Abstract
In this study, we used a previously described method of controlling gene expression with computer-based gene design and de novo DNA synthesis to attenuate the virulence of Streptococcus pneumoniae. We produced 2 S. pneumoniae serotype 3 (SP3) strains in which the pneumolysin gene (ply) was recoded with underrepresented codon pairs while retaining its amino acid sequence and determined their ply expression and pneumolysin production in vitro and their virulence in a mouse pulmonary infection model. Expression of ply and production of pneumolysin of the recoded SP3 strains were decreased, and the recoded SP3 strains were less virulent in mice than the wild-type SP3 strain or a Δply SP3 strain. Further studies showed that the least virulent recoded strain induced a markedly reduced inflammatory response in the lungs compared with the wild-type or Δply strain. These findings suggest that reducing pneumococcal virulence gene expression by altering codon-pair bias could hold promise for rational design of live-attenuated pneumococcal vaccines.
Codon-pair bias (CPB) defines a phenomenon whereby the codons that encode 2 sequential amino acids are found adjacent to one another with a higher or lower frequency than would be expected if their placement was random [1, 2]. CPB is observed across the kingdoms of life, can be quantified statistically [1, 2], and is independent of codon usage [3, 4]. Although the frequency of some of the 3,721 possible codon pairs is shared across species, species-specific codon-pair representations are distinct [5]. It has been shown elsewhere that synthetic recoding of adjacent codon pairs with underrepresented codon pairs decreases translation efficiency for poliovirus and influenza A virus, resulting in attenuation of their virulence in vivo [1, 6].
Streptococcus pneumoniae is the leading cause of pneumonia in adults and children in the United States and globally [7]. Use of the 7-valent pneumococcal capsular polysaccharide-protein conjugate vaccine has led to a dramatic reduction in the incidence of invasive pneumococcal disease in children and adults, due to herd immunity [8]. A reformulation with additional serotypes, including S. pneumoniae serotype 3 (SP3), was recently introduced to address the problem of serotype replacement [9]. However, reports of severe SP3 disease [10, 11], the failure of investigational conjugate vaccines to reliably protect against SP3 [12], and the emergence of drug-resistant SP3 strains [13] underscore the need for a vaccine that can protect against SP3. Given the efficacy of candidate experimental whole-cell vaccines against multiple serotypes in mice [14] and the fact that pneumolysin (PLY), the 54-kDa toxin of S. pneumoniae [15], has already been validated as a candidate vaccine antigen [16], we used CPB customization to decrease pneumolysin gene (ply) expression in a SP3 strain and determined the effect of modulating ply expression on virulence in vivo.
MATERIALS AND METHODS
Mice
Female BALB/c mice 6–8 weeks of age were obtained from the National Cancer Institute Mouse Repository (Fredrick, MD). All mouse experiments were performed according to the rules, regulations, and ethical standards for animal use of the Animal Institute of the Albert Einstein College of Medicine (AECOM).
SP3 Strains and Growth Conditions
The SP3 strains constructed in this study are shown in Table 1. The wild-type A66.1 and WU2 strains were originally provided by David Briles (University of Alabama at Birmingham, Birmingham, AL) [17, 18]. All cultures were begun with single colonies from a tryptic-soy agar–blood agar plate (TSA-BAP). Single colonies were inoculated into 15 mL of tryptic-soy broth (TSB) and grown at 37°C with .5% carbon dioxide (CO2) for 15 h, and then diluted 1:100 in TSB or Todd-Hewitt broth (THB) and grown at 37°C with .5% CO2.
Table 1.
Streptococcus pneumoniae Strains, Plasmids, and Pneumolysin Codon-pair Bias
| S. pneumoniae strain or plasmid | Genotype, description, or codon-pair bias value | Source, reference, or no. of nucleotide changes |
| S. pneumoniae strains | ||
| A66.1 | Serotype 3 S. pneumoniae | [17] |
| A66.1:PM2 | A66.1; synthetically modified pneumolysin via transformation with pPM2, Kmr | This study |
| A66.1:PM4 | A66.1; synthetically modified pneumolysin via transformation with pPM4, Kmr | This study |
| A66.1:Δply | A66.1; Δply via transformation with pΔPLY, Kmr | This study |
| A66.1::ply | A66.1; reconstituted ply via transformation with pPLY, Kmr | This study |
| Plasmids | ||
| pUCminus | Synthetic carrier plasmid | Blue Heron Biotechnology |
| pPM4 | Derivative of pUCminus that contains synthetic construct pPM4 (Figure 1) | This study |
| pPM2 | Derivative of pUCminus that contains synthetic construct pPM2 (Figure 1) | This study |
| pΔPLY | Derivative of pUCminus that contains synthetic construct pΔPLY (Figure 1) | This study |
| pPLY | Derivative of pUCminus that contains the wild-type pneumolysin and Kmr cassette (not shown) | This study |
| Strain (pneumolysin) | ||
| A66.1 (ply) | .095 | … |
| A66:PM2 (ply−.19) | −.194a | 275b |
| A66:PM4 (ply−.47) | −.472a | 360b |
NOTE. Kmr, kanamycin resistance; ply, pneumolysin gene; r, resistance.
The negative codon-pair bias value indicates pneumolysin genes encoded with codon pairs that are underrepresented in S. Pneumoniae genes. The more negative the value, the greater the density of underrepresented pairs.
All nucleotide changes to ply are silent mutations. The amino acid sequences are identical to those of the A66.1 ply gene.
Calculation of SP3 Codon-pair Representation and ply CPB Customization
The genomic sequence of strain SP3-BS71 (Center for Genomic Sciences, Allegheny-Singer Research Institute, Pittsburgh, PA; GenBank ID, AAZZ00000000.1) was used for calculations [19]. The CPB of the open reading frames of SP3 was calculated using previously defined computer algorithms, as described elsewhere [1]. Briefly, the statistical representation of all 3,721 codon pairs in all annotated SP3 open reading frames was defined by a codon-pair score (CPS), which quantifies individual codon-pair representation based on the log ratio of each pair's observed occurrence to its expected occurrence [1]. The CPB of a gene is the mean CPS of the codon pairs in the gene. A positive CPS indicates statistical overrepresentation and a negative CPS indicates underrepresentation of the codon pair relative to the expected frequency if codon pair representation was random. The CPB of ply was customized to use underrepresented codons with previously defined software, as described elsewhere [1]. Constructs with underrepresented codons were synthesized (Blue Heron Biotechnology) and used to produce recombinant SP3 strains.
Transformation of SP3 and Construction of Recoded Strains
Transformation of SP3 was performed as described elsewhere [20], using the plasmids in Table 1 with some modifications. A 1:100 dilution of A66.1 or WU2 was grown to an optical density at 550 nm (OD550) of .032 in THB supplemented with .16% bovine serum albumin and .5% yeast extract. Next, the pH of the culture was adjusted to 7.8 with 1 mol/L sodium hydroxide (NaOH), and a 1-mL aliquot was supplemented with 10 μL of 1 mol/L calcium chloride (CaCl2), 50 μL of a 50 μg/mL solution of Competence-Stimulating Peptide 2 (Anaspec Inc.), and 800 ng of plasmid DNA (containing ply constructs) and incubated at 30°C for 90 min, after which it was diluted 1:10 and plated on TSA-BAP containing kanamycin. Drug-resistant colonies were selected and genomic integration of synthetic ply was confirmed by sequencing and polymerase chain reaction (PCR) using the primers in Table 2. The kanamycin resistance gene was maintained in all constructed strains upstream of ply to ensure strain purity.
Table 2.
Primers Used in Sequencing of Pneumolysin Locus and Cloning of Plasmids
| Primer Name | |
| Primers for sequencing ply locusa | Sequence, 5′–3′ |
| HR5_Seq_1116(+) | GGATTTTATTAGCAAATCAAGCTAGG |
| HR5_Seq_1320(+) | GCACCTTTTGTTGACAGTCTACTCC |
| HR5_Seq_1526(+) | CTGACATGGTTTGAAGAGATTTTCG |
| HR5_Seq_1961(−) | CCGATTTGCCACTAGTGCGTAAGCG |
| PlySeq_IN_1969(+) | CCAAATAGAAATCGTCCGCTTACGC |
| PlySeq_IN_2391(−) | GCCCGAGTTGTAACAGGCAAGGTGG |
| PlySeq_IN_2221(+) | GCAACATAGGCACCACTATGATCCAGC |
| PlySeq_IN_2582(+) | GTTTCCAACTTGAGATAGACTTGGC |
| PlySeq_IN_2764(+) | CTGAATCTGCTTTTCGCCTGAATGG |
| PlyProSeq_3222(+) | GCTGATTACCCTCTTTGATGAAACG |
| HR3_Seq_3432(+) | CTCGTACAATTCTTCTGAGACATTGC |
| HR3_Seq_3721(+) | GCAAATAAAGAGCTACAACATCTCC |
| HR3_SeqO_4300(−) | CCAGCTACCTGTCGCCCTTGCTCTG |
| PLY_KOdetect_3409(−) | ACTATTAGTAGCGATTTGGC |
| SeqPLY_inKmr_(+) | TCCGGTCGATCAGGGAGGATATCG |
| PM2_Seq_3172(+) | CGTTGCGATATGCTGTAACTTTCG |
| PM4_Seq_3149(+) | ATCTAACAACAGATCACC |
| Primers for PCR of plya | |
| PCR_Ply_1830(+) | CATACTCCAATGGAAATCGCTAGGC |
| PCR_Ply_3354(-) | GGCTGATTTCGCTGAACAAGTCTG |
| Primers for cloning of pΔPLYb | |
| PstI_HR3(+) | GTctgcagCTAATCGTTCc |
| HR3_plyKO_(−) | TTGAATTCTCTTATTTGCTAG |
NOTE. PCR, polymerase chain reaction; ply, pneumolysin gene.
The numerical designation indicates the nucleotide position using the reference point 1,830,000 as nucleotide 1 in the SP3-BS71 genome (GenBank ID, AAZZ00000000.1) [19].
PCR of plasmid pPM4 (Table 1).
Italic and lowercase letters indicate the engineered restriction site PstI.
Hemolytic Assay
The hemolytic activity of the PLY produced by SP3 strains was determined as described elsewhere [21]. Hemolytic activity was expressed in hemolytic units (HUs) whereby 1 HU was the reciprocal of the dilution at which 100% of red blood cells lyse. A standard curve based on purified PLY, whereby 1 HU = 1.69 ng of PLY, was used to convert HUs to nanograms [22, 23].
Enzyme-linked Immunosorbent Assay (ELISA) Quantification and Western Blot Detection of PLY
PLY was detected in bacterial lysates by ELISA and Western blot analysis as described elsewhere [24, 25]. Anti-PLY antibodies (mouse monoclonal immunoglobulin G [IgG] anti-PLY and polyclonal rabbit IgG anti-PLY) were used for the ELISA, and mouse monoclonal IgG anti-PLY was used for Western blot analysis [25]. Lysates were obtained from bacterial pellets of S. pneumoniae cultures, which were centrifuged, re-suspended in phosphate-buffered saline, normalized to an OD550 of .4, incubated at 37°C for 60 min, and spun at 10,000 g for 1 min, after which the supernatants were concentrated by centrifugation through Amicon 30K centrifugal units (Millipore) and used for ELISA or Western blot analysis according to methods published elsewhere [24, 25]. Positive and negative controls for both assays were purified PLY and lysate from A66:Δply, respectively.
Survival of Mice Infected With SP3 Strains
The virulence of wild-type and recoded SP3 strains was determined in an intranasal infection model as described elsewhere [26]. Groups of 10 mice each were inoculated intranasally with 5 × 104 colony-forming units (CFUs) of S. pneumoniae in 40 μL of TSB and monitored daily for their clinical status.
Bacterial Burden of Mice Infected With SP3 Strains
Lung and blood CFUs were counted 48 h after infection. Separate groups of mice, infected as described above, were bled from the orbital sinus and killed, and lung tissue was removed and homogenized. Blood was diluted in TSB and plated. Serial dilutions of lung homogenates and blood were plated on TSA-BAP and CFUs were enumerated. The limit of detection of this method is 10 CFUs.
Lung Cellular Profiles of Mice Infected With SP3 Strains
Cellular profiles in the lungs of SP3-infected mice were determined by flow cytometry as described elsewhere [27] with separate groups of mice infected as described above. Polymorphoneutrophils (PMNs), B cells, and T cells were analyzed to determine the effect of PLY on cellular subsets previously found to contribute to resistance and susceptibility to A66.1 [27]. After 48 h, mice were killed, lungs were extracted and placed in 5 mL of digestion buffer, and samples that were obtained with the GentleMACS cell separation system (Miltenyi Biotec) were incubated at 37°C for 30 min, filtered, washed 3 times in staining buffer, and resuspended. Control mice were injected with TSB. Cellular Fc receptors were blocked with anti-CD16/32 (BD Pharmingen) for 15 min on ice and then incubated with cell surface receptor-specific antibodies for 30 min on ice, washed twice in cold staining buffer, fixed with cold 2% formaldehyde at 25°C for 30 min, washed, and analyzed. The antibodies used were CD45 Pacific for CD45+ cells, CD3 Alex 488 for CD3+ cells, CD Alex 700 for CD4+ cells, CD8 phycoerythrin-Cy7 for CD8+ cells, Ly6G fluorescein isothiocyanate for Ly6G+ cells, and CD19 phycoerythrin-Cy7 for CD19+ cells (BD Pharmingen). Events (105 events per sample) were collected on a LSR II cytometer using DiVa software (BD Biosciences, Version 4.1) and analyzed with FlowJo software (Tree Star, Version 9.2 for Mac) first by gating on live cells in the forward and side scatter and then with nested gating. Cell counts were defined as the percentage of CD45+CD3+CD4+ or CD8+ cells for T cells, CD45+CD19+ cells for B cells, and CD45+Ly6G+ cells for PMNs as a function of the total number of cells.
Cytokine Levels in Lungs of Mice Infected With SP3 Strains
Cytokine levels in the lungs of SP3-infected mice were determined 48 h after infection as described elsewhere [27] with separate groups of mice infected as described above. Mice were killed 48 h after infection and their lungs were removed, homogenized as described above, and centrifuged at 3,000 g for 30 min. The supernatants were spun at 10,000 g for 15 min and used for the detection of cytokines with the ELISA Duoset kit (R&D Systems) used according to the manufacturer's protocol.
In Vitro Dendritic Cell (DC) Gene Expression Induced by SP3 Strains
The effect of S. pneumoniae on DC gene expression was determined ex vivo as described elsewhere [28], with modification for lung DCs. DCs were isolated from naïve BALB/c mouse lungs with the Dendritic Cell Enrichment Set beads and iMag magnet (BD Biosciences) according to the manufacturer's protocol and incubated with S. pneumoniae strains at a multiplicity of infection of 10 for 3.5 h at 37°C with .5% CO2 as described elsewhere [28], after which 100 μg/mL gentamicin was added, leaving DCs in culture. After 18 h, DCs were lysed, RNA was isolated with the RNeasy kit (Qiagen), and complementary DNA was synthesized with the Maxima FirstStrand kit (Fermentas). Reverse-transcription PCR (RT-PCR) was performed as described elsewhere [29] using TaqMan Master Mix (Applied Biosystems) and primers for nitric oxide synthase 2 (NOS2), interleukin 12 (IL-12), CD86 antigen (CD86), indoleamine 2,3-dioxygenase 1 (IDO1), apoptosis-related cysteine peptidase (Casp1), and chemokine (C-X-C motif) ligand 9 (CXCL9) (SABiosciences). These genes were selected on the basis of previous studies of the effect of PLY on the DC response to S. pneumoniae [28, 30]. Samples were analyzed with the StepOnePlus system (Applied Biosystems). The glyceraldehyde-3-phosphate dehydrogenase gene (Gapdh) was used as an internal control.
Serum Pneumococcal Antibody Titers
Surviving mice were bled 28 d after infection, and recoded SP3 levels and serum titers of antibody to pneumococcal capsular polysaccharide (PPS) [31] and whole bacteria (A66.1) [14, 24] were determined by ELISA as described above.
Re-infection of Surviving Mice
Surviving mice that had been infected with recoded SP3 were re-infected intranasally with 105 CFUs of A66.1 (15 times the median lethal dose) 42 d after primary infection, and survival was monitored.
Statistical Analysis
Statistical analyses were performed with Prism software (version 5.0c for Macintosh; GraphPad). For all statistical evaluations, a 2-tailed P value of <.05 was considered significant. For normally distributed data, the Student t test was used. For mouse survival, the Kaplan-Meier log-rank test was used. A 1-way analysis of variance was used to compare the curves of HUs in culture supernatants.
RESULTS
Construction of Synthetically Recoded SP3 ply With Altered CPB
Two recoded DNA constructs with negative CPB, ply−.47 and ply−.19, were designed (Table 1). A negative CPB indicates that the gene is encoded primarily with underrepresented codon pairs. The synthetic gene ply−.47 had more underrepresented codon pairs, with a CPB of −.472, than did ply−.19, which had a CPB of −.194. Both genes had a more negative CPB than did the wild-type ST3 ply (CPB, .095) (Table 1). CPB customization maintained the primary amino acid sequence of ply.
SP3 strains expressing recoded ply genes were produced by transforming A66.1 with the synthesized DNA constructs pPM2 and pPM4 to produce 2 synthetically modified SP3 strains, A66:PM2 and A66:PM4, respectively (Table 1; Figure 1). The homologous regions HR5′ and HR3′ of each construct had a sequence identical to that of wild-type A66.1 ply and were 800 bp in length. A ply-deleted strain, A66:Δply, was produced by transformation of A66.1 with a synthesized knockout plasmid pΔPLY (Table 1; Figure 1), and a reconstituted ply strain was constructed using synthesized pPLY (Table 1). We also recoded ply in WU2, another SP3 strain that is commonly used in pneumococcal research [18, 26, 31], by transformation with pPM4 to produce WU2:PM4. All strains exhibited similar growth kinetics in vitro (data not shown).
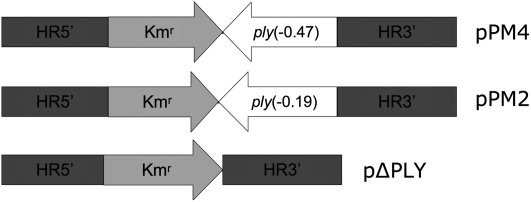
Figure 1.
Synthetic constructs for genomic integration of designed pneumolysin genes (ply) of Streptococcus pneumoniae. Synthetic DNA constructs used for transformation and recombination were inserted in plasmids designated pPM4, pPM2, and pΔPLY (Table 1). HR5′ and HR3′ are 800-bp sequences identically homologous to genomic regions (noncoding) of the wild-type ply locus, thus allowing for efficient recombination. The Kmr region is a kanamycin resistance cassette and remained in all strains.
Biological Activity of Recoded SP3 ply
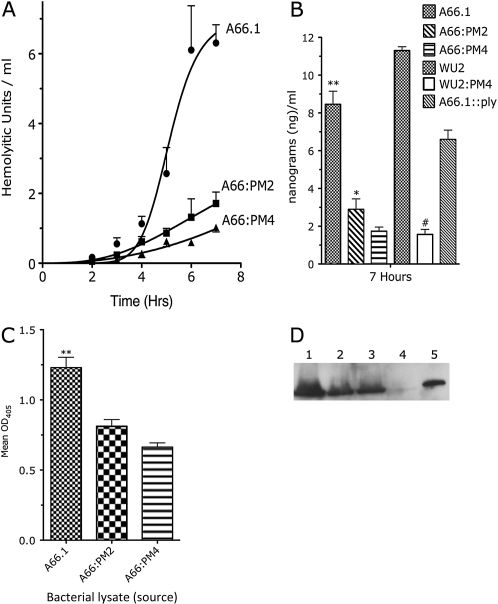
The impact of CPB alteration on ply expression was determined with an in vitro hemolytic assay (Figure 2A). A66:PM4 produced the fewest Hus, and A66:PM2 produced significantly more HUs than A66:PM4 but significantly fewer HUs than wild-type A66.1. A66:Δply did not produce detectible HUs (not shown). SP3 strain WU2 produced significantly more PLY than WU2:PM4 (Figure 2B). The higher amount of PLY produced by WU2 has been reported elsewhere [23]. The total PLY production of A66.1 was significantly greater than that of equally diluted lysates of A66:PM2 and A66:PM4 by ELISA (Figure 2C). A66.1, A66:PM2, and A66:PM4 produced PLY of equal size (52 kDa) on a Western blot (Figure 2D).
Figure 2.
Hemolytic activity and quantification of pneumolysin (PLY) produced by wild-type, ply-deleted, and ply-recoded Streptococcus pneumoniae serotype 3 strains. A, Hemolytic activity of PLY in the supernatant of growing S. pneumoniae strains measured over time. *P < .003 (1-way analysis of variance). All strains had similar growth kinetics. B, Concentration of PLY in growth media of the indicated strains 7 h after initiation of the culture, calculated from hemolytic units as described elsewhere [22]. *P < .001 comparing A66.1 and A66:PM4; *P < .002 comparing A66.1 and A66:PM2; **P < .004 comparing A66:PM2 and A66:PM4; #P < .001 comparing WU2 and WU2:PM4 (Student unpaired t test of 4–5 independent experiments). C, Total amount of PLY produced in vitro as measured by enzyme-linked immunosorbent assay from stationary phase cultures. *P < .01 comparing A66.1 and A66:PM4; *P < .04 comparing A66.1 and A66.1:PM2; **P = .051 comparing A66:PM2 and A66:PM4 (Student unpaired t test. D, Western blot of total PLY produced after in vitro growth reached the stationary phase. Lane 1, A66.1; lane 2, A66:PM2; lane 3, A66:PM4; lane 4, A66:Δply; lane 5, purified PLY (500 ng). All bands indicate similar molecular weights (52 kDa). OD405, optical density at 405 nm.
Mouse Virulence of SP3 Strains
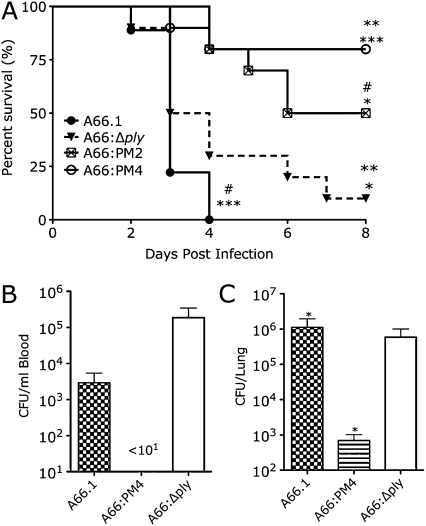
The lethality of the SP3 strains in BALB/c mice is shown in Figure 3A. A66:PM4 was significantly less lethal than A66.1 and A66:Δply. The lethality of A66:PM2 was statistically comparable to that of A66:PM4 (P = .09) but significantly less that of than A66.1 and A66:Δply. The lethality of 66:Δply and A66.1 was comparable.
Figure 3.
Survival and bacterial burden of mice infected with wild-type, pneumolysin gene (ply)–deleted, and ply-recoded Streptococcus pneumoniae serotype 3 (SP3) strains. A. Survival of BALB/c mice after intranasal infection with 5 × 104 colony-forming units (CFUs) of wild-type, ply-recoded, and ply-deleted SP3 A66.1 strains. **P < .001 comparing A66.PM4 and A66.1; **P < .002 comparing A66:PM4 and A66:Δply; *P < .02 comparing A66:PM2 and A66:Δply; *P < .001 comparing A66:PM2 and A66.1 (Kaplan-Meier log-rank test; curves depict results of 2 independent experiments with 5 mice per group; n = 10). B, CFUs in lungs of mice infected as in panel A, 48 h after infection. *P < .05 comparing A66.1 and A66:PM4; *P < .05 comparing A66.1 and A66:Δply (Student unpaired t test). C, CFUs in blood of mice infected as in panel A, 48 h after infection. No S. pneumoniae was detected in the blood of mice infected with A66:PM4 (lower limit of detection, 101). For panels B and C, bars represent the mean and error bars represent the standard deviation (n = 3).
Bacterial Burden of Mice Infected With SP3 Strains
A66:PM4-infected mice had no detectable CFUs in the blood and significantly fewer CFUs in the lungs than A66.1- or A66:Δply-infected mice 48 h after infection (Figures 3B and 3C). Lung and blood CFUs of A66.1- and A66:Δply-infected mice were comparable (Figures 3B and 3C).
Cellular Profiles in the Lungs of Mice Infected With SP3 Strains
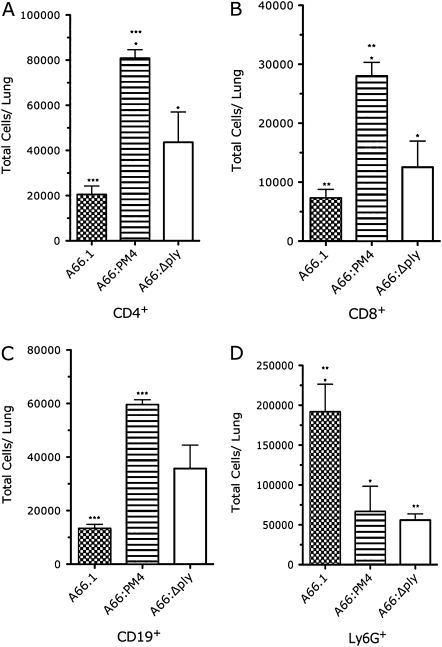
Lung cellular profiles exhibited strain-specific differences 48 h after infection. A66:PM4-infected mice had significantly more CD4+ and CD8+ T and B cells than either A66.1- or A66:Δply-infected mice (Figures 4A–4C). A66.1-infected mice had more PMNs (Ly6G+ cells) than either A66:PM4- or A66:Δply-infected mice (Figure 4D). Amounts of DCs were comparable in mice infected with each SP3 strain (data not shown). Hence, A66:PM4-infected mice had fewer lung neutrophils and more T and B cells than A66.1- or A66:Δply-infected mice.
Figure 4.
Cellular profiles of the lungs of mice infected with wild-type, pneumolysin gene (ply)–deleted, and ply-recoded Streptococcus pneumoniae serotype 3 strains, showing flow cytometric characterization of the indicated cells in the lungs of BALB/c mice 48 h after infection with 5 × 104 colony-forming units of the indicated strains or TSB. A, CD45+CD3+CD4+ T cells. *P < .001 comparing A66:PM4 and A66.1; *P < .02 comparing A66:PM4 and A66:Δply (Student unpaired t test; n = 3). B, CD45+CD3+CD8+ T cells. *P < .02 comparing A66:PM4 and A66.1; *P < .02 comparing A66:PM4 and A66:Δply (Student unpaired t test; n = 3). C, CD45+CD19+ B cells. *P < .001 comparing A66:PM4 and A66.1. D, CD45+Ly6G+ cells. *P < .02 comparing A66:PM4 and A66.1; **P < .01 comparing A66.1 and A66:Δply (Student unpaired t test; n = 6). Bars represent the mean and error bars represent the standard deviation.
Lung Inflammatory Response of Mice Infected With SP3 Strains
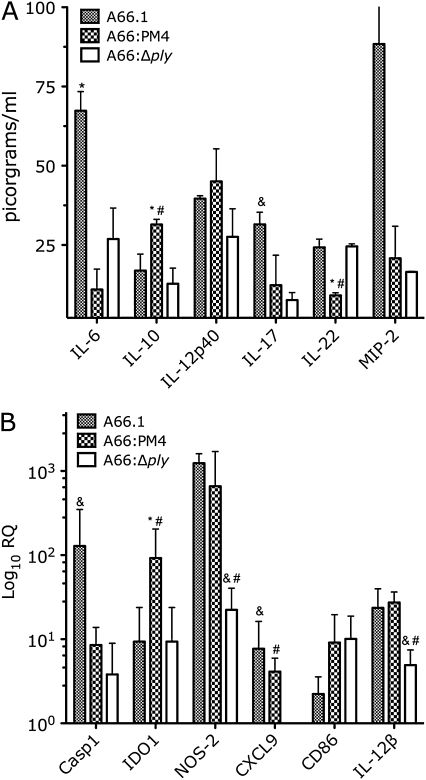
Levels of interleukin 6 (IL-6), interleukin 17 (IL-17), interleukin 22 (IL-22), interleukin 12p40 (IL-12p40), interleukin 10 (IL-10), and macrophage inflammatory protein 2 (MIP-2) in lung homogenates were determined by ELISA 48 h after infection to assess TH1, TH2, and TH17 immunity (Figure 5A). A66:PM4-infected mice had lower levels of IL-6 than A66.1-infected mice and lower levels of IL-22 and higher levels of IL-10 than A66.1- and A66:Δply-infected mice. MIP-2 levels were higher among A66.1-infected mice than among A66:Δply- and A66:PM4-infected mice, and IL-17 levels were higher among A66.1-infected mice than among A66:Δply-infected mice. Hence, A66:PM4-infected mice had higher levels of IL-10 and lower levels of IL-17, MIP-2, and IL-22 than A66.1- or A66:Δply-infected mice.
Figure 5.
Effect of wild-type, pneumolysin gene (ply)–deleted, and ply-recoded Streptococcus pneumoniae serotype 3 (SP3) strains on cytokine levels in the lungs of infected mice and lung dendritic cell (DC) gene expression in vitro. A, Concentration of the indicated cytokines in the lungs of BALB/c mice 48 h after infection with 5 × 104 colony-forming units of the indicated SP3 strains (n = 3). *P = .054 comparing macrophage inflammatory protein 2 (MIP-2) levels for A66.1 and A66:Δply (Student t test); *P < .04 comparing A66.1 and A66:PM4; #P < .04 comparing A66:PM4 and A66:Δply; &P < .04 comparing A66.1 and A66:Δply (Student t test). B, RNA levels of indicated genes in naïve lung DCs stimulated with the indicated SP3 strains as described in the text, expressed as logarithmic relative quantification (RQ) relative to naïve DCs from mouse lungs (n = 4–8). *P < .04 comparing A66.1 and A66:PM4; #P < .04 comparing A66:PM4 and A66:Δply; &P < .04 comparing A66.1 and A66:Δply (Mann-Whitney test). Casp1, apoptosis-related cysteine peptidase; IDO1, indoleamine 2,3-dioxygenase 1; IL-6, interleukin 6; IL-10, interleukin 10; IL-12β, interleukin 12 β; IL-12p40, interleukin 12p40; IL-17, interleukin 17; IL-22, interleukin 22; NOS2, nitric oxide synthase 2.
Gene Expression of Naïve BALB/c DCs Infected With SP3 Strains In Vitro
DC gene expression was analyzed in naïve DCs stimulated by A66.1, A66:PM4, and A66:Δply by means of quantitative RT-PCR (Figure 5B). Casp1 expression was higher in A66.1-stimulated DCs than in A66:Δply-stimulated DCs. NOS-2, IL-12β, and CXCL9 expression were each lower in A66:Δply-stimulated DCs than in A66.1- or A66:PM4-stimulated DCs; CXCL9 expression was undetectable in A66:Δply-stimulated DCs. IDO1 expression was higher in A66:PM4-stimulated DCs than in A66.1- or A66:Δply-stimulated DCs.
Antibody Response of A66:PM4-infected Mice
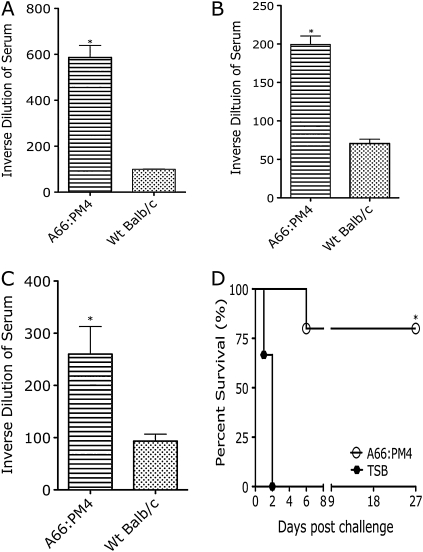
Eight (80%) of 10 A66:PM4-infected mice survived primary infection. Serologic analysis on 4 of these mice 28 d after infection showed they produced antibodies to whole bacteria (Figure 6A), PPS (Figure 6B), and PLY (Figure 6C). All surviving mice were infected with A66.1 on day 42 after primary infection with A66:PM4, and 4 (80%) of these 5 mice survived (Figure 6D).
Figure 6.
Antibody titers in serum samples of surviving A66:PM4-infected mice and their survival following challenge with A66.1. A, Serum levels of immunoglobulin G (IgG) and immunoglobulin M (IgM) to whole wild-type (Wt) A66.1 determined by whole-cell enzyme-linked immunosorbent assay (ELISA). *P < .001 comparing A66:PM4-infected surviving mice and uninfected mice (Student t test). B, Serum levels of IgG to pneumococcal capsular polysaccharide determined by ELISA. *P < .05 comparing A66:PM4-infected surviving mice and uninfected mice (Student t test). C, Serum levels of IgG to pneumolysin determined by ELSIA. *P < .001 comparing A66:PM4-infected surviving mice and uninfected mice (Student t test). D, Survival of surviving A66:PM4-infected mice challenged with wild-type A66.1 42 d after primary infection. *P < .003 comparing A66:PM4-infected and challenged mice and naïve mice (Kaplan-Meier test; n = 5). For panels A, B, and C, n = 5, bars represent the mean and error bars represent the standard deviation (n = 5).
DISCUSSION
In this study, synthetic gene design enabled modulation of SP3 ply expression and investigation of the effect of reduced, rather than absent, PLY production in vivo in experiments comparing the virulence of ply-expressing and ply-deleted SP3 strains in a mouse model of pneumococcal pneumonia. Heretofore, it has only been possible to evaluate the effect of PLY on the pathogenesis of S. pneumoniae with wild-type and ply-deleted strains [14, 32–34]. PLY production has been shown to stimulate innate immunity and bacterial clearance [35] and induce PMN and CD4+ T cell recruitment [36, 37]. Sublytic concentrations of PLY promote CD4+ T cell migration [37], pro-inflammatory cytokine secretion by mononuclear phagocytes [38], CXCL chemokine secretion by DCs [30], and mast cell antimicrobial peptide secretion [39]. The data herein suggest that the lower level of PLY production achieved by synthetic alteration of A66.1 ply resulted in beneficial immune stimulation, T cell recruitment, and bacterial clearance in vivo.
The synthetically modified SP3 strains A66:PM4 and A66:PM2 were less virulent in mice than either wild-type A66.1 or A66:Δply. The rate and level of PLY production and the resistance phenotype of the ply recoded strains was a function of the use of underrepresented codon pairs, with the caveat that although more A66:PM2 than A66.1-infected mice survived, the survival of A66:PM4- and A66:PM2-infected mice was comparable statistically. Interestingly, A66:Δply was as lethal as wild-type A66.1 in our model. Although a ply-deleted S. pneumoniae serotype 2 (SP2) strain was previously shown to be avirulent in mice [32], ply-deleted S. pneumoniae serotype 6A and serotype 8 strains were virulent [14, 33]. Consistent with our findings, PLY was shown to enhance resistance to SP3 (WU2) in mice [35]. A66:PM4 and wild-type A66.1 induced higher levels of DC pro-inflammatory gene expression in vitro than A66:Δply, but only A66:PM4-infected mice exhibited bacterial clearance. Hence our data suggest that the exuberant inflammatory response of wild-type A66.1-infected mice, as evidenced by their high lung levels of IL-17, IL-6, and MIP-2, and the absence of PLY-associated immune stimulation in A66:Δply-infected mice each result in an inability to promote bacterial clearance. Interestingly and relevant to our findings with A66:Δply, the Staphylococcus aureus toxin Panton-Valentine leukocidin was also recently shown to be required for bacterial clearance in mice [40].
PLY influenced IL-17 production in our model, as A66:PM4- and A66:Δply-infected mice each had fewer lung neutrophils and lower levels of IL-17 than wild-type A66.1-infected mice. These results underscore the previously reported importance of PLY in inducing neutrophil chemoattractants, PMN migration, and IL-17 in neutrophil recruitment [35, 41–43]. Although PLY and IL-17 were required for nasopharyngeal clearance of ST6B and ST23F in mice [34, 42], a recent study found that lung CFUs and survival were comparable in TH17-deficient and wild-type mice in an A66.1 pulmonary infection model [27]. Given that high levels of PLY inhibit neutrophil function [44], the lung bacterial burden in A66.1-infected mice could in part reflect a failure of neutrophil-mediated bacterial clearance.
The levels of CD4+ and CD8+ T cells in the lungs of A66:PM4-infected mice were higher than those of A66.1- or A66:Δply-infected mice. Although CD4+ T cells were shown to promote neutrophil recruitment and bacterial clearance in a SP2 infection model [37], they were recently shown to be detrimental in another SP2 model [45] and dispensable for survival in a SP3 (A66.1) model [27]. In the latter study, CD8+ T cells were required for resistance to lethal infection, which was associated with B cell recruitment, fewer lung PMNs, and a less exuberant TH17 response [27]. The data herein also show that A66:PM4-infected mice had a higher level of CD8+ T cells, fewer PMNs, and less IL-17 than wild-type A66.1-infected mice. A66:PM4-infected mice also had more lung B cells and higher levels of IL-10 than A66.1- and A66:Δply-infected mice. IL-10 was previously shown to improve survival in antibiotic-treated mice by dampening inflammation in a mouse SP3 pneumonia model [46]. Given the identification of an IL-10-producing regulatory B cell subset [47], it is possible that the lung IL-10 in A66:PM4-infected mice could originate from B cells. Although this hypothesis requires further study, our data link survival of A66:PM4-infected mice to production of this immunoregulatory mediator.
A66:PM4 and A66.1 induced DC expression of cellular activation (NOS-2) and chemokine (Il-12β and CXCL9) genes, whereas A66:Δply-stimulated DCs exhibited less pro-inflammatory gene expression. These results parallel those of previous studies that have linked PLY to pro-inflammatory and apoptotic mediator expression in human or bone-marrow-derived mouse DCs [30, 48]. Interestingly, A66:PM4 induced more DC IDO1 expression than either of the other strains. Given that IDO can modulate lung inflammation in mice [49], this is another mechanism by which A66:PM4 could have modulated the inflammatory response in our model. The decreased induction of Casp1 could enhance bacterial clearance in the setting of A66:PM4 infection, given that apoptosis of DCs has been linked to high levels of PLY [48]. Taken together, our findings suggest that the hypothesis that A66:PM4 stimulates lung DCs or other cells to produce immunoregulatory mediators that control S. pneumoniae–induced inflammation deserves further investigation.
The data herein provide proof of the principle that PLY production can be regulated by synthetic alteration of ply and suggest that synthetic gene customization could hold promise in the development of a universal vaccine for S. pneumoniae. By recoding rather than eliminating genes, wild-type epitopes are maintained. This is an advantage over knockout strategies, as demonstrated by our finding that A66:PM4-infected mice produced antibodies to PPS, PLY, and other bacterial antigens and were protected against wild-type A66.1 challenge. Anti-PLY antibodies, which can confer protection against S. pneumoniae in mice [50], would not be generated with a knockout vaccine strain. In our model, infection with A66:PM4 resulted in immunity to SP3, which was associated with lung T and B cell recruitment and IL-10 production. Expression of ply, albeit less than that of wild-type A66.1, was required for bacterial clearance. Either too much (wild-type) or the absence (A66:Δply) of ply expression was associated with a lethal phenotype in our model, with an intermediate amount (A66:PM4) being protective. Although we note that the effect of ply expression on virulence could be serotype-specific, the data herein suggest that modulation of virulence gene expression warrants further consideration as an approach to the design of a live-attenuated and/or universal S. pneumoniae vaccine.
Funding
This work was supported by the National Institutes of Health (NIH; grant numbers R01AI045459, R01AI044374 to L.P.); and the Ruth L. Kirschstein Molecular Pathogenesis Training Grant, NIH (grant number 5T32AI007506 to J.R.C.).
Acknowledgments
We appreciate the assistance of Berta Burd from the Laboratory for Macromolecular Analysis and Proteomics facility at AECOM for her assistance with Western blot resolution. We thank Catherine Manix for her assistance with the fluorescence-activated cell sorting (FACS) and cytokine sample preparation, Wendy Szymczak for her assistance with RT-PCR troubleshooting, and Shruti Gohil for her help with FlowJo analysis of FACS data.
References
- 1.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–7. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:3699–703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–74. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 5.Tats A, Tenson T, Remm M. Preferred and avoided codon pairs in three domains of life. BMC Genomics. 2008;9:463. doi: 10.1186/1471-2164-9-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller S, Coleman JR, Papamichail D, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–6. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (CDC) Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease–United States, 1998–2003. MMWR Morb Mortal Wkly Rep. 2005;54:893–7. [PubMed] [Google Scholar]
- 9.Hicks LA, Harrison LH, Flannery B, et al. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998–2004. J Infect Dis. 2007;196:1346–54. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 10.Li ST, Tancredi DJ. Empyema hospitalizations increased in US children despite pneumococcal conjugate vaccine. Pediatrics. 2009;125:26–33. doi: 10.1542/peds.2009-0184. [DOI] [PubMed] [Google Scholar]
- 11.Byington CL, Hulten KG, Ampofo K, et al. Molecular epidemiology of pediatric pneumococcal empyema from 2001 to 2007 in Utah. J Clin Microbiol. 2009;48:520–5. doi: 10.1128/JCM.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poolman J, Kriz P, Feron C, et al. Pneumococcal serotype 3 otitis media, limited effect of polysaccharide conjugate immunisation and strain characteristics. Vaccine. 2009;27:3213–22. doi: 10.1016/j.vaccine.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Abdelnour A, Soley C, Guevara S, Porat N, Dagan R, Arguedas A. Streptococcus pneumoniae serotype 3 among Costa Rican children with otitis media: clinical, epidemiological characteristics and antimicrobial resistance patterns. BMC Pediatr. 2009;9:52. doi: 10.1186/1471-2431-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roche AM, King SJ, Weiser JN. Live attenuated Streptococcus pneumoniae strains induce serotype-independent mucosal and systemic protection in mice. Infect Immun. 2007;75:2469–75. doi: 10.1128/IAI.01972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirst RA, Kadioglu A, O'Callaghan C, Andrew PW. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin Exp Immunol. 2004;138:195–201. doi: 10.1111/j.1365-2249.2004.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briles DE, Hollingshead SK, Nabors GS, Paton JC, Brooks-Walter A. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine. 2000;19(suppl 1):S87–95. doi: 10.1016/s0264-410x(00)00285-1. [DOI] [PubMed] [Google Scholar]
- 17.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med. 1944;79:137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briles DE, Nahm M, Schroer K, et al. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J Exp Med. 1981;153:694–705. doi: 10.1084/jem.153.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen K, Gladitz J, Antalis P, et al. Characterization, distribution, and expression of novel genes among eight clinical isolates of Streptococcus pneumoniae. Infect Immun. 2006;74:321–30. doi: 10.1128/IAI.74.1.321-330.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trzcinski K, Thompson CM, Lipsitch M. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol. 2003;69:7364–70. doi: 10.1128/AEM.69.12.7364-7370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paton JC, Lock RA, Hansman DJ. Effect of immunization with pneumolysin on survival time of mice challenged with Streptococcus pneumoniae. Infect Immun. 1983;40:548–52. doi: 10.1128/iai.40.2.548-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanclerski K, Mollby R. Production and purification of Streptococcus pneumoniae hemolysin (pneumolysin) J Clin Microbiol. 1987;25:222–5. doi: 10.1128/jcm.25.2.222-225.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benton KA, Paton JC, Briles DE. Differences in virulence for mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect Immun. 1997;65:1237–44. doi: 10.1128/iai.65.4.1237-1244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cima-Cabal MD, Mendez FJ, Vazquez F, del Mar Garcia-Suarez M, de los Toyos JR. A specific and ultrasensitive chemiluminescent sandwich ELISA test for the detection and quantitation of pneumolysin. J Immunoassay Immunochem. 2001;22:99–112. doi: 10.1081/IAS-100103223. [DOI] [PubMed] [Google Scholar]
- 25.Cima-Cabal MD, Mendez FJ, Vazquez F, et al. Immunodetection of pneumolysin in human urine by ELISA. J Microbiol Methods. 2003;54:47–55. doi: 10.1016/s0167-7012(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 26.Tian H, Weber S, Thorkildson P, Kozel TR, Pirofski LA. Efficacy of opsonic and nonopsonic serotype 3 pneumococcal capsular polysaccharide-specific monoclonal antibodies against intranasal challenge with Streptococcus pneumoniae in mice. Infect Immun. 2009;77:1502–13. doi: 10.1128/IAI.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber S, Tian H, Pirofski LA. CD8+ cells enhance resistance to pulmonary serotype 3 Streptococcus pneumoniae infection in mice. J Immunol. doi: 10.4049/jimmunol.1001963. Publish online before print 6 December 2010; doi:10.4049/jimmunol.1001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Littmann M, Albiger B, Frentzen A, Normark S, Henriques-Normark B, Plant L. Streptococcus pneumoniae evades human dendritic cell surveillance by pneumolysin expression. EMBO Mol Med. 2009;1:211–22. doi: 10.1002/emmm.200900025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymczak WA, Deepe GS., Jr The CCL7-CCL2-CCR2 axis regulates IL-4 production in lungs and fungal immunity. J Immunol. 2009;183:1964–74. doi: 10.4049/jimmunol.0901316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernatoniene J, Zhang Q, Dogan S, Mitchell TJ, Paton JC, Finn A. Induction of CC and CXC chemokines in human antigen-presenting dendritic cells by the pneumococcal proteins pneumolysin and CbpA, and the role played by Toll-like receptor 4, NF-kappaB, and mitogen-activated protein kinases. J Infect Dis. 2008;198:1823–33. doi: 10.1086/593177. [DOI] [PubMed] [Google Scholar]
- 31.Tian H, Groner A, Boes M, Pirofski LA. Pneumococcal capsular polysaccharide vaccine-mediated protection against serotype 3 Streptococcus pneumoniae in immunodeficient mice. Infect Immun. 2007;75:1643–50. doi: 10.1128/IAI.01371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berry AM, Yother J, Briles DE, Hansman D, Paton JC. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989;57:2037–42. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marks M, Burns T, Abadi M, et al. Influence of neutropenia on the course of serotype 8 pneumococcal pneumonia in mice. Infect Immun. 2007;75:1586–97. doi: 10.1128/IAI.01579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthias KA, Roche AM, Standish AJ, Shchepetov M, Weiser JN. Neutrophil-toxin interactions promote antigen delivery and mucosal clearance of Streptococcus pneumoniae. J Immunol. 2008;180:6246–54. doi: 10.4049/jimmunol.180.9.6246. [DOI] [PubMed] [Google Scholar]
- 35.Malley R, Henneke P, Morse SC, et al. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci U S A. 2003;100:1966–71. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun K, Salmon SL, Lotz SA, Metzger DW. Interleukin-12 promotes gamma interferon-dependent neutrophil recruitment in the lung and improves protection against respiratory Streptococcus pneumoniae infection. Infect Immun. 2007;75:1196–202. doi: 10.1128/IAI.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadioglu A, Coward W, Colston MJ, Hewitt CR, Andrew PW. CD4-T-lymphocyte interactions with pneumolysin and pneumococci suggest a crucial protective role in the host response to pneumococcal infection. Infect Immun. 2004;72:2689–97. doi: 10.1128/IAI.72.5.2689-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houldsworth S, Andrew PW, Mitchell TJ. Pneumolysin stimulates production of tumor necrosis factor alpha and interleukin-1 beta by human mononuclear phagocytes. Infect Immun. 1994;62:1501–3. doi: 10.1128/iai.62.4.1501-1503.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruse G, Fernandes VE, de Salort J, et al. Human lung mast cells mediate pneumococcal cell death in response to activation by pneumolysin. J Immunol. 2010;184:7108–15. doi: 10.4049/jimmunol.0900802. [DOI] [PubMed] [Google Scholar]
- 40.Yoong P, Pier GB. Antibody-mediated enhancement of community-acquired methicillin-resistant Staphylococcus aureus infection. Proc Natl Acad Sci U S A. 2010;107:2241–6. doi: 10.1073/pnas.0910344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Clarke TB, Weiser JN. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest. 2009;119:1899–909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu YJ, Gross J, Bogaert D, et al. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 2008;4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreland JG, Bailey G. Neutrophil transendothelial migration in vitro to Streptococcus pneumoniae is pneumolysin dependent. Am J Physiol Lung Cell Mol Physiol. 2006;290:L833–40. doi: 10.1152/ajplung.00333.2005. [DOI] [PubMed] [Google Scholar]
- 44.Paton JC, Ferrante A. Inhibition of human polymorphonuclear leukocyte respiratory burst, bactericidal activity, and migration by pneumolysin. Infect Immun. 1983;41:1212–6. doi: 10.1128/iai.41.3.1212-1216.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemessurier K, Hacker H, Tuomanen E, Redecke V. Inhibition of T-cells provides protection against early invasive pneumococcal disease. Infect Immun. 2010;78:5287–94. doi: 10.1128/IAI.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang E, Bergeron Y, Bergeron MG. Ceftriaxone pharmacokinetics in interleukin-10-treated murine pneumococcal pneumonia. J Antimicrob Chemother. 2005;55:721–6. doi: 10.1093/jac/dki085. [DOI] [PubMed] [Google Scholar]
- 47.Yanaba K, Bouaziz JD, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–50. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 48.McNeela EA, Burke A, Neill DR, et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog. 2010;6:e1001191. doi: 10.1371/journal.ppat.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson BA, 3rd, Kahler DJ, Baban B, et al. B-lymphoid cells with attributes of dendritic cells regulate T cells via indoleamine 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2010;107:10644–8. doi: 10.1073/pnas.0914347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton JC. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect Immun. 2007;75:350–7. doi: 10.1128/IAI.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]