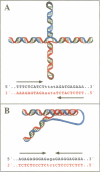
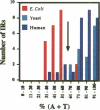
Abstract
We have used computer-assisted methods to search large amounts of the human, yeast and Escherichia coli genomes for inverted repeat (IR) and mirror repeat (MR) DNA sequence patterns. In highly supercoiled DNA some IRs can form cruciforms, while some MRs can form intramolecular triplexes, or H-DNA. We find that total IR and MR sequences are highly enriched in both eukaryotic genomes. In E. coli, however, only total IRs are enriched, while total MRs only occur as frequently as in random sequence DNA. We then used a set of experimentally derived criteria to predict which of the total IRs and MRs are most likely to form cruciforms or H-DNA in supercoiled DNA. We show that strong cruciform forming sequences occur at a relatively high frequency in yeast (1/19 700 bp) and humans (1/41 800 bp), but that H-DNA forming sequences are abundant only in humans (1/49 400 bp). Strong cruciform and H-DNA forming sequences are not abundant in the E.coli genome. These results suggest that cruciforms and H-DNA may have a functional role in eukaryotes, but probably not prokaryotes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaho J. A., Larson J. E., McLean M. J., Wells R. D. Multiple DNA secondary structures in perfect inverted repeat inserts in plasmids. Right-handed B-DNA, cruciforms, and left-handed Z-DNA. J Biol Chem. 1988 Oct 5;263(28):14446–14455. [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Influence of DNA sequence and supercoiling on the process of cruciform formation. J Mol Biol. 1988 Jul 5;202(1):35–43. doi: 10.1016/0022-2836(88)90516-5. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Dröge P. Protein tracking-induced supercoiling of DNA: a tool to regulate DNA transactions in vivo? Bioessays. 1994 Feb;16(2):91–99. doi: 10.1002/bies.950160205. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Fox K. R. Long (dA)n.(dT)n tracts can form intramolecular triplexes under superhelical stress. Nucleic Acids Res. 1990 Sep 25;18(18):5387–5391. doi: 10.1093/nar/18.18.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L. A., Garrard W. T. DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr. 1992;2(2):165–209. [PubMed] [Google Scholar]
- Gough G. W., Sullivan K. M., Lilley D. M. The structure of cruciforms in supercoiled DNA: probing the single-stranded character of nucleotide bases with bisulphite. EMBO J. 1986 Jan;5(1):191–196. doi: 10.1002/j.1460-2075.1986.tb04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. The ubiquitous potential Z-forming sequence of eucaryotes, (dT-dG)n . (dC-dA)n, is not detectable in the genomes of eubacteria, archaebacteria, or mitochondria. Mol Cell Biol. 1986 Aug;6(8):3010–3013. doi: 10.1128/mcb.6.8.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Klysik J., Wells R. D. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J Biol Chem. 1988 May 25;263(15):7386–7396. [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. II. Effect of base composition and noncentral interruptions on formation and stability. J Biol Chem. 1989 Apr 5;264(10):5950–5956. [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Detection of triple-helix related structures adopted by poly(dG)-poly(dC) sequences in supercoiled plasmid DNA. Nucleic Acids Res. 1991 Aug 11;19(15):4267–4271. doi: 10.1093/nar/19.15.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Ferl R. J. Homopurine/homopyrimidine sequences as potential regulatory elements in eukaryotic cells. Int J Biochem. 1993 Nov;25(11):1529–1537. doi: 10.1016/0020-711x(93)90508-c. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Kumarev V. P., Baranova L. V., Vologodskii A. V., Frank-Kamenetskii M. D. Energetics of the B-H transition in supercoiled DNA carrying d(CT)x.d(AG)x and d(C)n.d(G)n inserts. Nucleic Acids Res. 1989 Nov 25;17(22):9417–9423. doi: 10.1093/nar/17.22.9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H., Rao B. S., Martin R. G. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J Mol Evol. 1988;27(2):96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J Biol Chem. 1988 May 25;263(15):7370–7377. [PubMed] [Google Scholar]
- Mirkin S. M., Frank-Kamenetskii M. D. H-DNA and related structures. Annu Rev Biophys Biomol Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Bowater R., Aboul-ela F., Lilley D. M. Helix opening transitions in supercoiled DNA. Biochim Biophys Acta. 1992 May 7;1131(1):1–15. doi: 10.1016/0167-4781(92)90091-d. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Lilley D. M. Supercoiled DNA and cruciform structures. Methods Enzymol. 1992;211:158–180. doi: 10.1016/0076-6879(92)11010-g. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Burland V., Daniels D. L., Blattner F. R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993 Jul 25;21(15):3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A. DNA comes in many forms. Gene. 1993 Dec 15;135(1-2):99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Chou P. J., Ho P. S. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J Biol Chem. 1992 Jun 15;267(17):11846–11855. [PubMed] [Google Scholar]
- Senger D. R., Asch B. B., Smith B. D., Perruzzi C. A., Dvorak H. F. A secreted phosphoprotein marker for neoplastic transformation of both epithelial and fibroblastic cells. Nature. 1983 Apr 21;302(5910):714–715. doi: 10.1038/302714a0. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Hanvey J. C., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. I. Effect of loop size on formation and stability. J Biol Chem. 1989 Apr 5;264(10):5944–5949. [PubMed] [Google Scholar]
- Sinden R. R., Wells R. D. DNA structure, mutations, and human genetic disease. Curr Opin Biotechnol. 1992 Dec;3(6):612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Spiro C., Richards J. P., Chandrasekaran S., Brennan R. G., McMurray C. T. Secondary structure creates mismatched base pairs required for high-affinity binding of cAMP response element-binding protein to the human enkephalin enhancer. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4606–4610. doi: 10.1073/pnas.90.10.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N., Konopka A. K., Jovin T. M. Unusual frequencies of certain alternating purine-pyrimidine runs in natural DNA sequences: relation to Z-DNA. FEBS Lett. 1985 Jun 3;185(1):197–202. doi: 10.1016/0014-5793(85)80769-9. [DOI] [PubMed] [Google Scholar]
- Tripathi J., Brahmachari S. K. Distribution of simple repetitive (TG/CA)n and (CT/AG)n sequences in human and rodent genomes. J Biomol Struct Dyn. 1991 Oct;9(2):387–397. doi: 10.1080/07391102.1991.10507919. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Wittig B., Wölfl S., Dorbic T., Vahrson W., Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992 Dec;11(12):4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X., Kochel T., Hoepfner R. W., Timmons S. E., Sinden R. R. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J Mol Biol. 1991 Sep 5;221(1):107–122. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]
- Zheng G. X., Sinden R. R. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J Biol Chem. 1988 Apr 15;263(11):5356–5361. [PubMed] [Google Scholar]
- van Holde K., Zlatanova J. Unusual DNA structures, chromatin and transcription. Bioessays. 1994 Jan;16(1):59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]