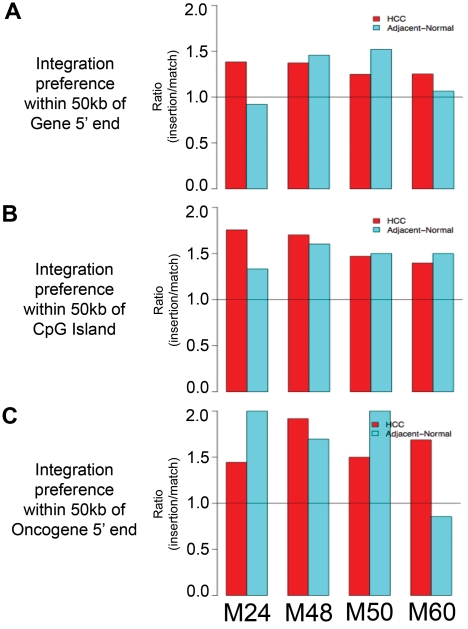
When integration sites from mice M24, M48, M50, and M60 were analyzed individually, similar trends were observed. In all 4 tumor samples and all 4 adjacent normal liver samples, the number of AAV vector integrants located within 50 kb of genes (Figure 3A), CpG islands (Figure 3B), and oncogenes (Figure 3C) was greater than the number of matched random control insertions within 50 kb of these features. The only exceptions were adjacent normal liver integrants within 50 kb of genes in mouse M24 and adjacent normal liver integrants within 50 kb of oncogenes in mouse M60.
Figure 3.
Vector integration site preferences in individual mice reflects trends of combined mice analysis. Ratio of number of vector integrants divided by number of random insertions showing likelihood over random for vector integrants to be located within 50 kb of (A) RefSeq genes, (B) CpG islands, and (C) oncogenes.
Effect of integrated vector on expression of nearby genes
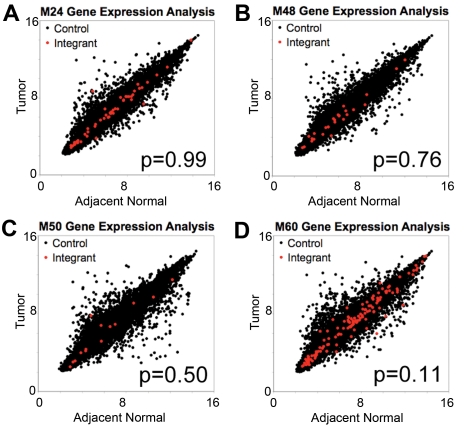
To determine whether AAV vector integration influences the expression of nearby genes, we compared steady-state mRNA levels in tumors and adjacent normal liver tissue of mice M24, M48, M50, and M60 by microarray analysis. To investigate whether the proximity to an integration site resulted in changes in mRNA accumulation, we quantified the change in expression of each gene on the array for tumor versus adjacent normal tissue in each mouse. We then compared the collection of all genes with the closest gene to each integration site mapped in the tumor samples. We asked whether the mean change in mRNA levels between tumor and adjacent normal tissue was greater for the group of genes near integration sites than for the collection of all genes assayed. None of the 4 mice showed a significant difference in the mean value (Figure 4A-D), thus failing to support the hypothesis of greater transcriptional changes near integrants.
Figure 4.
No difference in magnitude of expression change in adjacent normal tissue and in tumor tissue for genes near tumor integrants and genes distal to tumor integrants. Plots of log2-transformed gene expression levels in tumor tissue versus log2-transformed gene expression levels in adjacent normal for mice (A) M24, (B) M48, (C) M50, and (D) M60. Gene expression levels were determined from microarray with the use of the Mouse Gene 1.0ST Affymetrix chip. Red dots indicate the closest gene to an integrant cloned from tumor tissue, and black dots indicate all other genes on the array. P value from the Mann-Whitney U test compared the change in expression for genes near integrants with all other genes.
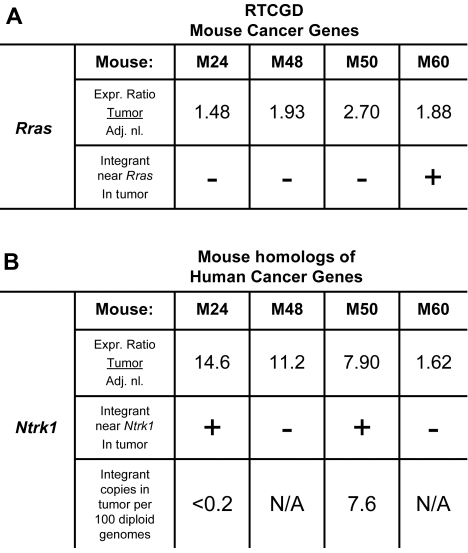
We next used the mouse Retrovirus Tagged Cancer Gene Database (RTCGD),44 which catalogs mouse genes implicated in tumor formation, to investigate possible association of AAV vector integration sites with cancer-associated genes. We queried the integration site dataset to identify RTCGD genes within 100 kb of an integrant cloned from tumor tissue that also showed at least a 50% increase in expression over adjacent normal tissue. One integration site met these criteria; the tumor sample from mouse M60 had an AAV2-hFIX16 integrant near Rras and a 1.88-fold increase in Rras expression over normal adjacent liver (Figure 5A). However, we also noted that the tumors from mice M24, M48, and M50, in which no AAV2-hFIX16 integrants near Rras were identified, also showed increases in Rras expression over adjacent normal tissue of 1.48-, 1.93-, and 2.7-fold, respectively, consistent with increased Rras expression as a result of transformation to HCC.
Figure 5.
Up-regulation of oncogenes near integrants cloned from tumor tissue occurs independently of vector integration. Expression analysis of up-regulated (A) mouse cancer-related genes and (B) mouse homologs of human cancer-related genes located within 100 kb of an AAV2-hFIX16 integrant cloned from tumor tissue. Expression ratio was obtained by dividing the absolute array signal from tumor tissue by the absolute array signal from adjacent normal tissue. Individual M24 and M50 Ntrk1 integrants were quantified by qPCR, performing 4 independent measurements on total DNA isolated from tumor tissue.
The same analysis was then applied to all mouse homologs of human oncogenes. These mouse genes have not been shown in the literature to cause tumor formation in mice, but their human homologs have been linked to cancer (for criteria, see http://microb230.med.upenn.edu/protocols/cancergenes). The only gene from this group that was also increased in expression was Ntrk1. Mouse M24 had a 14.6-fold and mouse M50 had a 7.9-fold increase in Ntrk1 expression over adjacent normal tissue, and both mice had AAV vector integrants near Ntrk1 in tumor tissue (Figure 5B). We were unable to identify integrated AAV vector near Ntrk1 in either mouse M48 or mouse M60; however, the tumor in mouse M48 had an 11.2-fold increase in Ntrk1 expression over adjacent normal tissue, whereas mouse M60 had a 1.62-fold increase.
If an integration event caused the malignant transformation of the hepatocyte that gave rise to the HCC by insertional activation, then the same integrant should be present in all HCC cells. To test this we performed qPCR on the M24 and M50 tumor samples for the specific integration junction between AAV2-hFIX16 and flanking genomic DNA at the Ntrk1 insertion site. We found that the M24 integrant was below the limit of detection of 0.2 copies/100 dge, and the M50 integrant was present at a rate of 7.6 copies/100 dge. The low frequency of the Ntrk1 integrant per diploid genome equivalent thus argues against insertional activation.
Discussion
To address concerns about the genotoxic potential of AAV vectors,19 our study aimed to determine whether liver-directed gene transfer to adult mice with the use of AAV vectors causes a significant increase in HCC risk. This issue warranted investigation because 2 current clinical trials are using liver-directed AAV-mediated gene transfer in adults. In addition, many therapeutic strategies that use AAV vectors in the liver are in preclinical development. Our study incorporated 2 features designed to bias the results in favor of detecting hepatocellular carcinomas. First, the mice were followed for a period of 18 months, so that latent effects could be detected. Second, we used very high doses, ≤ 50 times higher than those yet administered to humans, in an attempt to detect an effect. Our study of 132 mice did not show a statistically significant difference in HCC incidence between AAV-injected mice and control mice. A weak trend toward more HCC in the vector-injected mice was observed, but frequencies of HCC for all groups in our study were near or below the reported 8.8% HCC incidence rate in C57BL/6J mice (http://www.informatics.jax.org/mtbwi/index.do) (A caveat in comparing HCC incidence is the median survival of mice in the Jackson Laboratory data were 22-27 months, whereas our mice were 20-21 months of age at killing.) Thus, our study showed no significant association between HCC and AAV vector treatment.
To determine the genomic insertion sites of integrated AAV vector in the tumors, we had to develop a novel method of isolating junctions between AAV vector DNA and murine DNA. Inverse PCR has been the only method in the literature used to isolate ori-less integrated AAV vectors19; here we adapted the LM-PCR method for cloning retroviral and lentiviral integrants32,39,40 to AAV vector integrants. With the use of this technique we were able to map 1029 AAV vector insertion sites, the largest set of AAV integration sites published in the literature from a single study. This approach could also be used to analyze AAV vector integration patterns in clinical samples.
On the basis of the number of AAV vector integrants we cloned from normal liver, we estimated that 0.06% of diploid genome equivalents contain a detectable vector integrant. When determining the fraction of AAV vector copies within the liver that are integrated, 0.0006 integrated vector copies per diploid genome equivalent divided by 0.81 total vector copies per diploid genome equivalent yields that 0.07% of vector copies are integrated. In addition, because adult mice > 2 months of age average ∼ 5-6N hepatocyte DNA content,45,46 we calculated ≥ 1 of every 588 cells in the liver contains a detectable vector integrant. These integration rate calculations are lower-limit estimates because we probably have not cloned every integrant within each sample.
Our data about vector genome copy number and vector integration sites within tumors are consistent with the presence of nonneoplastic, vector transduced bystander hepatocytes entrapped within the tumor. The minimum number of integration sites identified in any of the 4 tumors was 14 integrants (M50). However, the maximum amount of vector genomes measured within any of the 4 tumors was 3.33 copies per diploid genome (M48). The main contribution to quantified vector genomes within a tumor should come from integrated vector because episomal vector genomes should be diluted out during clonal proliferation. Thus, our data suggest ≤ 3 vector integration events occurred per tumor and the remainder of identified integration sites occurred in transduced bystander hepatocytes entrapped within the tumor. By sequence analysis we cannot differentiate integrations in tumor cells from integrations in bystander cells, but the higher number of sequence reads for some integrants may suggest those integration events occurred in cells that subsequently underwent clonal expansion, as has recently been reported in hematopoietic stem cell–directed gene transfer.47,48
The profile of AAV vector integration sites in our study is consistent with previously described preferences for integration into genes and CpG islands. Thus, our LM-PCR technique validated the previous profiling studies done with plasmid rescue techniques.7,8 However, our study is unique in identifying G-C rich regions as preferred integration sites for AAV vectors. Although we did not find significantly different integration site profiles in tumor tissue compared with adjacent normal tissue, on the basis of previously published HIV and MLV insertion sites we did find both HIV and MLV vectors are more likely to integrate near genes than AAV vectors, and MLV vectors are more likely to integrate near oncogene 5′ ends. Thus, AAV vector integration targeting preferences may be relatively favorable for clinical gene transfer.
Expression profiling of genes near AAV vector insertion sites in tumors showed no significant changes compared with genes not near AAV vector insertion sites. This may be because integrated AAV vector genomes do not significantly alter the expression of nearby genes, but it is probably in part because many of the tumor integrants we identified were not clonal within the tumor. In addition, it may be possible that we were unable to detect integrants near some genes that had significant changes in expression. With regard to integration events occurring within genes, we found a significant number of integration events within both introns and exons. Phenotypic knockout of these genes would require a prior haploinsufficiency or a dominant negative mechanism created by vector integration.
Few cancer-related genes near integrants were up-regulated in the tumor from which the integrant was cloned. Rras is the only RTCGD gene that fulfilled these criteria, but our data suggest Rras overexpression may be associated with murine HCCs independent of AAV vector integration. Ntrk1 is the only mouse homolog of a human cancer-related gene up-regulated in tumors in which integrated AAV vector was mapped nearby. Although both tumors containing integrants near Ntrk1 exhibited Ntrk1 up-regulation, we found the 2 integrants near Ntrk1 were not present in every tumor cell, suggesting Ntrk1 up-regulation within tumor tissue was a hallmark of some murine HCCs. This is supported by previous studies that found overexpression of the Ntrk1 gene product, TrkA, in murine and human HCC tumors that are unrelated to AAV administration. In these tumors, TrkA expression has been localized to both HCC cells and endothelial cells lining the tumor vasculature.49,50
Our data document a low frequency of integration by AAV2 vectors after liver-directed gene transfer, but they provide no clear evidence supporting the idea that AAV vector insertional activation of oncogenes causes tumor formation. Although there is one study in the literature, in neonatal mice, that supports the oncogene insertional activation model for HCC development,19 our results are more consistent with long-term studies in canine14,15 and adult mouse16,17 models in which no correlation between AAV vectors and tumor formation was found. The differing results between these studies can potentially be explained by age at time of AAV administration and the effects of rapid cell division in the neonatal liver on AAV vector integration patterns. It would be of interest to know the expression levels of genes at the AAV-HCC locus during neonatal development and adulthood because we did not observe any integrants near the AAV-HCC locus (although we cannot definitively exclude this possibility). Future studies on the roles of the genes at the AAV-HCC locus and how dysregulation of these genes may interact with hepatocellular carcinogenesis will be useful, as would independent confirmation of the original results reported by Donsante et al.18,19
This study was designed to favor the detection of tumor formation by the use of high-vector doses and long periods of follow-up. Although we failed to establish definitive evidence for vector-mediated insertional activation of oncogenes, there were several shortcomings and intriguing findings that require additional study. First, the study was not sufficiently powered to detect small or modest differences in risk of tumor formation between vector-injected and control animals. Second, there was a trend toward higher incidence of tumor formation at higher doses. Although it seems unlikely that doses in this range will be used in human subjects, this is still a finding worth further investigation. Use of tumor-prone mouse models51 may shed further light on this. The finding that integration sites in tumors in our study were more likely to be located within 1 Mb of CpG islands, of expressed genes, and within RefSeq genes compared with integration sites in adjacent normal tissue may also be of interest. Our development of a method for recovering and characterizing large numbers of integrants will facilitate these important studies. Our data combined with that in the literature would suggest that single-stranded AAV vector-mediated gene transfer into adult liver is perhaps safer than that into neonatal livers, but this will probably continue to be an area for further study in the safety of AAV transduction of liver.
Supplementary Material
Acknowledgments
We thank Drs Bernd Hauck and Olga Zelenia for helpful discussion and manufacturing and characterization of the vector used in these studies, Dr Shu Li for assistance with statistical analysis, Dr John Tobias for assistance with microarray analysis, as well as Ms Jennifer Wellman and Mr Anand Bhagwat for helpful discussion.
This work was supported by the Howard Hughes Medical Institutes, National Institutes of Health (grants N01 HV78203-4-0-1, AI52845, and AI082020), and the Penn Genome Frontiers Institute with a grant with the Pennsylvania Department of Health.
The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.L., F.M., V.R.A., F.D.B., and K.A.H. designed research; H.L., N.M., S.R.H., A.S., G.B., S.E.E., and R.S. performed research and collected data; H.L., N.M., S.R.H., F.M., J.F.W., F.D.B., and K.A.H. analyzed and interpreted data; H.L., N.M., S.R.H., F.M., J.F.W., F.D.B., and K.A.H. wrote and edited the manuscript.
Conflict-of-interest disclosure: J.F.W. is an inventor on patents describing recombinant AAV chemistry, manufacturing, and controls and consults in the field of gene therapy. F.M. and K.A.H. hold patents related to AAV vector manufacturing and use. The remaining authors declare no competing financial interests.
Correspondence: Katherine A. High, 3501 Civic Center Blvd, Rm 5060, Philadelphia, PA 19104; e-mail: high@email.chop.edu.
References
- 1.Manno CS, Pierce GF, Arruda VR, et al. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat Med. 2006;12(5):342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 2.Nakai H, Yant SR, Storm TA, Fuess S, Meuse L, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75(15):6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutledge EA, Russell DW. Adeno-associated virus vector integration junctions. J Virol. 1997;71(11):8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakai H, Iwaki Y, Kay MA, Couto LB. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J Virol. 1999;73(7):5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36(7):767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- 6.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34(3):297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 7.Nakai H, Wu X, Fuess S, et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol. 2005;79(6):3606–3614. doi: 10.1128/JVI.79.6.3606-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R, Russell DW. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol. 2005;79(17):11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inagaki K, Lewis SM, Wu X, et al. DNA palindromes with a modest arm length of greater, similar 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice. J Virol. 2007;81(20):11290–11303. doi: 10.1128/JVI.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 11.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–50. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 13.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16(2):198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 14.Nichols TC, Dillow AM, Franck HW, et al. Protein replacement therapy and gene transfer in canine models of hemophilia A, hemophilia B, von Willebrand disease, and factor VII deficiency. ILAR J. 2009;50(2):144–167. doi: 10.1093/ilar.50.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemeyer GP, Herzog RW, Mount J, et al. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood. 2009;113(4):797–806. doi: 10.1182/blood-2008-10-181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell P, Moscioni AD, McCarter RJ, et al. Analysis of tumors arising in male B6C3F1 mice with and without AAV vector delivery to liver. Mol Ther. 2006;14(1):34–44. doi: 10.1016/j.ymthe.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Schuettrumpf J, Baila S, Khazi F, Liu J, Bunte R, Arruda VR. AAV vectors do not increase the risk of tumor formation in p53 deficient models [abstract]. Mol Ther. 2007;15(suppl 1) Abstract 2. [Google Scholar]
- 18.Donsante A, Vogler C, Muzyczka N, et al. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther. 2001;8(17):1343–1346. doi: 10.1038/sj.gt.3301541. [DOI] [PubMed] [Google Scholar]
- 19.Donsante A, Miller DG, Li Y, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317(5837):477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 20.Russell DW. AAV vectors, insertional mutagenesis, and cancer. Mol Ther. 2007;15(10):1740–1743. doi: 10.1038/sj.mt.6300299. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita T, Elliger S, Elliger C, et al. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5(7):938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 22.Kay MA, Manno CS, Ragni MV, et al. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 2000;24(3):257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 23.Kurachi S, Hitomi Y, Furukawa M, Kurachi K. Role of intron I in expression of the human factor IX gene. J Biol Chem. 1995;270(10):5276–5281. doi: 10.1074/jbc.270.10.5276. [DOI] [PubMed] [Google Scholar]
- 24.Le M, Okuyama T, Cai SR, et al. Therapeutic levels of functional human factor X in rats after retroviral-mediated hepatic gene therapy. Blood. 1997;89(4):1254–1259. [PubMed] [Google Scholar]
- 25.Miao CH, Ohashi K, Patijn GA, et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol Ther. 2000;1(6):522–532. doi: 10.1006/mthe.2000.0075. [DOI] [PubMed] [Google Scholar]
- 26.Wright JF, Le T, Prado J, et al. Identification of factors that contribute to recombinant AAV2 particle aggregation and methods to prevent its occurrence during vector purification and formulation. Mol Ther. 2005;12(1):171–178. doi: 10.1016/j.ymthe.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer JM, Smith PH, Parthasarathy S, et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol Ther. 2003;7(1):122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 29.Nakai H, Herzog RW, Hagstrom JN, et al. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood. 1998;91(12):4600–4607. [PubMed] [Google Scholar]
- 30.Kung SH, Hagstrom JN, Cass D, et al. Human factor IX corrects the bleeding diathesis of mice with hemophilia B. Blood. 1998;91(3):784–790. [PubMed] [Google Scholar]
- 31.Frith CH, Ward JM, Turusov VS. Tumours of the liver. IARC Sci Publ. 1994;111:223–269. [PubMed] [Google Scholar]
- 32.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann C, Minkah N, Leipzig J, Tebas P, Bushman FD. DNA bar coding and pyrophosphate sequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35(13):e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewinski MK, Bisgrove D, Shinn P, et al. Genome-wide analysis of chromosomal features repressing HIV transcription. J Virol. 2005;79(11):6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2(11):e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady T, Lee YN, Ronen K, et al. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 2009;23(5):633–642. doi: 10.1101/gad.1762309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- 39.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 40.Ciuffi A, Ronen K, Brady T, et al. Methods for integration site distribution analyses in animal cell genomes. Methods. 2009;47(4):261–268. doi: 10.1016/j.ymeth.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabriel R, Eckenberg R, Paruzynski A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15(12):1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 43.Paruzynski A, Arens A, Gabriel R, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5(8):1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 44.Retrovirus Tagged Cancer Gene Database. [Accessed October 15, 2010]. http://rtcgd.ncifcrf.gov/
- 45.Vinogradov AE, Anatskaya OV, Kudryavtsev BN. Relationship of hepatocyte ploidy levels with body size and growth rate in mammals. Genome. 2001;44(3):350–360. doi: 10.1139/g01-015. [DOI] [PubMed] [Google Scholar]
- 46.Lu P, Prost S, Caldwell H, Tugwood JD, Betton GR, Harrison DJ. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome. 2007;18(9):617–626. doi: 10.1007/s00335-007-9048-y. [DOI] [PubMed] [Google Scholar]
- 47.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang GP, Berry CC, Malani N, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115(22):4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishibe K, Yamada Y, Ogawa K. Production of nerve growth factor by mouse hepatocellular carcinoma cells and expression of TrkA in tumor-associated arteries in mice. Gastroenterology. 2002;122(7):1978–1986. doi: 10.1053/gast.2002.33581. [DOI] [PubMed] [Google Scholar]
- 50.Tokusashi Y, Asai K, Tamakawa S, et al. Expression of NGF in hepatocellular carcinoma cells with its receptors in non-tumor cell components. Int J Cancer. 2005;114(1):39–45. doi: 10.1002/ijc.20685. [DOI] [PubMed] [Google Scholar]
- 51.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24(6):687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 52.Marshall HM, Ronen K, Berry C, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2(12):e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.