Abstract
During hindbrain development, facial branchiomotor neurons (FBM neurons) migrate from medial rhombomere (r) 4 to lateral r6. In zebrafish, mutations in planar cell polarity genes celsr2 and frizzled3a block caudal migration of FBM neurons. Here, we investigated the role of cadherins Celsr1-3, and Fzd3 in FBM neuron migration in mice. In Celsr1 mutants (knock-out and Crash alleles), caudal migration was compromised and neurons often migrated rostrally into r2 and r3, as well as laterally. These phenotypes were not caused by defects in hindbrain patterning or neuronal specification. Celsr1 is expressed in FBM neuron precursors and the floor plate, but not in FBM neurons. Consistent with this, conditional inactivation showed that the function of Celsr1 in FBM neuron migration was non-cell autonomous. In Celsr2 mutants, FBM neurons initiated caudal migration but moved prematurely into lateral r4 and r5. This phenotype was enhanced by inactivation of Celsr3 in FBM neurons and mimicked by inactivation of Fzd3. Furthermore, Celsr2 was epistatic to Celsr1. These data indicate that Celsr1-3 differentially regulate FBM neuron migration. Celsr1 helps to specify the direction of FBM neuron migration, whereas Celsr2 and 3 control its ability to migrate.
Introduction
In the developing central nervous system, postmitotic neurons leave ventricular zones of precursor proliferation and migrate to their destinations. Several tightly regulated modes of migration have been described, including radial migration of excitatory neurons to the cortex, tangential migration of cortical interneurons, migration to the olfactory bulb in the rostral migratory stream, and migration of rhombic lip derivatives. A unique and intriguing example of neuronal migration is that of facial branchiomotor neurons (FBM neurons), which innervate muscles responsible for facial expression derived from the second pharyngeal arch (Garel et al., 2000; Chandrasekhar, 2004; Guthrie, 2007). FBM neurons are generated in rhombomere 4 (r4) and extend their axons from dorsal exit points toward muscle targets. Their cell bodies undergo a complex caudal, tangential migration from r4 to r6. They migrate in the subventricular region and pass medial to the nucleus abducens (nVI) in r5 (Song et al., 2006), before moving laterally and dorsally in r5-r6. Finally, they undergo a radial, gliophilic, Reelin- and Cdk5-dependent migration in r6 to reach their terminal, subpial location (Goffinet, 1984; Ohshima et al., 2002; Chandrasekhar, 2004). While migrating, FBM neurons leave their axons behind to form the “genu” of the facial nerve, with facial motor axons looping around nVI and the medial longitudinal fasciculus. Caudal translocation of the soma of FBM neurons with looping of axons is conserved from fish to mammals (except chicken), with some species-specific differences (Gilland and Baker, 2005).
In zebrafish, several planar cell polarity (PCP) genes such as van gogh-like 2 (vangl2), frizzled3a (fzd3a), celsr2, prickle1a, and prickle1b are necessary for the caudal migration of FBM neurons (Bingham et al., 2002; Jessen et al., 2002; Carreira-Barbosa et al., 2003; Wada et al., 2006; Rohrschneider et al., 2007). In mice, some Wnt/PCP genes (Fzd3;Vangl2, Wnt5a) and downstream signaling events have recently been implicated in regulating FBM neuron movement (Vivancos et al., 2009). To understand better the role of PCP-related mechanisms in FBM neuron migration in mice, we studied the role of cadherins Celsr1-3. Mouse Celsr1-3 are orthologs of Drosophila flamingo/starry night and are expressed in the developing nervous system in complementary patterns (Formstone and Little, 2001; Shima et al., 2002; Tissir et al., 2002). Celsr and Fzd proteins have established roles in PCP-related processes such as neural tube closure, organization of stereocilia of hair cells in the inner ear (Curtin et al., 2003; Montcouquiol and Kelley, 2003; Wang and Nathans, 2007), and skin hair patterning (Guo et al., 2004; Devenport and Fuchs, 2008; Ravni et al., 2009), as well as dendritic growth (Shima et al., 2004) and axon guidance (Wang et al., 2002; Zhou et al., 2008). Using Celsr1Crsh (Curtin et al., 2003), Celsr1KO (Ravni et al., 2009), Celsr2KO (Tissir et al., 2010), Celsr3KO (Tissir et al., 2005), and Fzd3 (Wang et al., 2002) mutant mice, we demonstrate specific and different roles for Celsr1 on one hand and Celsr2,3 and Fzd3 on the other hand in regulating the direction and the rostrocaudal extent of FBM neuron migration.
Materials and Methods
All animal procedures were carried out in accord with European and American guidelines and approved by the animal ethics committee of the University of Louvain and the animal care and use committee of the University of Missouri (Columbia, MO).
Mouse lines.
The Celsr1KO, Celsr1Crsh, Celsr3, and Fzd3 mutants and the SE1::GFP (where GFP is green fluorescent protein) transgenics were described previously (Wang et al., 2002; Curtin et al., 2003; Tissir et al., 2005; Shirasaki et al., 2006; Ravni et al., 2009). The Celsr2 mutant is described by Tissir et al. (2010). For timed mating, noon on the day of plugging was defined as embryonic day 0.5 (E0.5). Embryos were dissected in ice-cold PBS and the appropriate embryonic stage was determined using morphological criteria before fixation.
In situ hybridization.
Hindbrains were dissected and cut at the dorsal edge of the neural tube and fixed in 4% paraformaldehyde in PBS at 4°C overnight. For the Isl1, Celsr1, and Hb9 genes, the following primer sets were used to amplify cDNA fragments, which were used as templates for synthesis of digoxigenin-labeled probe: Isl1 cDNA [GenInfo Identifier (GI): 162287064], nucleotides 435-1383, primers 5′-TTCCCACTTTCTCCAACAGG-3′ and 5′-ACGTGCTTTGTTAGGGATGG-3′; Celsr1 cDNA (GI: 3800735), nucleotides 7812-8257, primers 5′-GACTGGCTGTTGGCTTGGAC-3′ and 5′-CGTTAGCAGAGTGGCCCGAG-3′; Hb9 cDNA (GI: 5733508), nucleotides 1176-1992, primers 5′-GAAGACGGAAGAGGAGCTGA-3′ and 5′-GGGGGATGGGAAAGCTAAAT-3′. The other probes were as follows: Fzd3 (Tissir and Goffinet, 2006); EphA4, Hoxb1 (Garel et al., 2000); Tbx20 (Coppola et al., 2005); Krox20 (Marin and Charnay, 2000); Gata3 (Karis et al., 2001). Hindbrains were permeabilized with proteinase K treatment, postfixed with 4% paraformaldehyde and 0.2% glutaraldehyde in PBS, and hybridized with the probe at 70°C overnight. Samples were washed in 50% formamide, 5× SSC, and 1% SDS at 70°C and then washed in 50% formamide, 2× SSC at 65°C and incubated with anti-digoxigenin alkaline phosphatase-conjugated antibody (Roche) overnight at 4°C. The hybridization signal was revealed with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium chloride substrates. After bleaching, hindbrains were flat mounted on a slide glass for visualization and photographed; images were processed with Photoshop (Adobe).
β-Galactosidase staining.
Embryos were fixed with 2% paraformaldehyde, 0.1% glutaraldehyde in PBS on ice for 20 min and stained with a solution containing 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 5 mm K3Fe(CN)6, 5mMK4Fe(CN)6, and 2 mm MgCl2 in PBS.
Immunohistochemistry.
For neurofilament staining, samples were processed as described previously (Lee et al., 1995; Tiveron et al., 2003; Tissir et al., 2005) using anti-neurofilament-160 (NF-160; Sigma catalog #N5264), or 2H3 [Developmental Studies Hybridoma Bank (DSHB)] monoclonal antibodies. For immunofluorescence, the following primary antibodies were used: mouse anti-Isl1 (1/5000, DSHB), guinea pig anti-Celsr1 (1:200, a gift from Elaine Fuchs, Rockefeller University, New York NY), rabbit anti-Celsr1 C-terminal (1:400, this work), and rabbit anti-cleaved caspase-3 (1:200, Cell Signaling). Secondary antibodies were goat anti-mouse IgG Alexa Fluor 594 (1:800, Invitrogen), goat anti-rabbit IgG Alexa Fluor 488 (1:800, Invitrogen), and goat anti-guinea pig IgG DyLight 488 (1:400, Jackson ImmunoResearch Laboratories). Preparations were examined with a Zeiss Axio Scope microscope and photographed with an Eclipse system (Nikon), and montages were edited with Photoshop (Adobe).
Celsr1 N-terminal (extracellular) and C-terminal (intracellular) antibodies.
For the N-terminal antibody raised in rabbit, a 906 bp cDNA fragment encoding amino acids 1663–1964 of the Celsr1 protein (GI: 22095546) was cloned into the His-tagged QE30 plasmid. The C-terminal antibody was raised in rabbit against the most membrane proximal region of the Celsr1 cytoplasmic tail (Formstone et al., 2010).
Retrograde labeling of hindbrain neurons.
To visualize cranial motor neurons, nylon filters coated with NeuroVue lipophilic dyes (Molecular Targeting Technologies) were applied to peripheral nerve projections as described previously (Fritzsch et al., 2005). NeuroVue Jade (emission maximum, 508 nm), Maroon (667 nm), and Red (588 nm) were used in this study. Confocal images were captured on a Zeiss LSM 510 Meta NLO system and processed using Zeiss software.
Results
Expression of Celsr1-3 in the hindbrain
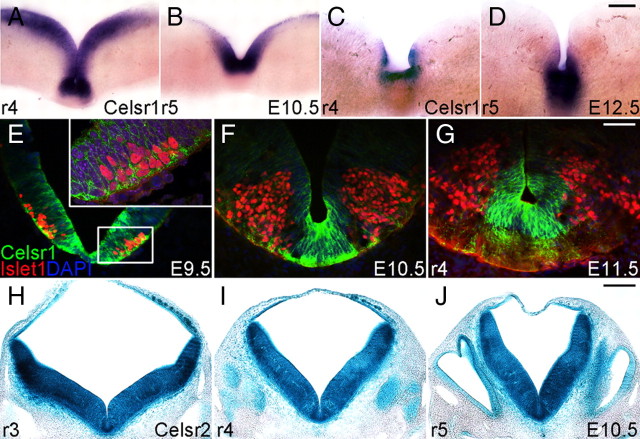
Celsr1 expression was studied by in situ hybridization (ISH) with a digoxigenin-labeled riboprobe and immunohistochemistry with anti-Celsr1 antibodies (Fig. 1). At E10.5, Celsr1 mRNA was expressed in the ventricular zone and floor plate cells at all axial levels (6/6 embryos) (Fig. 1A,B). By E12.5, Celsr1 expression in floor plate cells was downregulated in r4 and more anterior regions but maintained at high levels in and caudal to r5, adjacent to the migrating FBM neurons (4/6 embryo) (Fig. 1C,D). At the protein level, Celsr1 was strongly expressed in radial neuroepithelial cells located in the paramedian ventricular zone at E9.5–E10.5, with high signal in their pial end feet (Fig. 1E). Celsr1-positive cells were intermingled with emerging Islet1-positive FBM neurons in r4. Expression was also seen in radial cells in the alar plate. Using confocal imaging (Fig. 1E, inset), we found that Celsr1 protein may colocalize with Isl1 in some FBM neurons at E9.5. From E10.5 to E11.5, Celsr1 expression was downregulated in neuroepithelial cell, but remained high in floor plate cells (Fig. 1F–G). At this stage, we did not find any Celsr1 and Isl1 colocalization in FBM neurons. Expression of Celsr2, studied using the knocked-in β-galactosidase, was broader than that of Celsr1. The Celsr2-related signal was detected in the floor and alar plates in the neuroepithelium, as well as in newly generated FBM neurons (Fig. 1H–J). Celsr3 expression, previously studied by ISH, is found in all postmitotic neurons but not in precursors, including in the hindbrain (Song et al., 2006; Tissir and Goffinet, 2006). Altogether, these results show that, in the developing hindbrain, Celsr1 is expressed in neuroepithelial cells, Celsr2 in both precursors and postmitotic neurons, and Celsr3 in postmitotic neurons, a pattern similar to that reported in other regions of the nervous system (Formstone and Little, 2001; Shima et al., 2002; Tissir et al., 2002).
Figure 1.
Expression of Celsr1 and 2 in the developing hindbrain. A–D, ISH of Celsr1 digoxigenin-labeled probe on hindbrain coronal sections. At E10.5 (A, B), Celsr1 mRNA is expressed in the neuroepithelium and the floor plate. At E12.5 (C, D), Celsr1 expression is downregulated in r4 but remains high in the floor plate of r5. E–G, Immunofluorescence using anti-Celsr1 (green) and anti-Isl1 (red) in r4. Celsr1 protein expression is found in FBM neuron precursors in ventricular zones at E9.5 and restricted to floor plate from E10.5. Confocal analysis shows that Celsr1 (green) may colocalize with Isl1 (red) in FBM neurons at E9.5. H–J, Coronal sections in the mouse hindbrain processed for LacZ histochemistry to examine Celsr2 expression in r4–r6. The Celsr2-related signal is detected in the basal and alar plates and in the neuroepithelium, as well as in all postmitotic neurons. Scale bars: (in D), A–D, 100 μm; (in G), E–G, 50 μm; (in J), H–J, 200 μm.
Inactivation of Celsr1 perturbs the direction of FBM neuron migration
Given the intriguing pattern of Celsr1 expression in the developing hindbrain, we tested its potential role in FBM neuron migration by analyzing Celsr1 knock-out (Ravni et al., 2009) (Fig. 2) and Celsr1Crsh mice that harbor an N-ethyl-N-nitrosurea-induced allele (Curtin et al., 2003) (Fig. 3).
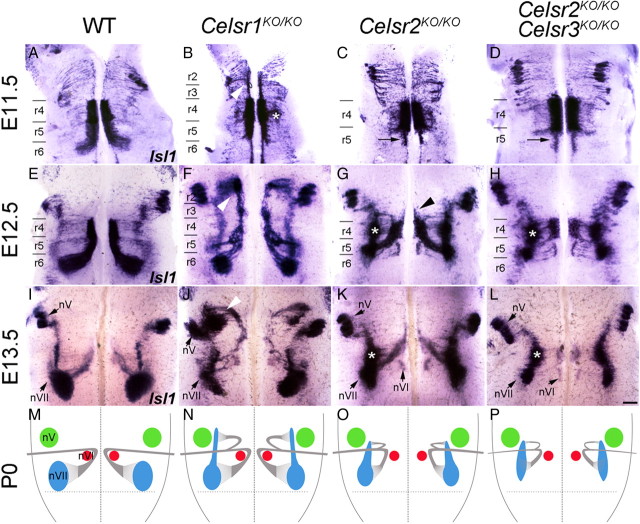
Figure 2.
Defective FBM neuron migration in Celsr1,2,3 KO mice. A–L, FBM neurons were visualized by ISH of flat-mounted hindbrain preparations with an Isl1 probe at E11.5 (A–D), E12.5 (E–H), and E13.5 (I–L) in WT, Celsr1KO/KO, Celsr2KO/KO, and double Celsr2KO/KO;Celsr3KO/KO mice. M–P, Drawings summarize the phenotypes at P0 (H&E and neurofilament staining) (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Celsr1KO/KO mutants are characterized by an ectopic, rostral migratory stream (B, arrowhead), and abnormal migration in lateral r4 (B, asterisk), whereas Celsr2KO/KO and Celsr2KO/KO;Celsr3KO/KO FBM neurons engage in premature lateral migration in r4–r5, forming lateral heterotopias (G, H, K, L, asterisks), and very few neurons migrate rostrally (G, arrowhead). At P0, the facial nerve genu is reduced in Celsr1KO/KO and completely abnormal in Celsr2 KO/KO and Celsr2KO/KO;Celsr3KO/KO mice in which axons do not loop around nVI (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). FBM neurons migrate normally in Celsr3KO/KO mice. Scale bar (in L) A–L, 200 μm.
Figure 3.
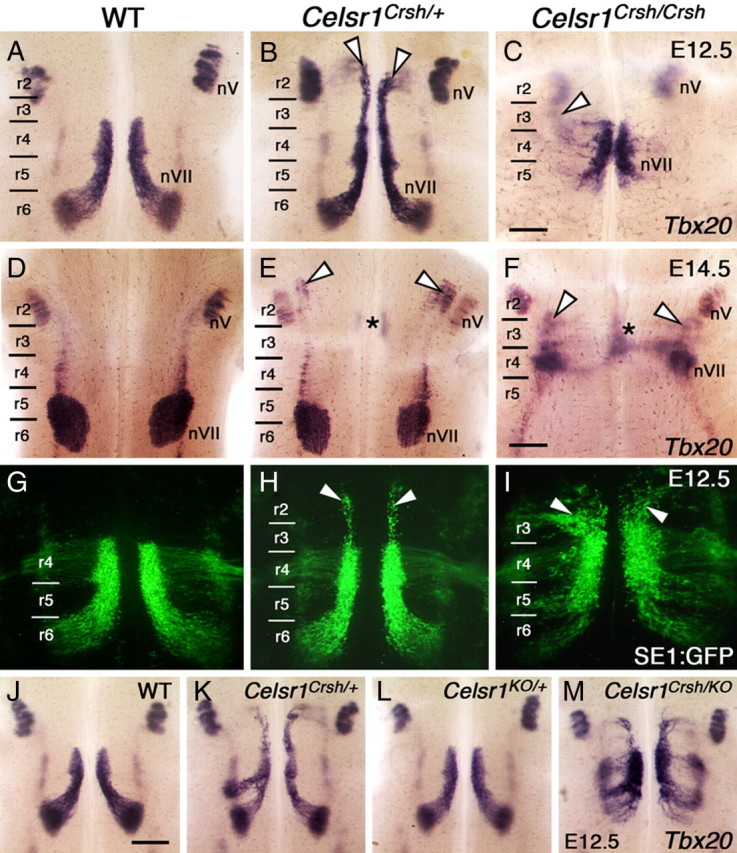
FBM neurons migrate rostrally in Celsr1Crsh embryos. A–M, Ventricular side (A–C, G–M) and pial side (D–F) views of flat-mounted hindbrains processed for ISH using Tbx20 (A–F, J–M) and imaging of neurons in SE1::GFP embryos (G–I). A, In a WT embryo at E12.5, FBM neurons are distributed throughout their migratory pathway spanning r4, r5, and r6. Within r6, the neurons migrate radially (laterally in this view) toward the pial surface to form the facial (nVII) nucleus. B, In a Celsr1Crsh/+ embryo, a large number of putative FBM neurons (arrowheads) migrate rostrally into r2 and r3. C, In a Celsr1Crsh/Crsh embryo, putative FBM neurons mostly remain in r4 or migrate into r5. A few Tbx20-expressing cells are found in r3 (out of focus, arrowhead). The trigeminal nucleus (nV) develops normally in all embryos. D, In a WT embryo at E14.5, the facial (nVII) nucleus spans r5 and r6, and the trigeminal (nV) nucleus is found in r2. E, In a Celsr1Crsh/+ embryo, the nVII nucleus forms but is smaller than in Celsr1+/+ siblings. In addition, ectopic Tbx20-expressing cells are found in r3 (asterisk) and r2 (arrowheads) adjacent to the nV neurons. F, In a Celsr1Crsh/Crsh embryo, a putative facial (nVII) nucleus forms in r4. Ectopic Tbx20-expressing cells are also found in medial (asterisk) and lateral (arrowheads) locations in the r2–r3 region. G, H, In SE1::GFP transgenic WT and Celsr1Crsh/+ embryos, the organization of FBM neurons is identical to that seen by Tbx20 in situs (A, B), with rostral migration in heterozygotes (arrowheads). I, In a Celsr1Crsh/Crsh embryo, the SE1::GFP reporter reveals extensive rostral FBM neuron migration into r3 and r2 (arrowheads), and few neurons migrate into r6. J–M, Siblings from a Celsr1KO /+ × Celsr1Crsh /+ cross. J, L, FBM neurons migrate normally in WT (J) and Celsr1KO/+ (L). K, Rostral neuron migration in Celsr1Crsh/+. M, In a Celsr1Crsh/KO transheterozygote, most FBM neurons remain in r4 and r5, with smaller numbers migrating into r2/r3 and r6. This phenotype is very similar to the migration pattern seen in Celsr1Crsh/Crsh (I) and Celsr1KO/KO (Fig. 2F). Scale bars: (in C), A–C, G–I, 300 μm; (in F) D–F, 375 μm; (in J), J–M, 375 μm.
We examined FBM neuron migration in E11.5–E13.5 embryos using ISH with an Islet1 (Isl1) probe, an established marker for FBM neurons (Garel et al., 2000; Pattyn et al., 2003). In wild-type (WT) embryos (n = 16) (Fig. 2A,E,I,M) at E11.5 (Fig. 2A), FBM neurons formed one longitudinal column at each side of the hindbrain in r4, where the facial nerve exits laterally. The rostral edge of the column was sharp, whereas FBM neurons migrated across the caudal border of r4 into r5 and r6. At E12.5 (Fig. 2E), FBM neurons were aligned in a migratory stream from r4 to r6, where they were engaged in lateral-dorsal and then radial migration, events that were essentially complete by E13.5, when a prominent facial motor nucleus (nVII) was clearly defined in lateral r6 (Fig. 2I).
In Celsr1 knock-out (Celsr1KO/KO) mice (n = 12) (Ravni et al., 2009) (Fig. 2B,F,J, N) at E11.5, some FBM neurons were confined to r4 and r5. Intriguingly, some also migrated rostrally into r3 and r2, a phenotype not seen previously in any vertebrate (Fritzsch, 1998; Chandrasekhar, 2004; Gilland and Baker, 2005). In addition, a cohort often migrated laterally in r4 (Fig. 2B, asterisk). While the rostral migration phenotype was fully penetrant, its expressivity varied among animals and even between the left and right sides of the body (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), perhaps reflecting stochastic variations as proposed for Fzd6 mutant skin phenotypes (Wang et al., 2006). At E12.5 (Fig. 2F), while the abnormal lateral and rostral migratory streams persisted, caudally directed FBM neurons moved through r5, medially to the abducens (nVI) nucleus, before migrating laterally in r6 like their WT counterparts. Hence, a facial (nVII) nucleus formed in its normal location in lateral r6 by E13.5. In addition, rostrally migrating FBM neurons subsequently migrated laterally to form an ectopic nucleus adjacent to the trigeminal (nV) nucleus (Fig. 2J, arrowhead). The nVII nucleus was examined at P0 and in adults using neurofilament and Nissl staining (supplemental Figs. S2, S3, available at www.jneurosci.org as supplemental material). In WT mice at postnatal day 0 (P0), axons of all FBM neurons loop around the abducens (supplemental Fig. S2A, available at www.jneurosci.org as supplemental material). In Celsr1KO/KO, only a part of the axons loop around the abducens (Fig S2B). At P21, a facial nerve (nVII) nucleus was found in normal position in the pons (data not shown), and clusters of FBM-like neurons were found ectopically at the level of the trigeminal (nV) motor nucleus (supplemental Fig. S3E, available at www.jneurosci.org as supplemental material), indicating that the rostrally migrated FBM neurons may be integrated into local circuits.
FBM neurons also migrated aberrantly in Celsr1Crsh mutants, which carry a missense mutation in the Celsr1 coding sequence resulting in one amino acid substitution in the eighth cadherin repeat (Curtin et al., 2003). Importantly, the defects were very similar to those observed in Celsr1KO embryos. In WT embryos, Tbx20 (Coppola et al., 2005; Song et al., 2006) was expressed by FBM neurons throughout their migratory pathway from r4 to r6 from E10.5 to E12.5 (3/3 embryos) (Fig. 3A) (data not shown) as defined by Isl1 expression (Fig. 2A,E). By E14.5, FBM neurons had completely migrated and formed the facial (nVII) nucleus in lateral-dorsal r6 (9/9 embryos) (Fig. 3D). In E12.5 Celsr1Crsh/+ heterozygotes, Tbx20 ISH revealed abnormal, rostrally migrating columns of putative FBM neurons in r2 and r3, in addition to caudally migrating neurons in r5 and r6 (6/6 embryos) (Fig. 3B). By E14.5, the caudally migrating FBM neurons had formed the nVII nucleus in dorsal r6, and the rostrally migrating cells also formed a putative nVII nucleus in dorsolateral r2, adjacent to the trigeminal (nV) nucleus (6/8 embryos) (Fig. 3E). In Celsr1Crsh/Crsh homozygotes, FBM neuron migration was greatly reduced compared to heterozygotes at E12.5 (2/2 embryos) (Fig. 3C) and E14.5 (5/5 embryos) (Fig. 3F). Nevertheless, by E14.5 an ectopic putative nVII nucleus was formed in r3 (2/5 embryos) (Fig. 3F) or a facial nucleus was seen in r5/r6 (3/5 embryos) (data not shown), indicating that FBM neurons in Celsr1Crsh/Crsh can migrate out of r4 in both directions in a similar fashion as Celsr1Crsh/+ and Celsr1KO/KO mutants.
We used additional tools and reagents to characterize the FBM neuron migration defects in Celsr1Crsh mutants. Neurofilament (NF-160) staining (Tiveron et al., 2003) revealed abnormal, rostrally migrating columns of putative FBM neurons in r2 and r3, in addition to caudally migrating neurons in r5 and r6 in Celsr1Crsh/+ and nonmigrated cells in r4 of Celsr1Crsh/Crsh (supplemental Fig. S4A–C). Despite aberrant neuronal migration, motor innervation patterns in the periphery were unaffected (supplemental Fig. S4D–F). Retrograde labeling of FBM neurons from the second arch using NeuroVue dyes (Fritzsch et al., 2005) stained cell bodies in r2 and r3, confirming the aberrant rostral migration of FBM neurons in Celsr1Crsh/+ (4/6 embryos) (supplemental Fig. S4H, available at www.jneurosci.org as supplemental material). In some Celsr1Crsh/Crsh, FBM neuron migration appeared completely blocked, with cells remaining confined to r4 (3/3 embryos) (supplemental Fig. S4I, available at www.jneurosci.org as supplemental material). To investigate further whether FBM neurons migrated in Celsr1Crsh/Crsh homozygotes, we examined mutant phenotypes in the SE1::GFP background, where all cranial motor neurons express GFP (Song et al., 2006). As expected, rostrally migrating FBM neurons were found in Celsr1Crsh/+ (12/12 embryos) (Fig. 3H; supplemental Fig. S5A,B, available at www.jneurosci.org as supplemental material). In Celsr1Crsh/Crsh, FBM neurons migrated rostrally and caudally out of r4 (5/5 embryos) (Fig. 3I; supplemental Fig. S5C,F, available at www.jneurosci.org as supplemental material), but they failed to form orderly streams of migrating cells as in WT (supplemental Fig. S5D, available at www.jneurosci.org as supplemental material) and Celsr1Crsh/+ embryos (supplemental Fig. S5E, available at www.jneurosci.org as supplemental material). Despite migration into ectopic locations, Celsr1 mutant FBM neurons did not undergo any significant apoptosis (supplemental Fig. S6, available at www.jneurosci.org as supplemental material).
Nature of the Celsr1KO and Celsr1Crsh alleles
The Celsr1Crsh/+ phenotype is similar to that of homozygous Celsr1KO/KO mutant mice, suggesting that either Celsr1KO encodes a hypomorphic allele or Celsr1Crsh has dominant-negative effect. In Celsr1 KO/KO mice, the mRNA level is reduced by 80%, indicating that the mutant RNA may be unstable (Ravni et al., 2009). Western blot analysis with an N-terminal antibody (supplemental Fig. S7, available at www.jneurosci.org as supplemental material) detected a positive band in WT and heterozygous (Celsr1KO/+), but not in homozygous (Celsr1KO/KO) brain extracts, clearly demonstrating that Celsr1KO is a null allele.
Given that the Celsr1KO allele is null, the Celsr1Crsh/+ phenotype could result from haploinsufficiency or a dominant-negative effect. To further investigate this, we crossed the Celsr1Crsh and Celsr1KO strains to examine the FBM neuron migration phenotype of Celsr1Crsh/KO transheterozygotes at E12.5 by using Tbx20 ISH (Fig. 3J–M). Rostrally and caudally migrating cells were seen in Celsr1Crsh/KO embryos (5/5), and the phenotype was very similar to that of Celsr1KO/KO mutants (compare Figs. 2F, 3M). Thus, while the Crsh allele behaves like a null in trans with the KO allele, only Celsr1Crsh/+, but not Celsr1KO/+, embryos exhibit migration defects, suggesting that the Celsr1Crsh allele is dominant negative.
FBM neurons migrate prematurely into lateral r4-r5 in Celsr2, Celsr2;Celsr3, and Fzd3 mutant mice
In zebrafish, knockdown of celsr1a,b has little effect on FBM neuron migration, whereas celsr2 loss of function leads to a severe phenotype (Wada et al., 2006). Therefore, we tested the roles of mouse Celsr2, and Celsr3 in FBM neuron migration. In Celsr2KO/KO (n = 27) (Fig. 2C,G,K,O), the caudal migration stream was severely truncated. From E12.5, the majority of mutant FBM neurons migrated prematurely into lateral r4 and r5 (Fig. 2G). In contrast to WT neurons, Celsr2KO/KO FBM neurons that migrated into r5 turned laterally rostral to the abducens nucleus and formed facial nuclei in lateral r5. Consequently, the neurons that migrated laterally within r4 and those that prematurely migrated laterally in r5 formed elongated facial nuclei spanning r4 and r5 (Fig. 2G, K). At postnatal stages, the facial nerve did not form a genu around nVI, since it looped laterally without turning around nVI (supplemental Fig. S2A,C).
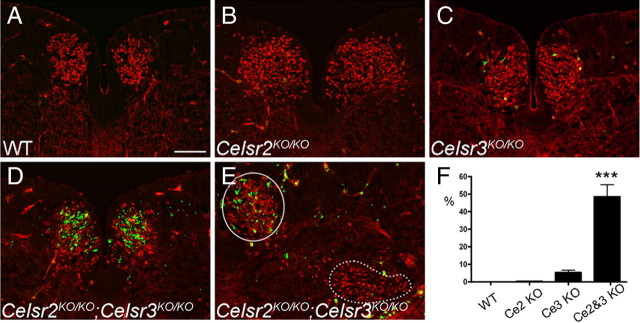
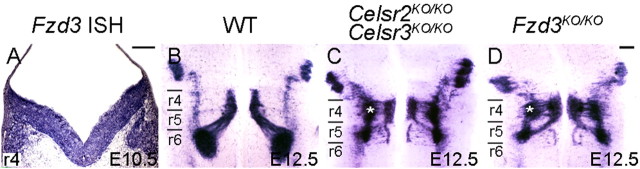
In Celsr3KO/KO mice (n = 8), FBM neurons migrated normally (data not shown). However, in Celsr2KO/KO;Celsr3KO/KO (n = 16), in addition to abnormalities seen in Celsr2KO/KO embryos, the facial nuclei were greatly reduced in size by E13.5 due to a marked reduction in the number of FBM neurons compared to their WT and Celsr2KO/KO counterparts (Fig. 2, compare L to I and K). To test whether apoptosis was involved, we performed activated caspase-3 immunostaining at E12.5, 1 d before atrophy of the Celsr2KO/KO;Celsr3KO/KO facial nucleus became evident. The number of apoptotic profiles in medial r4/r5 was considerably increased in Celsr2KO/KO;Celsr3KO/KO embryos compared with WT and Celsr2KO/KO embryos (Fig. 4A–D), and no apoptotic cells were detected among post-migrated FBM neurons in the facial nucleus (Fig. 4E, dotted area). This result suggests that the combined loss of Celsr2 and Celsr3 leads to cell death. To test whether neuronal death in double mutants could be due to axonal pathfinding defects, we carried out whole-mount staining of E10.5 embryos with anti-neurofilament antibodies (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). Cranial axon outgrowth and cranial ganglia development in Celsr2KO/KO;Celsr3KO/KO embryos were indistinguishable from those in WT and Celsr2KO/KO mutant embryos, suggesting that cell death is not due to defective axonal outgrowth. Hematoxylin and eosin (H&E) and neurofilament staining at P0 confirmed that FBM neuron loss in Celsr2KO/KO;Celsr3KO/KO mice resulted in a diminutive nVII nucleus that was displaced to a rostral position (supplemental Fig. S3C, available at www.jneurosci.org as supplemental material). Given the importance of Fzd3 in PCP, we examined its expression using ISH. Fzd3 was widely expressed in the hindbrain, both in neuroepithelial and postmitotic cells, including FBM neurons (Fig. 5A). In homozygous Fzd3 mutant (Fzd3KO/KO) mice, FBM neurons migrated prematurely in lateral r4 and r5 and formed a very diminutive nVII, as seen in Celsr2KO/KO;Celsr3KO/KO mice (Fig. 5B–D) (Vivancos et al., 2009). The observation that Celsr3KO alone generates no phenotype but enhances the Celsr2 KO phenotype, together with the observation that this phenotype is mimicked by Fzd3KO, suggests strongly that Celsr2 and Celsr3 act redundantly during FBM neuron migration through mechanisms that probably involve Fzd3.
Figure 4.
Celsr2 and Celsr3 are required for FBM neuron survival. A–F, FBM neurons shown with anti-Isl1 (red) were stained for activated (cleaved) caspase-3 (green) (A–E) at E12.5. No apoptotic cells were found in WT (A) and Celsr2KO/KO mice (B). In Celsr3KO/KO (C), a few FBM neurons were positive for activated caspase-3. In Celsr2KO/KO;Celsr3KO/KO (D–F), the number of caspase-3 positive cells increased dramatically in FBM neurons located in the medial region (circle), but not when they finish lateral migration (dotted contour). F, Cell death, estimated as the ratio of green to red signals (ImageJ software), was 5% in Celsr3KO/KO and 50% in Celsr2KO/KO;Celsr3KO/KO samples. ***p < 0.001, Student's t test, Bonferroni correction. Error bars are SD. Scale bar (in A) A–E, 100 μm.
Figure 5.
FBM neuron migration in Fzd3 KO mice. A, Coronal section of E10.5 mouse hindbrain at the level of r4 processed for ISH with a Fzd3 probe. Fzd3 mRNA is found in both neuroepithelial cells and postmitotic cells, including FBM neurons. B–D, E12.5 hindbrains hybridized with an Isl1 riboprobe. Abnormalities of FBM neurons migration in Fzd3KO/KO mutant mice (D) phenocopy those in double Celsr2KO/KO;Celsr3KO/KO mice (C). Note the premature lateral migration in r4 (asterisk). Scale bars, A (for A), D (for B–D), 200 μm.
Mutations in Celsr genes do not affect rhombomere identity or neuronal specification
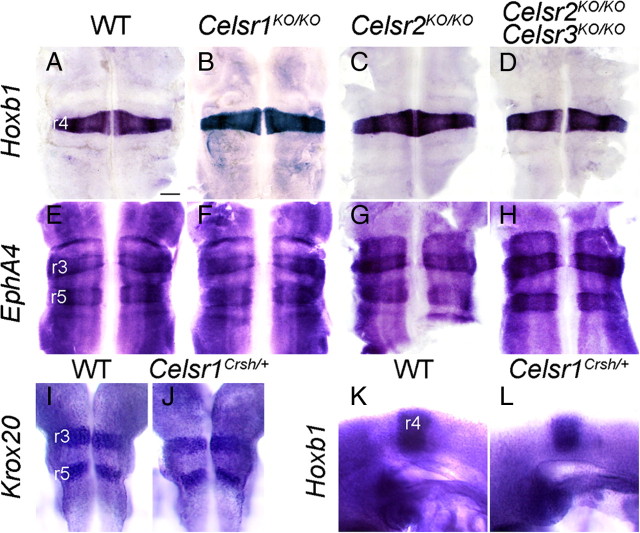
To test whether mutations in Celsr genes affect rhombomere specification and patterning, we performed ISH at E10.5 with Hoxb1 (Fig. 6A–D) and Eph4 (Fig. 6E–H), which label r4 and r3 plus r5, respectively (Murphy et al., 1989; Gilardi-Hebenstreit et al., 1992). The specification of r3-r6 and rhombomeric borders appeared normal in all mutants studied. Similarly, expression of Krox20 in r3 and r5, and of Hoxb1 in r4, was unaffected in Celsr1Crsh/+ mice (Krox20, n = 5; Hoxb1, n = 3) (Fig. 6I–L). Together, these results indicate that defective migration of FBM neurons in the Celsr mutants does not result from aberrant specification of rhombomere identity.
Figure 6.
Normal hindbrain patterning in Celsr mutant mice. A–H, Hindbrains at E10.5 were processed by ISH with Hoxb1 (A–D) to label r4 and Eph4A (E–H) to label r3 and r5. As compared with the WT (A, E), no difference was seen in Celsr1KO/KO (B, F), Celsr2 KO/KO (C, G), or Celsr2KO/KO;Celsr3KO/KO (D, H) mice. I–J, Dorsal views of E9.5 hindbrains from WT (I) and Celsr1Crsh/+ (J) hybridized with Krox20 probe. K, L, Lateral view of whole-mount embryos WT (K) and Celsr1Crsh/+ (L) hybridized with Hoxb1 probe. As in Celsr1KO/KO, Celsr2 KO/KO, and Celsr2KO/KO;Celsr3KO/KO, the patterning of r3–r5 is preserved in Celsr1Crsh/+. Scale bar (in A) A–L, 200 μm.
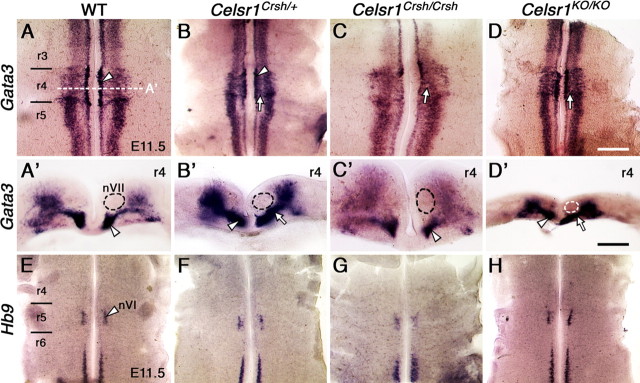
Another possibility is that many FBM neurons fail to migrate in Celsr mutants because they become misspecified as nonmigratory neurons. To test this, we examined the expression of Gata3, which marks inner ear efferent neurons that are specified in but do not migrate out of r4 (Karis et al., 2001), and Hb9, which identifies somatic motor neurons such as the abducens (nVI) neurons in r5 (Thaler et al., 1999). In r4 of E11.5 WT embryos, there was a medial domain of high Gata3-expressing cells adjacent to the floor plate, a lateral domain of weaker expressing cells, and a narrow domain ventral to the FBM neurons connecting the two (Fig. 7A, A′). In all three backgrounds with defective FBM neuron migration (Celsr1Crsh/+, Celsr1Crsh/Crsh, and Celsr1KO/KO mutants), there was a marked increase in the number of Gata3-expressing cells in r4 (Fig. 7B–D). However, cross-sections revealed that the excess cells were located either in the lateral domain or the connecting domain and not within the FBM neuron domain (Fig. 7B′–D′), suggesting that non-migratory FBM neurons are not misspecified as Gata3-expressing cells. Similarly, Hb9 was not expressed ectopically in r4 in nonmigratory FBM neurons in any of the mutant backgrounds (Fig. 7E–H), suggesting that the neuronal migration defects are not caused by transdifferentiation of FBM neurons into non-migratory cell types.
Figure 7.
Normal neuronal specification in Celsr1Crsh and Celsr1KO mutants. Ventricular side views (A–H) and cross-sections (A′–D′) of flat-mounted E11.5 hindbrains processed for ISH of Gata3 (A–D, A′–D′) and Hb9 (E–H) expression. A′–D′ show cross-sections in r4 of the respective embryos (A–D) at the approximate level indicated in A. Thickness of the hindbrain tissue varied considerably between embryos (compare C′ and D′) due to genotype and slight stage differences. A, In a WT embryo two domains of Gata3 expression are evident, a high expression medial domain (arrowhead) and a weaker-expressing lateral domain. The cross-section at the r4 level (A′) shows the inner ear efferent (IEE) neurons (arrowhead) with the position of FBM (nVII) neurons outlined in black, adjacent to the Gata3-expressing cells. B, In a Celsr1Crsh/+ embryo, many Gata3-expressing cells (arrow) bridge the gap between the two expression domains in r4. Cross-section in r4 (B′) reveals a continuous column (arrow) connecting the IEE domain (arrowhead) to the lateral domain, but these cells are outside the region of FBM neurons (black outline). C, D, In Celsr1Crsh/Crsh and Celsr1KO/KO embryos, Gata3-expressing cells (arrows) found in between the two expression domains are located outside the region of the FBM neurons (outlines in C′ and D′). E–H, There is a bilateral cluster of Hb9-expressing cells (arrowhead) in r5, corresponding to the abducens motor (nVI) nucleus in WT (E) and Celsr1-deficient embryos (F–H). Importantly, there are no ectopic Hb9-expressing cells in r4 in the mutants (G, H), even though there are a large number of nonmigrated FBM neurons in these embryos. Scale bars: (in D) A–H, 400 μm; (in D′), A′–D′, 200 μm.
Celsr1 and Celsr2 and 3 define a dual control of FBM neuron migration
The observation that Celsr1 is expressed in neuroepithelial cells, including FBM neuron precursors, but not in postmitotic neurons, suggests that the function of Celsr1 may be extrinsic to FBM neurons. To test this, we crossed conditional Celsr1fl mutants (Ravni et al., 2009) with Isl1-Cre mice (Isl1tm1(cre)Sev) (Yang et al., 2006) that express the Cre recombinase very early and efficiently in postmitotic motor neurons to generate Celsr1|Isl1 embryos in which Celsr1 should be inactivated in FBM neurons as soon as they exit the cell cycle. None of the ectopic migration streams evident in constitutive Celsr1KO/KO mutants were present in Celsr1|Isl1 embryos in which FBM neurons migrated normally (Fig. 8A–C), suggesting that Celsr1 regulates FBMN migration in a non-cell-autonomous manner.
Figure 8.
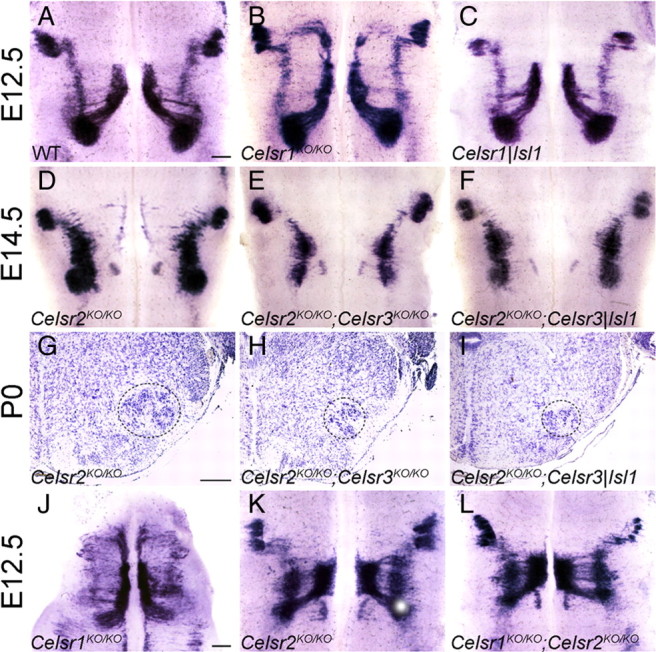
Cell autonomy and epitasis in the Celsr gene family. A–C, In Celsr1|Isl1 conditional KO mice (C), no rostral migration of FBM neurons was seen, a situation similar to that of WT (A) but unlike Celsr1KO/KO constitutive mutants (B) (E12.5, Isl1 ISH). Scale bar (in A) A–F, 200 μm. D–I, Compared to Celsr2 KO/KO (D) hindbrain, the number of FBM neurons is similarly reduced in double Celsr2 KO/KO;Celsr3 KO/KO (E) and double Celsr2 KO/KO;Celsr3|Isl1 (F) (E14.5, Isl1 ISH). This is confirmed in Nissl-stained coronal sections at P0. The number of FBM neurons (dotted contour) is diminished in double Celsr2 KO/KO;Celsr3 KO/KO (H), and double Celsr2KO/KO;Celsr3|Isl1 (I) as compared with single Celsr2 KO/KO (G). Scale bar: (in G) G–I, 200 μm. J–L, Unlike in Celsr1 KO/KO (J), no rostral migration occurs in Celsr1 KO/KO;Celsr2 KO/KO double-mutant mice (L). Instead, premature FBM neuron lateral migration is seen as in Celsr2 KO/KO (K) (E12.5, Isl1 ISH). Scale bar: (in J) J–L, 200 μm.
Similarly, to determine the cell autonomy of the Celsr3KO phenotype, we produced Celsr2KO/KO;Celsr3|Isl1 mutant mice in which Celsr2 is inactivated constitutively and Celsr3 is inactivated only in FBM neurons upon Isl1-CRE expression. Comparative histological examination at E14.5 (Fig. 8D–F) and P0 (Fig. 8G–I) showed that the facial nucleus of Celsr2KO/KO; Celsr3|Isl1 mutant mice was atrophic and morphologically indistinguishable from that in Celsr2KO/KO;Celsr3KO/KO and different from that of Celsr2KO/KO mutants, which was dispersed along the rostrocaudal axis but not atrophic. This indicated that the action of Celsr3 is cell autonomous. Both the phenotypic differences between Celsr1KO/KO and Celsr2KO/KO;Celsr3KO/KO mice and the differences in cell autonomy suggest that Celsr1 on one hand and Celsr2 and Celsr3 on the other hand regulate two different aspects of FBM neuron migration.
Do Celsr1 and Celsr2 act on FBM neurons in independent manner? To test for a putative interaction between the two proteins, we produced Celsr1KO/KO;Celsr2 KO/KO double-mutant embryos and studied the migration of FBM neurons and the development of facial motor nucleus. FBM neurons underwent premature migration in lateral r4-r5, but almost no ectopic rostral migration was seen (n = 8) (Fig. 8L). This phenotype was indistinguishable from that in Celsr2KO/KO embryos (Fig. 8K) but unlike that observed in Celsr1KO/KO (Fig. 8J). These data show that the phenotype of Celsr2 mutation masks that of Celsr1 mutation, indicating that Celsr2 is epistatic to Celsr1.
Discussion
Our results show that Celsr1, Celsr2, Celsr3, and Fzd3 play critical roles in FBM neuron migration in mice. In zebrafish, celsr2, celsr1a, and celsr1b regulate FBM neuron migration out of r4 such that combined loss of function results in a block of caudal FBM neurons migration, with all neurons confined to r4 (Wada et al., 2006). In contrast, we show that in the mouse Celsr1 on one hand and Celsr2,3 and Fzd3 on the other regulate FBM neuron migration in two different ways: Celsr1 helps specify the caudal direction of FBM neuron migration in a non-cell-autonomous manner, whereas Celsr2,3 and Fzd3 regulate the trajectory that FBM neurons follow from r4 to r6, as well as their survival. Celsr3 acts in a cell-autonomous manner and Celsr2 is epistatic to Celsr1.
Celsr1 regulates directionality of FBM neuron migration
When Celsr1 function is compromised, FBM neurons can still migrate caudally out of r4, but many move rostrally. Isl1- and Tbx20-expressing cells display a continuous distribution in medial r2-r4. The size of the facial nucleus in r6 is diminished when the number of these cells increases, whereas that of the trigeminal motor nucleus is comparable to its wild-type counterpart. Altogether, these data suggest that Isl1- or Tbx20-positive cells located in medial r2-r4 are not transfated trigeminal motor neurons. All mouse and zebrafish mutants studied thus far exhibit complete or partial block of FBM neuron caudal migration, but never direction reversal (Chandrasekhar, 2004; Cooper et al., 2005; Wada et al., 2005; Nambiar et al., 2007; Rohrschneider et al., 2007; Song, 2007). Interestingly, only a subset of FBM neurons migrates rostrally in Celsr1 mutants. One possibility is that Celsr1 functions redundantly with other molecules to specify directionality. Alternatively, the Celsr1 mutant phenotype may reflect heterogeneity within the FBM neuron population such that only a subset is depends on Celsr1 for specification of caudal direction of migration. Intriguingly, in zebrafish only the earliest migrating FBM neurons express Tag1 protein (Sittaramane et al., 2009), suggesting that there may be molecular heterogeneity among FBM neurons. Whether such heterogeneity could underlie differential responses to Celsr1 remains to be explored. Nevertheless, the role for Celsr1 in regulating the caudal direction of migration is particularly significant, given that no other gene has been shown to perform a similar function.
Our observations suggesting that the Celsr1Crsh allele encodes a protein with a dominant-negative activity is supported by analysis of the skin and cultured keratinocytes in Celsr1Crsh mutant mice (Devenport and Fuchs, 2008). In the skin, homophilic interactions between Celsr1 molecules appear to drive PCP signaling. The mutant protein encoded by Celsr1Crsh is produced but no longer polarized along the anterior posterior axis, which perturbs the subcellular distribution of the PCP proteins Stbm/Vangl2 and Fzd6. Furthermore, Celsr1 and Vangl2 interact physically (Devenport and Fuchs, 2008), and this interaction is abolished by the Looptail mutation in Vangl2, which disrupts PCP (Kibar et al., 2001; Murdoch et al., 2001; Montcouquiol et al., 2003), and blocks FBM neuron migration (Vivancos et al., 2009) (D. M. Glasco, B. Fritzsch, J. N. Murdoch, and A. Chandrasekhar, manuscript in preparation). Altogether, our observations in Celsr1 mutants suggest that PCP genes of the Celsr, Fzd, and Vangl families may regulate interactions between cells and thereby specify directionality in cell migration along the rostrocaudal axis (Goodrich, 2008). It would be instructive to examine the subcellular distribution of PCP proteins in FBM neuron precursors in WT and Celsr1 mutant mice, as well as the role of other PCP genes such as Fzd6 in FBM neuron migration.
Celsr1 regulates migration of FBM neurons in a non-cell-autonomous manner
Celsr1 is expressed broadly in ventricular zone precursors of the developing hindbrain (Tissir et al., 2002), and our data demonstrate a particularly high expression in the ventricular zone where FBM neurons are generated. To probe the cell autonomy of Celsr1 function in FBM neuron migration, we used the Isl1-Cre line to specifically inactivate Celsr1 in these cells. In contrast to Celsr1KO/KO mice, FBM neurons migrated normally in Celsr1|Isl1 mice. Although an FBM neuron-autonomous effect of residual Celsr1 protein inherited from precursors cannot be formally excluded, the fact that no Celsr1 protein was detected in early FBM neurons clearly indicates non-cell-autonomous action. Thus, Celsr1 specifies caudal movement of FBM neurons indirectly by acting in neuroepithelial precursors or in radial cells, both of which have their cell bodies in ventricular zones. In line with this, in zebrafish PCP proteins Frizzled3a and Celsr2 also regulate migration of FBM neurons non-cell autonomously, probably through their function in the neuroepithelium (Wada et al., 2006). Although the role of radial neuroepithelial cells as a substrate to guide radially migrating neurons is well known, it is more difficult to figure out how they could regulate tangential migration. The non-FBM neuron-autonomous action of Celsr1 implies that Celsr1, expressed in radial cells but not in FBM neurons, would mediate heterophilic interactions, which runs counter to evidence implicating Celsr and Flamingo cadherins in homophilic interactions (Takeichi, 2007). We deem it more likely that Celsr1 acts at the level of FBM neuron precursors, even though the mechanism whereby an action in precursors can impact the migration of daughter cells is unclear. Crosses to inactivate Celsr1 specifically in FBM neurons precursors (Nkx6.2-Cre) or in r4, where FBM neurons are generated (Hoxb1-Cre), might be useful to test this hypothesis. In the zebrafish neural tube, rostrocaudal PCP is lost during and re-established after division of precursors, and this transient loss of planar polarity is considered important for neural tube closure (Ciruna et al., 2006). FBM neurons are generated almost synchronously at E9.5–E10.5 (Taber-Pierce, 1973; Goffinet, 1984). One may hypothesize that rostrocaudal PCP, acting during the last division, may impact the migration of daughter cells. For example, the orientation of the mitotic spindle could influence the intracellular location of the centrosome or the Golgi in daughter cells, which would in turn specify the direction of migration. Consistent with this view, the asymmetric distribution of Leu-Gly-Asn and Numa, two proteins involved in spindle orientation, is lost in neuron progenitors that are defective in the PCP protein Vangl2 (Lake and Sokol, 2009). It would be interesting to compare the planar orientation of mitotic spindles in r4 and/or the localization of the Golgi or centrosome in early FBM neurons at E9.5–E10.5 in WT and Celsr1 mutant mice.
Roles of Celsr2, Celsr3 and Fzd3 in FBM neuron migration
Inactivation of Celsr2 reduces severely the caudal movement of FBM neurons, and added inactivation of Celsr3 impacts further on their migration and survival, indicating redundancy between the two proteins. The phenotype of the double Celsr2,3 KO mice closely resembles that of Fzd3KO, as shown also by others (Vivancos et al., 2009), and of Mash1,Math3 double KO mice (Ohsawa et al., 2005). The redundant role of Celsr2 and Celsr3 in FBM neuron migration and the probable association with Fzd3 fits in with similar roles proposed in zebrafish (Wada et al., 2006). It is also consistent with their synergistic role in axon guidance (Wang et al., 2002; Tissir et al., 2005, 2010). The Celsr2,3-Fzd3 system regulates FBM neuron migration by promoting their locomotion along the midline until they pass the abducens nucleus, after which they initiate their lateral displacement. Importantly, mutant neurons that fail to reach r5-r6 remain able to move laterally and differentiate locally into neurons with similar features as FBM neurons. The mediolateral displacement may be a default behavior of differentiated FBM neurons in response to as yet unidentified attractive cues expressed laterally, such as Wnt or Bmp proteins, and/or repulsive signals from the midline region, such as Shh or Slit factors (Guthrie, 2007).
As in Celsr2 mutant mice, almost no caudal FBM neuron migration occurs in chick and some other nonmammalian vertebrates where the facial (nVII) nucleus forms normally but facial axons do not loop around nVI nucleus (Studer, 2001; Gilland and Baker, 2005). Intriguingly, the chicken genome contains Celsr1 and Celsr3 but lacks Celsr2, indicating that loss of Celsr2 might be the origin of the peculiar anatomy of nVII and its axons in birds.
Whereas Celsr2 and Celsr3 act redundantly, combined mutations of Celsr1 and Celsr2 phenocopy the Celsr2KO/KO phenotype. The fact that Celsr2 is epistatic to Celsr1 provides additional support to the view that two genes regulate FBM neurons migration in specific, sequential manners. The Celsr1 system may regulate planar orientation of precursor divisions, which could provide a directionality cue to the FBM neurons. The second system, involving Celsr2, Celsr3, and Fzd3 would be required for the acquisition of a rostrocaudal migratory phenotype and for maintenance of FBM neurons. For example, activation of this pathway may allow FBM neurons to respond to Wnt proteins (Vivancos et al., 2009). Studies of FBM neuron migration in other PCP mutants and the use of in vitro systems that faithfully recapitulate migration events (Vivancos et al., 2009) may provide ways to test these models further.
Footnotes
This work was supported by grants from the following: Actions de Recherches Concertées (ARC-186), Fonds de la Recherche Fondamentale Collective (FRFC 2.4504.01), Fonds de la Recherche Scientifique Médicale (FRSM 3.4501.07), Interuniversity Poles of Attraction (SSTC, PAI p6/20), Région Wallonne, and the Fondation Médicale Reine Elisabeth, all from Belgium, (to F.T. and A.M.G.); University of Missouri Research Board (RB07-03) and National Institutes of Health (NS040449) (to A.C.). Y.Q. has a FRIA PhD fellowship and F.T. is a research associate at the Belgian National Fund for Scientific Research. We thank Jeremy Nathans for providing Fzd3 mutant mice, Sonia Garel for providing Hoxb1 and EphA4 probes, Michele Studer for providing Tbx20 probe, and Elaine Fuchs for anti-Celsr1 antibody. D.M.G. and A.C. are indebted to Angelo Iulianella and Paul Trainor (Stowers Institute, Kansas City, MO) for training in several mouse embryological techniques and for providing Hoxb1 and Krox20 probes. D.G. thanks the Molecular Cytology Core facility (University of Missouri, Columbia, MO) for imaging assistance.
References
- Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. The Zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Concha ML, Takeuchi M, Ueno N, Wilson SW, Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–224. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Armstrong J, Moens CB. Zebrafish foggy/spt 5 is required for migration of facial branchiomotor neurons but not for their survival. Dev Dyn. 2005;234:651–658. doi: 10.1002/dvdy.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola E, Pattyn A, Guthrie SC, Goridis C, Studer M. Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 2005;24:4392–4403. doi: 10.1038/sj.emboj.7600897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM, Nolan PM, Steel KP, Brown SD, Gray IC, Murdoch JN. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol. 2003;13:1129–1133. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- Devenport D, Fuchs E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol. 2008;10:1257–1268. doi: 10.1038/ncb1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone CJ, Little PF. The flamingo-related mouse Celsr family (Celsr1-3) genes exhibit distinct patterns of expression during embryonic development. Mech Dev. 2001;109:91–94. doi: 10.1016/s0925-4773(01)00515-9. [DOI] [PubMed] [Google Scholar]
- Formstone CJ, Moxon C, Murdoch J, Little P, Mason I. Basal enrichment within neuroepithelia suggests novel function(s) for Celsr1 protein. Mol Cell Neurosci. 2010;44:210–222. doi: 10.1016/j.mcn.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. Of mice and genes: evolution of vertebrate brain development. Brain Behav Evol. 1998;52:207–217. doi: 10.1159/000006564. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Muirhead KA, Feng F, Gray BD, Ohlsson-Wilhelm BM. Diffusion and imaging properties of three new lipophilic tracers, NeuroVue Maroon, NeuroVue Red and NeuroVue Green and their use for double and triple labeling of neuronal profile. Brain Res Bull. 2005;66:249–258. doi: 10.1016/j.brainresbull.2005.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Garcia-Dominguez M, Charnay P. Control of the migratory pathway of facial branchiomotor neurones. Development. 2000;127:5297–5307. doi: 10.1242/dev.127.24.5297. [DOI] [PubMed] [Google Scholar]
- Gilardi-Hebenstreit P, Nieto MA, Frain M, Mattei MG, Chestier A, Wilkinson DG, Charnay P. An Eph-related receptor protein tyrosine kinase gene segmentally expressed in the developing mouse hindbrain. Oncogene. 1992;7:2499–2506. [PubMed] [Google Scholar]
- Gilland E, Baker R. Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav Evol. 2005;66:234–254. doi: 10.1159/000088128. [DOI] [PubMed] [Google Scholar]
- Goffinet AM. Abnormal development of the facial nerve nucleus in reeler mutant mice. J Anat. 1984;138:207–215. [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV. The plane facts of PCP in the CNS. Neuron. 2008;60:9–16. doi: 10.1016/j.neuron.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Hawkins C, Nathans J. Frizzled6 controls hair patterning in mice. Proc Natl Acad Sci U S A. 2004;101:9277–9281. doi: 10.1073/pnas.0402802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8:859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Topczewski J, Bingham S, Sepich DS, Marlow F, Chandrasekhar A, Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, Fritzsch B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::aid-cne8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Vogan KJ, Groulx N, Justice MJ, Underhill DA, Gros P. Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nat Genet. 2001;28:251–255. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- Lake BB, Sokol SY. Strabismus regulates asymmetric cell divisions and cell fate determination in the mouse brain. J Cell Biol. 2009;185:59–66. doi: 10.1083/jcb.200807073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- Marin F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127:4925–4935. doi: 10.1242/dev.127.22.4925. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Kelley MW. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurosci. 2003;23:9469–9478. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- Murdoch JN, Doudney K, Paternotte C, Copp AJ, Stanier P. Severe neural tube defects in the loop-tail mouse result from mutation of Lpp1, a novel gene involved in floor plate specification. Hum Mol Genet. 2001;10:2593–2601. doi: 10.1093/hmg/10.22.2593. [DOI] [PubMed] [Google Scholar]
- Murphy P, Davidson DR, Hill RE. Segment-specific expression of a homoeobox-containing gene in the mouse hindbrain. Nature. 1989;341:156–159. doi: 10.1038/341156a0. [DOI] [PubMed] [Google Scholar]
- Nambiar RM, Ignatius MS, Henion PD. Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech Dev. 2007;124:682–698. doi: 10.1016/j.mod.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R. Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci. 2005;25:5857–5865. doi: 10.1523/JNEUROSCI.4621-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Ogawa M, Takeuchi K, Takahashi S, Kulkarni AB, Mikoshiba K. Cyclin-dependent kinase 5/p35 contributes synergistically with Reelin/Dab1 to the positioning of facial branchiomotor and inferior olive neurons in the developing mouse hindbrain. J Neurosci. 2002;22:4036–4044. doi: 10.1523/JNEUROSCI.22-10-04036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Sander M, Ericson J. Complementary roles for Nkx6 and Nkx2 class proteins in the establishment of motoneuron identity in the hindbrain. Development. 2003;130:4149–4159. doi: 10.1242/dev.00641. [DOI] [PubMed] [Google Scholar]
- Ravni A, Qu Y, Goffinet AM, Tissir F. Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J Invest Dermatol. 2009;129:2507–2509. doi: 10.1038/jid.2009.84. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Shima Y, Copeland NG, Gilbert DJ, Jenkins NA, Chisaka O, Takeichi M, Uemura T. Differential expression of the seven-pass transmembrane cadherin genes Celsr1-3 and distribution of the Celsr2 protein during mouse development. Dev Dyn. 2002;223:321–332. doi: 10.1002/dvdy.10054. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kengaku M, Hirano T, Takeichi M, Uemura T. Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev Cell. 2004;7:205–216. doi: 10.1016/j.devcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Lewcock JW, Lettieri K, Pfaff SL. FGF as a target-derived chemoattractant for developing motor axons genetically programmed by the LIM code. Neuron. 2006;50:841–853. doi: 10.1016/j.neuron.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Sittaramane V, Sawant A, Wolman MA, Maves L, Halloran MC, Chandrasekhar A. The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev Biol. 2009;325:363–373. doi: 10.1016/j.ydbio.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR. Moving cell bodies: understanding the migratory mechanism of facial motor neurons. Arch Pharm Res. 2007;30:1273–1282. doi: 10.1007/BF02980268. [DOI] [PubMed] [Google Scholar]
- Song MR, Shirasaki R, Cai CL, Ruiz EC, Evans SM, Lee SK, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M. Initiation of facial motoneurone migration is dependent on rhombomeres 5 and 6. Development. 2001;128:3707–3716. doi: 10.1242/dev.128.19.3707. [DOI] [PubMed] [Google Scholar]
- Taber-Pierce E. Time of origin of neurons in the brainstem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Expression of planar cell polarity genes during development of the mouse CNS. Eur J Neurosci. 2006;23:597–607. doi: 10.1111/j.1460-9568.2006.04596.x. [DOI] [PubMed] [Google Scholar]
- Tissir F, De-Backer O, Goffinet AM, Lambert de Rouvroit C. Developmental expression profiles of Celsr (Flamingo) genes in the mouse. Mech Dev. 2002;112:157–160. doi: 10.1016/s0925-4773(01)00623-2. [DOI] [PubMed] [Google Scholar]
- Tissir F, Bar I, Jossin Y, De Backer O, Goffinet AM. Protocadherin Celsr3 is crucial in axonal tract development. Nat Neurosci. 2005;8:451–457. doi: 10.1038/nn1428. [DOI] [PubMed] [Google Scholar]
- Tissir F, Qu Y, Montcouquiol M, Zhou L, Komatsu K, Shi D, Fujimori T, Labeau J, Tyteca D, Coutoy P, Poumay Y, Uemura T, Goffinet AM. Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat Neurosci. 2010;13:700–707. doi: 10.1038/nn.2555. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Pattyn A, Hirsch MR, Brunet JF. Role of Phox2b and Mash1 in the generation of the vestibular efferent nucleus. Dev Biol. 2003;260:46–57. doi: 10.1016/s0012-1606(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Vivancos V, Chen P, Spassky N, Qian D, Dabdoub A, Kelley M, Studer M, Guthrie S. Wnt activity guides facial branchiomotor neuron migration, and involves the PCP pathway and JNK and ROCK kinases. Neural Dev. 2009;4:7. doi: 10.1186/1749-8104-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Iwasaki M, Sato T, Masai I, Nishiwaki Y, Tanaka H, Sato A, Nojima Y, Okamoto H. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 2005;132:2273–2285. doi: 10.1242/dev.01810. [DOI] [PubMed] [Google Scholar]
- Wada H, Tanaka H, Nakayama S, Iwasaki M, Okamoto H. Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development. 2006;133:4749–4759. doi: 10.1242/dev.02665. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nathans J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 2007;134:647–658. doi: 10.1242/dev.02772. [DOI] [PubMed] [Google Scholar]
- Wang Y, Thekdi N, Smallwood PM, Macke JP, Nathans J. Frizzled-3 is required for the development of major fiber tracts in the rostral CNS. J Neurosci. 2002;22:8563–8573. doi: 10.1523/JNEUROSCI.22-19-08563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Badea T, Nathans J. Order from disorder: Self-organization in mammalian hair patterning. Proc Natl Acad Sci U S A. 2006;103:19800–19805. doi: 10.1073/pnas.0609712104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Bar I, Achouri Y, Campbell K, De Backer O, Hebert JM, Jones K, Kessaris N, de Rouvroit CL, O'Leary D, Richardson WD, Goffinet AM, Tissir F. Early forebrain wiring: genetic dissection using conditional Celsr3 mutant mice. Science. 2008;320:946–949. doi: 10.1126/science.1155244. [DOI] [PMC free article] [PubMed] [Google Scholar]