Abstract
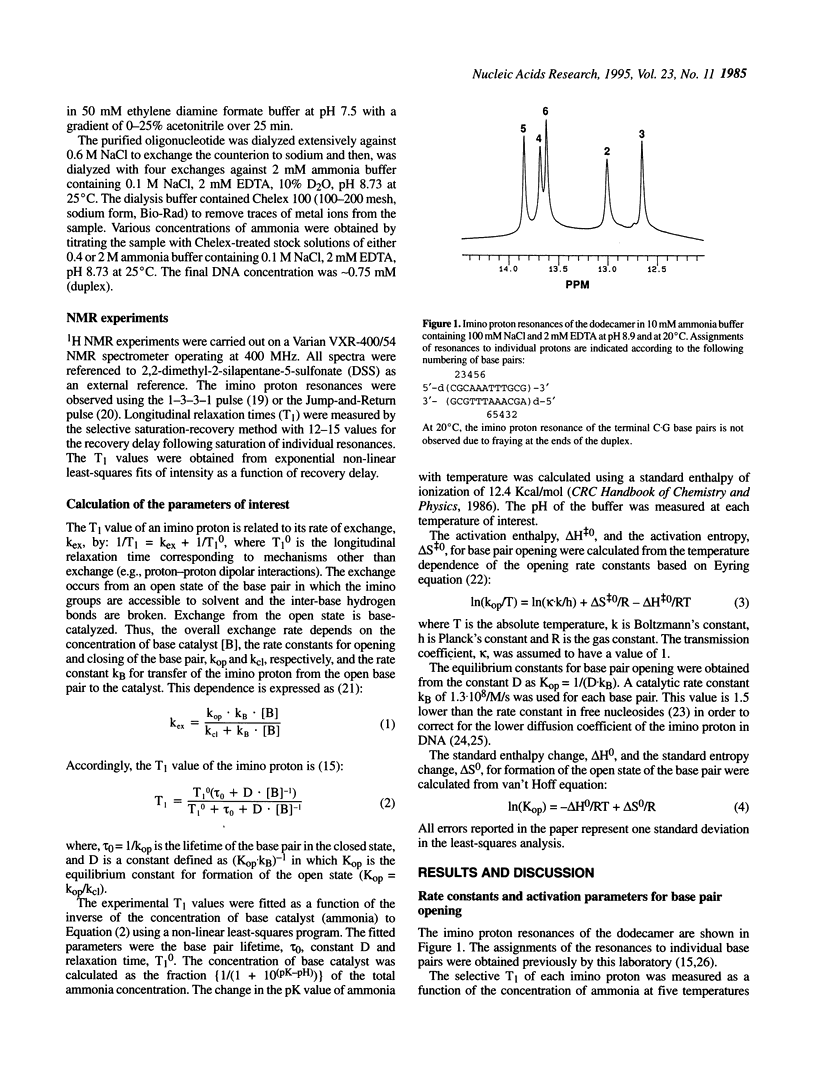
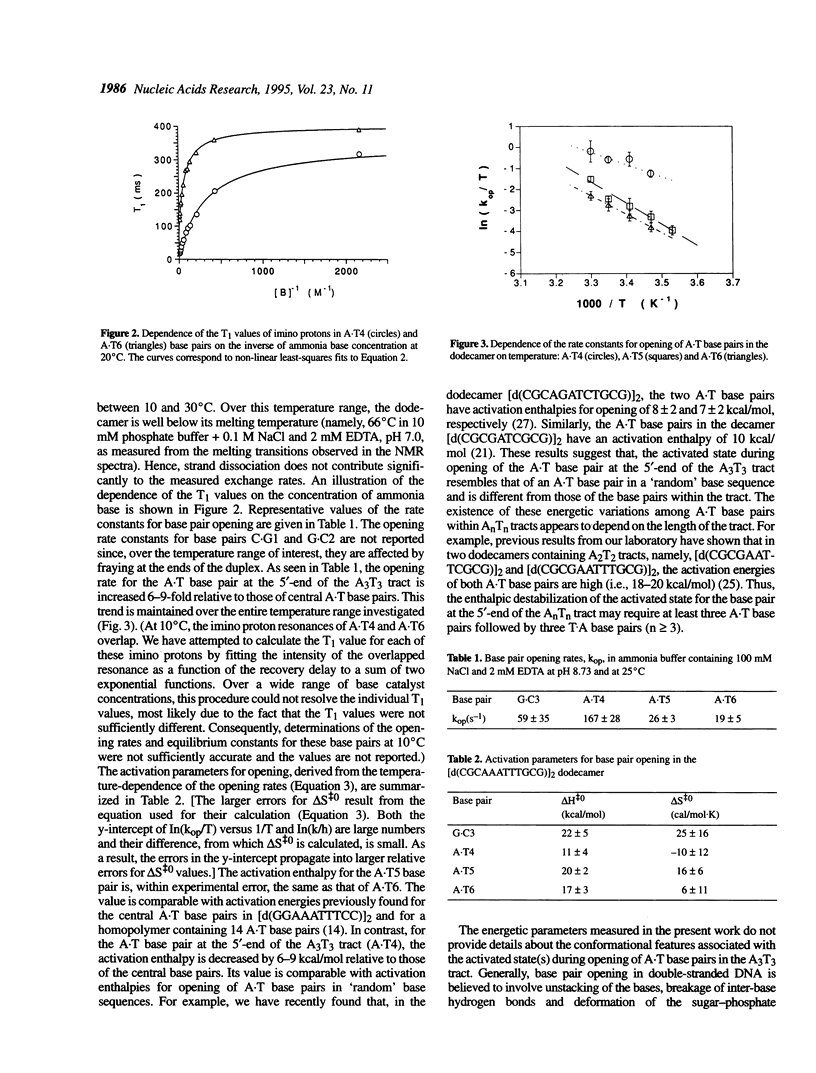
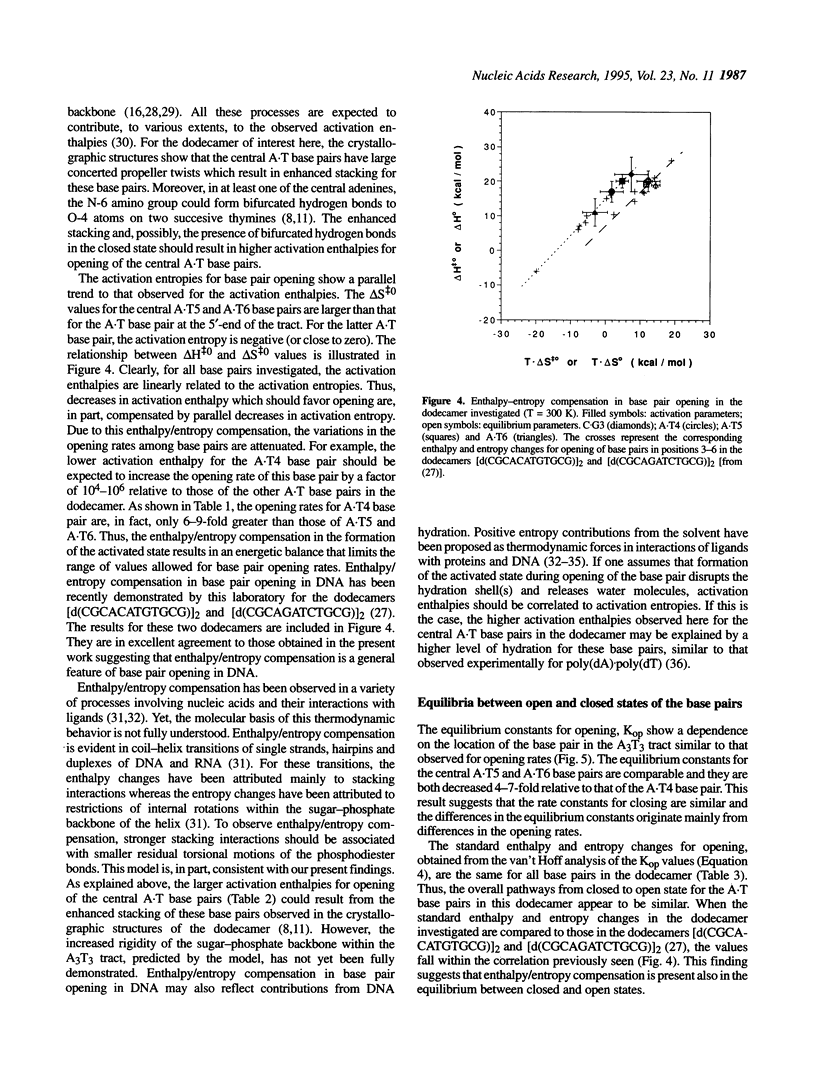
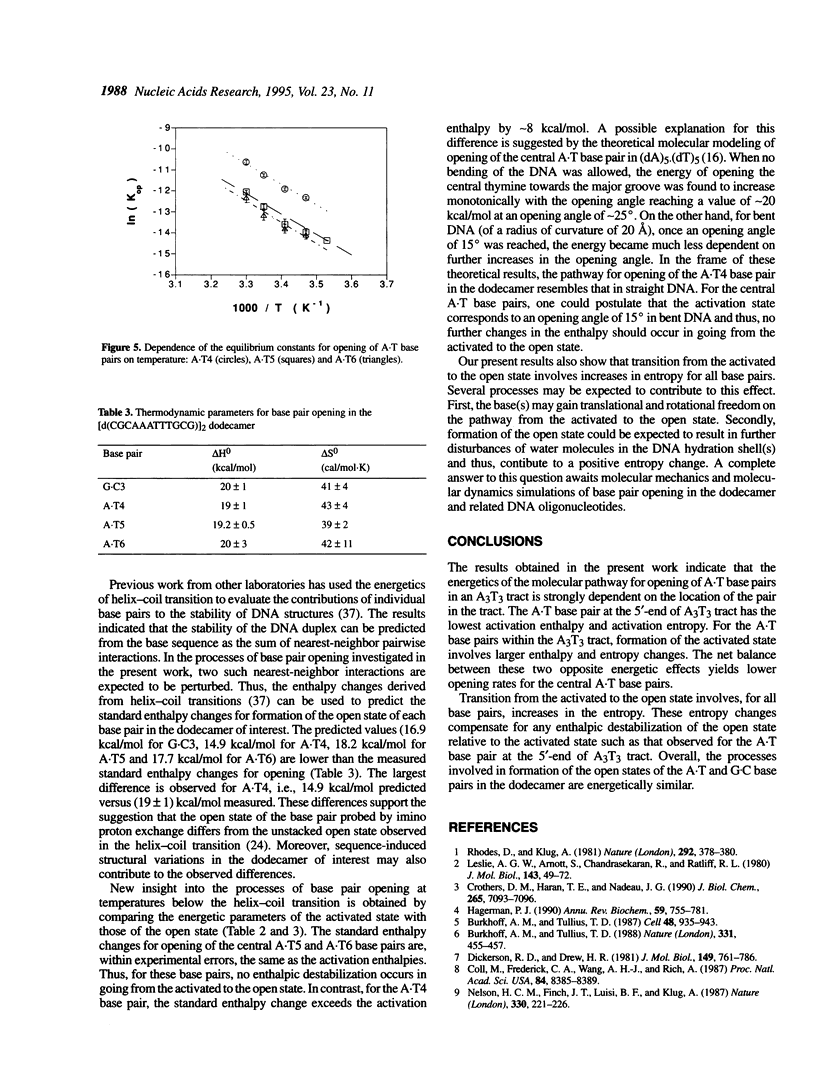
Nuclear magnetic resonance spectroscopy has been used to characterize the kinetics and energetics of opening of base pairs in the DNA dodecamer [d(CGCAAATTTGCG)]2. The dodecamer contains an A3T3 tract that induces intrinsic curvature of the helix axis. Previous studies from this and other laboratories have shown that the kinetics of base pair opening in AnTn tracts is unique: the opening rates of the A.T base pairs in the interior of the tract are much lower than that of the A.T base pair at the 5'-end of the tract. In the present work, we have investigated the energetics of the pathways for opening of the A.T base pairs in the A3T3 tract. The energetic parameters of the activated state(s) are obtained from the temperature dependence of the opening rate constants. The lower opening rates for the A.T base pairs situated in the interior of the tract are shown to originate from higher activation enthalpies which are compensated, in part, by increases in the activation entropies. We have also obtained an energetic characterization of the open state(s) of the A.T base pairs in the dodecamer by measuring the equilibrium constants for base pair opening and their temperature dependence. The results suggest that the transitions from closed to open state(s) in the A.T base pairs of the A3T3 tract are energetically similar.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benight A. S., Schurr J. M., Flynn P. F., Reid B. R., Wemmer D. E. Melting of a self-complementary DNA minicircle. Comparison of optical melting theory with exchange broadening of the nuclear magnetic resonance spectrum. J Mol Biol. 1988 Mar 20;200(2):377–399. doi: 10.1016/0022-2836(88)90248-3. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K. J., Remeta D. P., Chou W. Y., Ferrante R., Curry J., Zaunczkowski D., Snyder J. G., Marky L. A. Enthalpy-entropy compensations in drug-DNA binding studies. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8922–8926. doi: 10.1073/pnas.84.24.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Heinemann U., Nurislamov A. A., Zielenkiewicz P., Dickerson R. E., Saenger W. Molecular dynamics simulation of the hydration shell of a B-DNA decamer reveals two main types of minor-groove hydration depending on groove width. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):593–597. doi: 10.1073/pnas.88.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Mazzarelli J. M., McLaughlin L. W., von Kitzing E., Travers A. A. DNA curvature does not require bifurcated hydrogen bonds or pyrimidine methyl groups. J Mol Biol. 1992 Jun 5;225(3):729–738. doi: 10.1016/0022-2836(92)90397-3. [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Brown D. G., Spink N., Skelly J. V., Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992 Aug 20;226(4):1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E., Russu I. M. Sequence dependence of base-pair opening in a DNA dodecamer containing the CACA/GTGT sequence motif. Biochemistry. 1994 Sep 13;33(36):11016–11024. doi: 10.1021/bi00202a022. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Keepers J. W., Kollman P. A., Weiner P. K., James T. L. Molecular mechanical studies of DNA flexibility: coupled backbone torsion angles and base-pair openings. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5537–5541. doi: 10.1073/pnas.79.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy J. L., Charretier E., Kochoyan M., Guéron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988 Dec 13;27(25):8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Kochoyan M., Huynh-Dinh T., Guéron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J Mol Biol. 1988 Mar 20;200(2):223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- Leslie A. G., Arnott S., Chandrasekaran R., Ratliff R. L. Polymorphism of DNA double helices. J Mol Biol. 1980 Oct 15;143(1):49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9(10):1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Origins of netropsin binding affinity and specificity: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4359–4363. doi: 10.1073/pnas.84.13.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczyk R., Russu I. M. Determination of distances involving exchangeable protons from NOESY spectra of two DNA dodecamers. FEBS Lett. 1993 Oct 4;331(3):217–222. doi: 10.1016/0014-5793(93)80340-z. [DOI] [PubMed] [Google Scholar]
- Moe J. G., Russu I. M. Kinetics and energetics of base-pair opening in 5'-d(CGCGAATTCGCG)-3' and a substituted dodecamer containing G.T mismatches. Biochemistry. 1992 Sep 15;31(36):8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- Moe J. G., Russu I. M. Proton exchange and base-pair opening kinetics in 5'-d(CGCGAATTCGCG)-3' and related dodecamers. Nucleic Acids Res. 1990 Feb 25;18(4):821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet J., Blicharski J., Brahms J. Conformations and structural transitions in polydeoxynucleotides. Biochemistry. 1975 May 6;14(9):1869–1876. doi: 10.1021/bi00680a011. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Searle M. S., Williams D. H. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993 May 11;21(9):2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]



