Abstract
Purpose
To investigates the impact of non-rigid motion correction on pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI in patients with solitary pulmonary nodules (SPNs). Misalignment of focal lesions due to respiratory motion in free-breathing dynamic contrast-enhanced MRI (DCE-MRI) precludes obtaining reliable time-intensity curves, which are crucial for pharmacokinetic analysis for tissue characterization.
Materials and Methods
Single-slice 2D DCE-MRI was obtained in 15 patients. Misalignments of SPNs were corrected using non-rigid B-spline image registration. Pixel-wise pharmacokinetic parameters Ktrans, ve and kep were estimated from both original and motion-corrected DCE-MRI by fitting the two-compartment pharmacokinetic model to the time-intensity curve obtained in each pixel. The “goodness-of-fit” was tested with χ2-test in pixel-by-pixel basis to evaluate the reliability of the parameters. The percentages of reliable pixels within the SPNs were compared between the original and motion-corrected DCE-MRI. In addition, the parameters obtained from benign and malignant SPNs were compared.
Results
The percentage of reliable pixels in the motion-corrected DCE-MRI was significantly larger than the original DCE-MRI (p=4×10−7). Both Ktrans and kep derived from the motion-corrected DCE-MRI showed significant differences between benign and malignant SPNs (p=0.024, 0.015).
Conclusion
The study demonstrated the impact of non-rigid motion correction technique on pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI in SPNs.
Keywords: dynamic contrast-enhanced MRI, pharmacokinetic analysis, non-rigid image registration, motion correction, solitary pulmonary nodule
INTRODUCTION
With the progress in fast MRI technology, dynamic contrast-enhanced MRI (DCE-MRI) is becoming one of practical diagnostic tools for tissue characterization. Unlike conventional T1- or T2-weighted MRI, DCE-MRI allows evaluation of information of the response to administration of contrast agents in localized lesions as changes of signal intensity over time, referred as time-intensity curves. According to the two-compartment pharmacokinetic model (1), the time-intensity curves in DCE-MRI are considered to reflect tissue properties that potentially correlate with the malignancy of localized lesions e.g. microvascular vessel wall permeability and extravascular-extracellular volume fraction. This hypothesis has been motivating researchers to investigate pharmacokinetic analysis of DCE-MRI, which estimates parameters representing the abovementioned tissue properties by fitting the two-compartment model to the time-intensity curves. Tissue characterization based on the pharmacokinetic analysis has been demonstrated in various clinical applications (2-6).
However, application of DCE-MRI with pharmacokinetic analysis to the lung is still challenging because of the large displacements of localized lesions due to respiratory motion during several minutes of periodic imaging, where the breath-holding technique is not feasible. Inter-frame misalignment of focal tissue in free-breathing DCE-MRI inhibits reliable measurement of signal intensity change. Frame-by-frame manual selection of the region of interests (ROIs) requires tedious effort by radiologists and may not be practical for larger cohort studies and potential clinical routines. Misalignment of focal tissue also prohibits the pixel-wise pharmacokinetic analysis to generate parameter maps, which should reflect the heterogeneity. To the best of our knowledge, only a limited number of studies have investigated the pharmacokinetic analysis of pulmonary DCE-MRI (7), and none of them have investigated the impact of image registration on pharmacokinetic parameters, despite the correlation of time-intensity curve patterns in DCE-MRI and the tumor malignancy in solitary pulmonary nodules (SPNs) in the literature (8-12).
The contribution of this study is, for the first time, to demonstrate the impact of retrospective non-rigid motion correction on pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI in SPNs. The underlying hypothesis is that the motion correction of DCE-MRI improves the reliability of pharmacokinetic parameters, in terms of ‘goodness-of-fit’ between the pharmacokinetic model and the time-intensity curve in each pixel. This hypothesis was tested by first comparing the percentages of pixels accepted in a pixel-wise ‘goodness-of-fit’ test of pharmacokinetic parameter maps generated from DCE-MRI before and after motion correction. In addition, the pharmacokinetic parameters derived from benign and malignant SPNs were compared.
MATERIALS AND METHODS
Data Acquisition
The study protocol was approved by the institutional review board prior to the experiment. Informed consent for DCE-MRI of the lung was obtained from the subjects after the nature of the procedures and potential hazards had been fully explained. Fifteen patients (age 26 – 82, 2 males and 13 females) with SPNs measuring 15-30 mm, scheduled for surgical resection, underwent DCE-MRI examinations of the lung. The SPNs studied included 3 benign tumors (hamartoma, intrapulmonary solitary fibrous tumor, sclerosing pneumocytoma) and 12 malignant tumors (9 adenocarcinomas, 2 squamous cell carcinomas, 1 carcinoid), which were histologically determined prior to the image analysis. DCE-MRI was carried out on a 3.0 T whole-body scanner (Trio Tim, Siemens, Erlangen, Germany) using a 2D turbo fast low-angle shot (turbo FLASH) sequence combined with the generalized auto-calibrating partially parallel acquisition (GRAPPA) technique. The images were obtained from a single slice in each patient. The imaging plane was set in oblique sagittal so that the target SPN moved along the imaging plane based on a method developed in our previous work (13). The images were acquired after intravenous administration of a standard dose of 0.1 mmol per kg of body weight of Gd-DTPA (Magnevist, Schering AG, Germany) over a 4 s period with subjects in the supine position, breathing normally during the scan. The imaging parameters used were: TR/TE: 2.79/1.59 ms; flip angle: 10 degrees; receiver bandwidth: 1560 Hz/pixel; field of view: 320 × 300 mm; acquisition matrix: 192 × 180; pixel size: 1.66 × 1.66; slice thickness: 5.00 mm; scan time 500 ms/image. One hundred and twenty-four image frames were acquired every 2 seconds for each DCE-MRI.
Image Registration for Motion Correction of DCE-MRI
Misalignments of the SPNs among the image frames in the free-breathing DCE-MRI series were corrected by registering each frame to a reference image frame selected from the same series. Prior to the image registration process, the ROI in the images was manually cropped and masked so that anatomical structures that moved independently from the target nodules were excluded from the ROI. The same ROI and the mask were applied to all images in the same series of DCE-MRI. The reference image frame was manually selected among expiratory frames after contrast enhancement. The image registration process consisted of 2D rigid image registration as initial alignment followed by 2D non-rigid B-spline registration (14). For both rigid and non-rigid image registration, the transformation parameters were estimated by maximizing the mutual information (15) of each frame and the reference image. The abovementioned cropping, masking and registration were performed using 3D Slicer (16), a medical image processing and visualization software package, on a Linux-based workstation (OS: Fedora 10; 4 Xeon E7440 quad-core 2.4 GHz CPUs (total 16 cores); 128 GB memory). The computation time was measured using the internal clock of the workstation. In the following experimental steps, the DCE-MRI before the motion-correction (original DCE-MRI) and after the motion-correction (motion-corrected DCE-MRI) were analyzed.
Pharmacokinetic Analysis
Pharmacokinetic parameters, including the volume transfer constant Ktrans, the fractional volume of extravascular extracellular space of the target tissue ve, and the rate constant kep, were estimated by fitting a pharmacokinetic model to the actual time-intensity curves obtained from the DCE-MRI. In this study, the concentration of tracer Ct(t) after bolus injection was assumed to obey the model proposed by Tofts and Kermode(1):
| [1] |
where vp is the blood plasma volume per unit volume of the tissue. In this study, vp was ignored. Cp(t) is the tracer concentration in the blood plasma, which is a biexponential decay expected from the compartmental theory as:
| [2] |
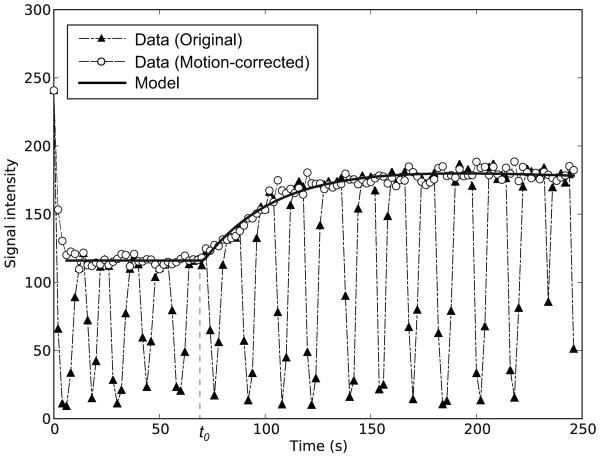
where D is the dose and t0 is the duration from the bolus injection of tracer to the beginning of the contrast change. To fit the abovementioned model to the actual time-intensity curve S(t), S(t) was converted to the enhancement curve E(t) = S(t)/S(0) −1. The enhancement was assumed to be proportional to the concentration as E(t) = R1T10 Ct(t), where R1 and T10 are relaxation rate and native T1 respectively. A least-square algorithm was used to optimize free parameters Ktrans, ve, and t0 to fit Ct(t) to E(t). Once Ktrans and ve were estimated, the rate constant kep was given by Ktrans / ve. Figure 1 shows representative curves obtained from original and motion corrected DCE-MRI with fitted model curve.
Figure 1.
Representative time-intensity curves obtained from original and motion-corrected DCE-MRI are shown with the model curve.
Evaluations
Reliable Area Percentage in Pharmacokinetic Parameter Maps
First, pharmacokinetic parameter maps of the SPNs were generated based on pixel-wise pharmacokinetic analyses of the original and motion-corrected DCE-MRI. Two sets (from original and motion-corrected DCE-MRI) of Ktrans, ve and kep maps were generated per patient. To examine the reliability of the parameters in those maps, ‘goodness-of-fit’ between the measured time-intensity curve and the pharmacokinetic model curve was evaluated pixel by pixel based on the χ2 test (17,18). When the model curve is y = Ct(t), the χ2 statistics is given by
| [3] |
where ti is time point to measure the concentration, yi is the concentration estimated from the enhancement measured on the DCE-MRI, and σi is the standard deviation of the error for the i-th measurement. The χ2 test is used to evaluate the null hypothesis stating that the measured curve is consistent with the theoretical model curve. The pharmacokinetic parameter in a given pixel was considered to be reliable only if the confidence level is more than 50 % in the χ2 test. The percentages of reliable pixels within the SPN region was then calculated for each SPN for both original and motion-corrected DCE-MRI. The SPN region was manually segmented on the reference frame in the original DCE-MRI. The Mann-Whitney U test was performed to compare the percentage of reliable pixels in the SPN between the original and motion-corrected DCE-MRI.
Parameter Comparison Between Benign and Malignant
To further investigate the impact of the motion-correction of free-breathing DCE-MRI on tissue characterization, the average Ktrans, ve, and kep in the manually segmented region of the SPN and in the reliable area in the region of the SPN on the parameter maps were compared between benign and malignant SPNs. For comparison with a conventional approach, Ktrans, ve, and kep estimated from manually-measured curves were also correlated with the tissue types. The manually-measured curves were sampled from ROIs, which are manually segmented within the SPNs frame by frame in the original DCE-MRI by a radiologist (H.M.). The Mann-Whitney U test was performed to examine if the distributions of the parameters were different between benign and malignant.
RESULTS
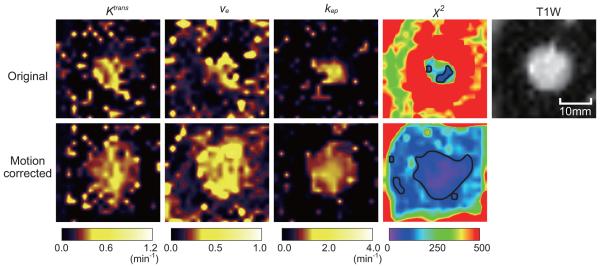
The average processing time for rigid correction per patient (124 frames) as preprocessing of non-rigid motion correction was 176 ± 51 s and that of non-rigid motion correction was 407 ± 73 s. Table 1 summarizes the percentages of acceptable pixels within the area of target SPNs for all 15 patients. The percentage of acceptable pixels was significantly improved after motion correction in all patients (p=4×10−7). Figure 2 shows the representative color-coded Ktrans, ve and kep maps generated from the pixel-wise pharmacokinetic analysis of the original and motion-corrected DCE-MRI, along with the color-coded χ2 maps and the T1-weighted image from the same region. The color-coded χ2 maps demonstrate that the reliable area in the motion-corrected DCE-MRI is larger than the original DCE-MRI. In addition, the Ktrans, ve and kep maps from the motion-corrected DCE-MRI show better reflections of the boundary of the SPN in the T1-weighted image.
Table 1.
Percentage of the acceptable pixels within the tumor region on the χ2 maps derived from the original DCE-MRI (Original) and the motion-corrected DCE-MRI (Motion-corrected).
| Patient ID |
Original (%) | Motion-corrected (%) |
|---|---|---|
| 1 | 0.0 | 95.0 |
| 2 | 0.0 | 47.0 |
| 3 | 0.0 | 48.9 |
| 4 | 0.0 | 21.0 |
| 5 | 10.7 | 89.3 |
| 6 | 1.9 | 86.0 |
| 7 | 0.0 | 63.4 |
| 8 | 1.0 | 46.5 |
| 9 | 0.0 | 54.8 |
| 10 | 0.0 | 30.4 |
| 11 | 0.9 | 45.0 |
| 12 | 0.0 | 81.1 |
| 13 | 0.0 | 9.7 |
| 14 | 0.0 | 73.7 |
| 15 | 11.1 | 75.9 |
|
| ||
| P = 0.0000004* | ||
Mann-Whitney U test
Figure 2.
Representative maps of Ktrans, ve and kep generated from the original and motion-corrected DCE-MRI from one patient. The χ2 maps and T1-weighted image are also shown. The contours of the reliable pixel areas are indicated by the black line on the χ2 maps. The χ2 map generated from the motion-corrected DCE-MRI shows a broadened acceptable pixel area covering the greater area of the SPN, when compared to the map from the original DCE-MRI. The Ktrans, ve and kep maps from the motion-corrected DCE-MRI show better reflections of the boundary of the nodule than those from the original DCE-MRI.
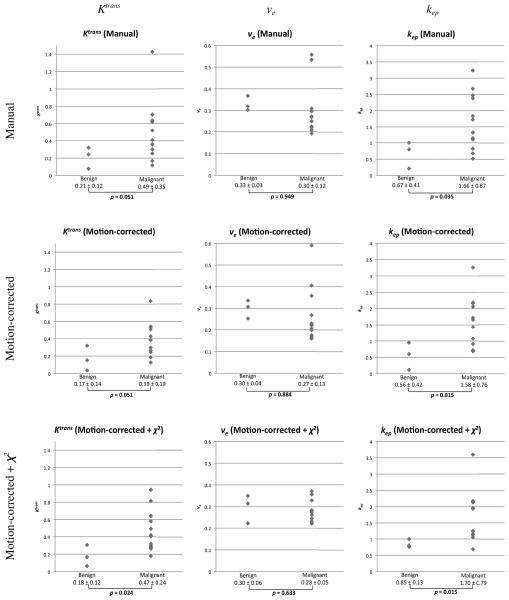
Figure 3 shows the distributions of Ktrans, ve and kep in benign and malignant tumors (benign: n=3, malignant: n=12). In the figure, the average Ktrans, ve and kep among the pixels in the lesions on the parameter maps generated from the motion-corrected DCE-MRI with and without the χ2 test and those from the pharmacokinetic analysis of the manually-measured curves were compared. The graphs show that the overlaps of the distributions of Ktrans and kep in the motion-corrected DCE-MRI with and without χ2 test are smaller than the manual measurement. Both Ktrans and kep from the motion-corrected DCE-MRI demonstrated significant difference of distributions (p = 0.024 and 0.015 respectively), whereas only kep showed significant difference in the motion-corrected DCE-MRI without the χ2 test (p = 0.015) and in the analyses of the manually-measured curves (p = 0.035). The p-values for the parameters from the motion-corrected DCE-MRI with and without the χ2 test tended to be smaller when compared to those from the analysis of the manually-measured curves.
Figure 3.
The distributions of Ktrans, ve, and kep derived from the manually-measured curves (Manual) and mean Ktrans, ve and kep in the reliable pixels in the parameter maps estimated from the motion-corrected DCE-MRI without χ2 test (Motion-corrected) and with χ2 test (Motion-corrected + χ2) are plotted for benign (n=3) and malignant (n=12). P-values derived from two-sample Mann-Whitney U test between benign and malignant are presented below each plots.
DISCUSSION
This study demonstrates the impact of the retrospective non-rigid motion correction on pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI in SPNs. Since the proposed method does not involve any frame-by-frame manual ROI selection requiring tedious effort of radiologists, it is practical for larger cohort studies and clinical characterization of SPNs.
For validation of motion correction of DCE-MRI, most related studies in the literature took a morphological approach such as detection of anatomical landmarks by observers (19,20), similarity measures (17,21), dice similarity coefficients, and Hausdorff distances (22). In contrast, our study employed the χ2 test, which is a statistical approach to evaluate the reliability of pixel-wise pharmacokinetic analysis, in addition to using similarity measures for the motion correction process. Although the χ2 test does not validate the pharmacokinetic parameters based on physiological information, it still gives an indication of how the observed time-intensity curves reflect the theoretical model from the statistical point of view. The advantage of using χ2 over those morphological approaches is, first of all, that it can be calculated solely from a pair of time-intensity curve and model curve and does not require any segmentation of images to calculate the distance of boundaries. Second, χ2 maps allow validation of the effect of motion correction inside the region, while most morphological evaluations only evaluate misalignment of boundaries or landmarks.
In the correlation of the pharmacokinetic parameters with tissue types, the average parameters among the reliable pixels in the parameter maps generated based on the pixel-wise pharmacokinetic analysis of the motion-corrected DCE-MRI showed better discrimination of the parameter distribution between benign and malignant than those estimated from the manually-measured time-intensity curves. Although this study still does not have enough statistical power to be conclusive, the result implies the potential advantage of pixel-wise pharmacokinetic analysis for tissue characterization over the conventional manual measurement of time-intensity curves. The study also suggests the potential usage of pixel-wise χ2 test to exclude unreliable parameters, which may help improve sensitivity and specificity in tissue characterization.
The pixel-wise pharmacokinetic analysis may provide more detailed information about target nodules to a clinician. For example, the distribution of the pixel-wise kinetic parameters within a single nodule could imply inhomogeneous distribution of tissue types. In fact, the histological findings in 3 patients among 15 patients showed mixed histological subtypes of tumors. Conventional curve pattern analysis based on manually-measured time-intensity curves may average signals from different types of tissues. The pixel-wise analysis, in theory, allows fitting the model to time-intensity curves from different subregions, and thus discriminates tissue types within a heterogeneous SPN.
One technical limitation in this study was that the images were acquired from a single slice. Although the imaging slice was carefully chosen so that the lesion moved along it using the method described in our previous work (13), the out-of-slice motion of the lesion due to drift or sudden motion of the patient might cause inaccurate pharmacokinetic parameter estimation. To solve this issue, either multi-slice or 3D image should be acquired for 3D motion correction in future work. Although the technical feasibility of the non-rigid motion correction for pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI is demonstrated, the potential of tissue characterization discriminating between benign and malignant SPNs has to be tested in a larger cohort. Further investigation should be made on the overlap of the parameter values between benign and malignant to improve the clinical utility of the method. Future investigation would include more comprehensive analysis of DCE-MRI, which combines the presented technique with other types of methods including curve patterns analysis (10,23).
In conclusion, we demonstrated the impact of the retrospective non-rigid motion correction on pixel-wise pharmacokinetic analysis of free-breathing DCE-MRI in SPNs. The study showed the motion correction technique increased the area of reliable pixels in the parameter maps. Since the method does not require any manual segmentation for time-intensity curves, it potentially provides a practical way to perform tissue characterization of SPNs based on free-breath DCE-MRI. Further evaluation of the technique in a larger cohort is required. This approach will also be applied to evaluation of therapy response for lung cancer.
Acknowledgments
Grant Support:
This study is supported by NIH 5R21CA116271-02, 5P01CA067165-10, 5U41RR019703-04, 2P41RR013218-11 and 5U54EB005149-05.
REFERENCES
- 1.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 2.Brix G, Semmler W, Port R, Schad LR, Layer G, Lorenz WJ. Pharmacokinetic parameters in CNS Gd-DTPA enhanced MR imaging. J Comput Assist Tomogr. 1991;15(4):621–628. doi: 10.1097/00004728-199107000-00018. [DOI] [PubMed] [Google Scholar]
- 3.Knopp MV, Brix G, Junkermann HJ, Sinn HP. MR mammography with pharmacokinetic mapping for monitoring of breast cancer treatment during neoadjuvant therapy. Magn Reson Imaging Clin N Am. 1994;2(4):633–658. [PubMed] [Google Scholar]
- 4.Turnbull LW, Buckley DL, Turnbull LS, Liney GP, Knowles AJ. Differentiation of prostatic carcinoma and benign prostatic hyperplasia: correlation between dynamic Gd-DTPA-enhanced MR imaging and histopathology. J Magn Reson Imaging. 1999;9(2):311–316. doi: 10.1002/(sici)1522-2586(199902)9:2<311::aid-jmri24>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 5.Egmont-Petersen M, Hogendoorn PC, van der Geest RJ, et al. Detection of areas with viable remnant tumor in postchemotherapy patients with Ewing's sarcoma by dynamic contrast-enhanced MRI using pharmacokinetic modeling. Magn Reson Imaging. 2000;18(5):525–535. doi: 10.1016/s0730-725x(00)00144-2. [DOI] [PubMed] [Google Scholar]
- 6.Rosen MA, Schnall MD. Dynamic contrast-enhanced magnetic resonance imaging for assessing tumor vascularity and vascular effects of targeted therapies in renal cell carcinoma. Clin Cancer Res. 2007;13(2 Pt 2):770s–776s. doi: 10.1158/1078-0432.CCR-06-1921. [DOI] [PubMed] [Google Scholar]
- 7.Ashton E, Raunig D, Ng C, Kelcz F, McShane T, Evelhoch J. Scan-rescan variability in perfusion assessment of tumors in MRI using both model and data-derived arterial input functions. J Magn Reson Imaging. 2008;28(3):791–796. doi: 10.1002/jmri.21472. [DOI] [PubMed] [Google Scholar]
- 8.Hittmair K, Eckersberger F, Klepetko W, Helbich T, Herold CJ. Evaluation of solitary pulmonary nodules with dynamic contrast-enhanced MR imaging--a promising technique. Magn Reson Imaging. 1995;13(7):923–933. doi: 10.1016/0730-725x(95)02010-q. [DOI] [PubMed] [Google Scholar]
- 9.Ohno Y, Hatabu H, Takenaka D, Adachi S, Kono M, Sugimura K. Solitary pulmonary nodules: potential role of dynamic MR imaging in management initial experience. Radiology. 2002;224(2):503–511. doi: 10.1148/radiol.2242010992. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer JF, Vollmar J, Schick F, et al. Solitary pulmonary nodules: dynamic contrast-enhanced MR imaging--perfusion differences in malignant and benign lesions. Radiology. 2004;232(2):544–553. doi: 10.1148/radiol.2322030515. [DOI] [PubMed] [Google Scholar]
- 11.Kono R, Fujimoto K, Terasaki H, et al. Dynamic MRI of solitary pulmonary nodules: comparison of enhancement patterns of malignant and benign small peripheral lung lesions. AJR Am J Roentgenol. 2007;188(1):26–36. doi: 10.2214/AJR.05.1446. [DOI] [PubMed] [Google Scholar]
- 12.Zou Y, Zhang M, Wang Q, Shang D, Wang L, Yu G. Quantitative investigation of solitary pulmonary nodules: dynamic contrast-enhanced MRI and histopathologic analysis. AJR Am J Roentgenol. 2008;191(1):252–259. doi: 10.2214/AJR.07.2284. [DOI] [PubMed] [Google Scholar]
- 13.Kino A, Takahashi M, Ashiku SK, Decamp MM, Lenkinski RE, Hatabu H. Optimal breathing protocol for dynamic contrast-enhanced MRI of solitary pulmonary nodules at 3T. Eur J Radiol. 2007;64(3):397–400. doi: 10.1016/j.ejrad.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. Medical Imaging, IEEE Transactions on. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 15.Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. Medical Imaging, IEEE Transactions on. 2003;22(1):120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- 16.Gering DT, Nabavi A, Kikinis R, et al. An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J Magn Reson Imaging. 2001;13(6):967–975. doi: 10.1002/jmri.1139. [DOI] [PubMed] [Google Scholar]
- 17.Noseworthy MD, Haider MA, Sussman MS, Wright GA. Free-breathing motion compensation using template matching: a technique allowing for tracer kinetic modeling of liver metastases. J Comput Assist Tomogr. 2007;31(2):193–197. doi: 10.1097/01.rct.0000235069.50055.30. [DOI] [PubMed] [Google Scholar]
- 18.Martel AL, Froh MS, Brock KK, Plewes DB, Barber DC. Evaluating an optical-flow-based registration algorithm for contrast-enhanced magnetic resonance imaging of the breast. Phys Med Biol. 2007;52(13):3803–3816. doi: 10.1088/0031-9155/52/13/010. [DOI] [PubMed] [Google Scholar]
- 19.Torheim G, Amundsen T, Rinck PA, Haraldseth O, Sebastiani G. Analysis of contrast-enhanced dynamic MR images of the lung. J Magn Reson Imaging. 2001;13(4):577–587. doi: 10.1002/jmri.1081. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann KH, Wurdinger S, Fischer DR, et al. Application and assessment of a robust elastic motion correction algorithm to dynamic MRI. Eur Radiol. 2007;17(1):259–264. doi: 10.1007/s00330-006-0224-3. [DOI] [PubMed] [Google Scholar]
- 21.Hayton PM, Brady M, Smith SM, Moon M. A non-rigid registration algorithm for dynamic breast MR imaging. Artificial Intelligence. 1999;114(1-2):125–156. [Google Scholar]
- 22.Bottger T, Grunewald K, Schobinger M, et al. Implementation and evaluation of a new workflow for registration and segmentation of pulmonary MRI data for regional lung perfusion assessment. Phys Med Biol. 2007;52(5):1261–1275. doi: 10.1088/0031-9155/52/5/004. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer JF, Schneider V, Vollmar J, et al. Solitary pulmonary nodules: association between signal characteristics in dynamic contrast enhanced MRI and tumor angiogenesis. Lung Cancer. 2006;53(1):39–49. doi: 10.1016/j.lungcan.2006.03.010. [DOI] [PubMed] [Google Scholar]