Abstract
The activation of natural killer (NK) cells is modulated by surface molecules. We analyzed NK cells in human immunodeficiency virus (HIV)–exposed seronegative (HESN) individuals by means of molecular typing of HLA B, Cw, and killer cell immunoglobulin-like receptor (KIR) molecules. In HESN individuals, compared with HIV patients, the frequency of the inhibitory KIR3DL1 allele and of the KIR3DL1+/Bw4+ inhibitory complex was reduced, whereas that of the activatory KIR3DS1+ ligand and the activatory Bw4+/3DL1–/3DS1+ complex was increased, resulting in a statistically significant diversion from Hardy-Weinberg equilibrium (KIR3DS1 homozygote) in HESN individuals. The reciprocal equilibrium between inhibitory and activatory NK receptors and their ligands favors NK activation in HESN individuals.
The effect of host genetic variation on the outcome of human immunodeficiency virus (HIV) infection is well documented; studies have recently focused on genes encoding for HLA class I molecules whose polymorphism influences disease progression [1, 2]. The crucial role of such molecules in determining interindividual levels of susceptibility to diseases was highlighted by the discovery of HLA class I as ligands for killer cell immunoglobulin-like receptors (KIRs), a set of molecules that modulate natural killer (NK) cell activity [3]. NK cells play an important role in innate immunity by providing protection against viruses and tumor cells [4]; these cells are functionally stimulated or suppressed by the integration of activating and inhibitory signals transmitted through cell surface receptors, including KIRs [3, 5]. Recent evidence has shown that the net result of such integration plays an important role in modulating differential susceptibility to diseases [6, 7]. In the case of HIV infection, in particular, the functional interaction between HLA-B Bw4–Ile80 and KIR3DS1 leads to activation of NK cells, resulting in a more efficient elimination of HIV type 1 (HIV-1)–infected cells [8, 9].
Resistance to HIV infection in individuals who have been repeatedly exposed to HIV but do not become infected (HIV–exposed seronegative [HESN] individuals) is a multifactorial event that includes both a particular genetic background and the activation of multiple effector mechanisms belonging to innate and acquired immunity [reviewed in 10, 11]. Notably, NK cell activity has been shown to be increased in HESN individuals [12]. To assess whether genetic interactions between specific KIR genes and their HLA class I ligands could justify this observation and may contribute to resistance to HIV infection, we examined activatory and inhibitory KIR molecules and their putative HLA ligands in Italian HESN individuals, comparing the results with those obtained in a homogeneous group of HIV-infected patients and with those obtained in healthy individuals without known risk factors for HIV infection.
METHODS
HESN Individuals and Control Participants
Three hundred thirty-nine unrelated white Italian individuals were enrolled in the study: 44 HESN individuals, 192 HIV-infected patients, and 103 healthy control participants without known risk factors for HIV infection. The HESN cohort has been followed since 1997 and is described in detail elsewhere [13]. Briefly, inclusion criteria for HESN individuals were a history of multiple unprotected sexual episodes for more than 4 years at the time of enrollment, with at least 3 episodes of at-risk intercourse within 4 months before study entry, and an average of 30 (range, 18 to >100) reported unprotected sexual contacts per year. Epidemiological and clinical analyses of the HESN individuals were performed at the Department of Obstetrics and Gynecology, Santa Maria Annunziata Hospital (Florence, Italy). A group of 192 consecutively enrolled and unrelated patients with chronic HIV infection but no diagnosis of AIDS was also included in the study. Patients had seroconverted at least 4 years before the study period and had progressed to category 2–3 according to the Centers for Disease Control and Prevention 1993 definition [14] (ie, CD4+ count, <500 cells/mL); most (89%) were receiving antiretroviral therapy (ART). HIV-infected patients displayed a median nadir CD4+ count of 138 cells/μL (range, 5–498 cells/mL) and had a median pre-ART viremia of 4.8 log10 HIV - RNA (range, 2.7 -– 5.9). Finally, 103 age-matched HIV-seronegative healthy control participants, without known risk factors for HIV infection, were studied to account for possible bias in the selection of the HIV-infected population. All the participants were included in the study upon signing an informed consent release approved by the institutional ethical committee of the enrolling centers.
HLA and KIR Genotyping
Genomic DNA was isolated from peripheral blood using standard procedures. Molecular typing of HLA B (22 alleles), Cw (14 alleles), and KIR (18 alleles) was performed by polymerase chain reaction (PCR) using the sequence-specific primer (SSP) method with commercial kits according to the manufacturers’ instructions (BAG-Lich and Invitrogen Corporation). Detection of the alleles recognized by the SSPs was performed after amplification in a GeneAmp PCR 9700 thermocycler (Applied Biosystems) by means of gel electrophoresis on 2% agarose gel.
KIR Haplotype and Ligands
Genotypes can be resolved into 2 broad haplotypes, termed A and B, on the basis of KIR genes. The B haplotype is defined by the presence of 1 or more of the following genes: 2DL2, 2DL5, 3DS1, 2DS1, 2DS2, 2DS3, or 2DS5. If none of these genes is present, an AA genotype is identified. In the A haplotype, variability in gene content is usually absent but allele variability is high. To classify A and/or B haplotypes, we referred to the criteria adopted by Middleton (http://www.allelefrequencies.net), which do not distinguish between AB and BB genotypes, calling any of these Bx [15]. Ligand groups were defined as follows:
KIRs 2DL1 and 2DS1 have as their ligand the C2 epitope (asparagine at position 77, lysine at position 80) present in HLA-Cw2, -Cw4, -Cw5, -Cw6, -Cw17, and -Cw18.
KIRs 2DL2, 2DL3, and 2DS2 have as their ligand the C1 epitope (serine at position 77, asparagine at position 80) present in HLA-Cw1, -Cw3, -Cw7, -Cw8, -Cw13, -Cw14, and -Cw16.
HLA-Bw4 was considered the ligand for 3DL1 (and 3DS1) [16].
Statistical Analysis
Differences between groups were assessed by means of the χ2 test for categorical variables. P values were calculated with the Yates correction (py) or Fisher exact test (pf) applied when appropriate. The Bonferroni correction for multiple tests (pc) was applied by multiplying the P value by the number of degrees of freedom (dof) obtained by calculating n – 1, where n is the number of alleles analyzed. The association of each polymorphism with disease (condition of HIV infection) was measured by the odds ratio (OR) with 95% confidence interval (CI). For each independent variable, crude and adjusted ORs and 95% CIs were calculated. Analyses were carried out using SPSS, version 16.0 for Windows.
RESULTS
HLA and KIR Distribution in the Cohort Study
Initial results indicated that HLA-B and -Cw allelic frequencies are comparable in HESN individuals, HIV-infected patients, and healthy control participants (data not shown). Frequencies of KIR gene distribution were not different in HIV-infected individuals, compared with healthy control individuals, either, confirming the lack of a selection bias in the cohort of HIV-infected individuals.
In contrast with this result, whereas the frequency of the inhibitory KIR3DL1 allele was comparable in HIV-infected individuals (93.2%) and healthy control participants (94.2%), this allele was significantly less frequent in HESN individuals (77.2%) (vs HIV-infected individuals: pf = .003, pc = .05 [17 dof]; OR, 4.05 [95% CI, 1.5–10.9]; vs healthy controls: pf = .006, pc = .10 [17 dof]). This difference was confirmed both by Fisher exact text for low number comparisons (pf) and by Bonferroni correction for multiple tests (pc) (Table 1). No statistically significant differences were detected between HESN individuals and HIV-infected patients in the frequency of either KIR haplotypes (AA and Bx) (data not shown) or HLA B (Bw4 and Bw6) (Table 1) and C1/C2 KIR ligands (data not shown).
Table 1.
KIR and KIR Ligands in Human Immunodeficiency Virus-Exposed Seronegative Individuals, Human Immunodeficiency Virus-Infected Participants, and Healthy Unexposed Individuals (Control Participants)
| KIR gene and ligands | HESN individuals, no. (%)‐ (N = 44) | HIV-infected participants, no. (%) (N = 192) | P | OR (95% CI) | Healthy controls, no. (%) (N = 103) |
| 3DL1 | 34 (77.2) | 179 (93.2) | pf = .003a | 4.05 (1.50–10.88) | 97 (94.2) |
| 3DS1 | 20 (45.5) | 69 (35.9) | py = .32 | 0.67 (0.33–1.37) | 43 (41.7) |
| Bw4+ | 29 (65.9) | 118 (61.5) | py = .71 | 0.82 (0.39–1.72) | 64 (62.1) |
| Bw4– | 15 (34.1) | 74 (38.5) | 39 (37.9) | ||
| 3DL1+/Bw4+ | 22 (75.9) | 111 (94.1) | pf = .007b | 5.05 (1.41–18.19) | 62 (96.9) |
| 3DL1–/Bw4+ | 7 (24.1) | 7 (5.9) | 2 (3.1) | ||
| 3DL1+/Bw4– | 13 (86.7) | 68 (91.9) | pf = .61 | 1.74 (0.22–11.41) | 35 (89.7) |
| 3DL1–/Bw4– | 2 (13.3) | 6 (8.1) | 4 (10.3) | ||
| 3DS1+/Bw4+ | 13 (44.8) | 38 (32.2) | py = .29 | 0.58 (0.24–1.45) | 26 (40.6) |
| 3DS1–/Bw4+ | 16 (55.2) | 80 (67.8) | 38 (59.4) | ||
| 3DS1+/Bw4– | 7 (46.7) | 31 (41.9) | py = .95 | 0.82 (0.24–2.87) | 17 (43.6) |
| 3DS1–/Bw4– | 8 (53.3) | 43 (58.1) | 22 (56.4) | ||
| 3DL1+/3DS1+ | 11 (25.0) | 56 (29.2) | py= .71 | 1.24 (0.55–2.81) | 38 (36.9) |
| 3DL1+/3DS1– | 24 (54.5) | 123 (64.0) | py = .32 | 1.49 (0.73–3.03) | 59 (57.3) |
| 3DL1–/3DS1+ | 9 (20.4) | 13 (6.7) | pf = .009c | 0.28 (0.10–0.78) | 6 (5.8) |
| Bw4+/3DL1+/3DS1+ | 6 (20.7) | 31 (26.3) | py = .70 | 1.37 (0.47–4.15) | 24 (37.5) |
| Bw4+/3DL1+/3DS1– | 16 (55.2) | 80 (67.8) | py = .28 | 1.71 (0.69–4.22) | 38 (59.4) |
| Bw4+/3DL1–/3DS1+ | 7 (24.1) | 7 (5.9) | pf = .007c | 0.20 (0.05–0.71) | 2 (3.1) |
| Bw4–/3DL1+/3DS1+ | 5 (33.3) | 25 (33.8) | py = .79 | 1.02 (0.28–3.89) | 14 (35.9) |
| Bw4–/3DL1+/3DS1– | 8 (53.3) | 43 (58.1) | py = .95 | 1.21 (0.35–4.20) | 21 (53.8) |
| Bw4–/3DL1–/3DS1– | 2 (13.3) | 6 (8.1) | pf = .61 | 0.57 (0.09–4.64) | 4 (10.3) |
NOTE. Data are no. (%) of participants unless otherwise specified. Statistical significance is indicated by a P value of <.05. Bw4– (Bw4–/– aka Bw6+/+); Bw4+, (Bw4+/+ Bw4+/–). CI, confidence interval; OR, odds ratio; pf, P value with Fisher correction; pc, P value corrected for degrees of freedom (dof = n – - 1, where n is the number of alleles analyzed) calculated between Human Immunodeficiency Virus (HIV) group and HIV-exposed seronegative (HESN) group.
apc= .05 corrected for 17 dof.
bpc = .01 corrected for 3 dof.
cpc = .02 corrected for 2 dof.
Because the activity of KIR molecules is secondary to their interaction with HLA ligands, we next evaluated the frequency of all the possible KIR3DL1-HLABw4 ligand complexes in HESN individuals and HIV-infected individuals. A statistically significantly different distribution was observed for the inhibitory KIR3DL1/Bw4 complex (pc = .02 [3 dof]). This result was due to a different distribution of the KIR3DL1 molecule when Bw4 carriers among HESN individuals and HIV-infected patients were compared. Thus, the KIR3DL1+/Bw4+ complex was less frequent in HESN than in HIV-infected individuals (pf = .007; pc = .02 [3 dof]; OR, 5.05 [95% CI, 1.41–18.19]).
The segregation of the activatory KIR3DS1+ with its putative ligand Bw4+ was evaluated as well; this complex was observed in 48.8% of HESN individuals and 32.2% of HIV-infected patients (py = .29).
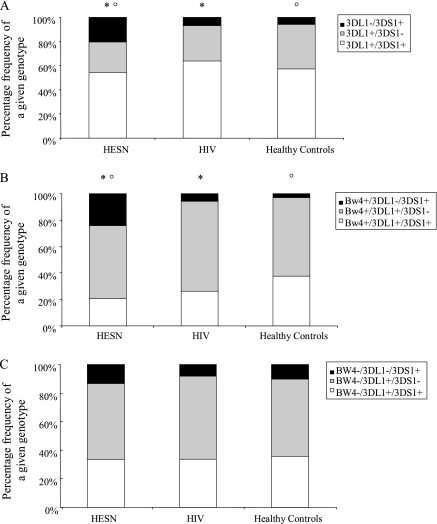
The distribution of the KIR3DL1/3DS1 genotype was evaluated next. Whereas the KIR3DS1 and KIR3DL1 homozygotes, as well as the KIR3DL1/3DS1 heterozygote, did not deviate statistically from Hardy-Weinberg equilibrium at this locus in HIV-infected individuals (P = .07), the same distribution in HESN individuals significantly diverged from Hardy-Weinberg equilibrium (P = .004). A different distribution of KIR3DL1/3DS1 was revealed between the HESN group and both the HIV-infected group and the healthy control group (pc = .02 [2 dof]) (Figure 1). This result was mostly associated with a higher frequency of KIR3DS1 homozygous (3DL1–/3DS1+) genotype in HESN individuals (20.4% vs 6.7% in HIV-infected individuals) (pf = .009; OR, 0.28 [95% CI, 0.10 -–0 .78]).
Figure 1.
A, KIR3DS1/3DL1 genotypes in human immunodeficiency virus–exposed seronegative individuals (Human Immunodeficiency Virus-exposed seronegative [HESN]; n = 44), HIV-infected patients (Human Immunodeficiency Virus [HIV]; n = 192), and unexposed healthy control participants ( n = 103). *pc = .02 between HESN and HIV-infected individuals; pc = .02 between HESN individuals and healthy controls. B, KIR3DS1/3DL1 genotypes in Bw4 carriers (HESN, n = 29; HIV, n = 118; healthy controls, n = 64). *pc = .01 between HESN and HIV; pc = .04 between HESN individuals and healthy controls. C, KIR3DS1/3DL1 genotypes in Bw4-negative participants (HESN, n = 15; HIV, n = 74; healthy controls, n = 39).
Genotypes for both KIR3DL1 and KIR3DS1 alleles in Bw4-positive and Bw4-negative HESN and HIV-infected participants were also analyzed. No differences were detected in Bw4-negative participants, whereas a different distribution of KIR3DL1/3DS1 was observed in Bw4 carriers (pc = .01 [2 dof]). In particular, the activatory Bw4+/3DL1–/3DS1+ complex was more frequent in HESN individuals (24.1%), compared with HIV-infected individuals (5.9%) (pf = .007; OR, 0.20 [95% CI, 0.05– - 0.71]), whereas the inhibitory Bw4+/3DL1+/3DS1– complex was observed in 53.1% of HESN individuals and 67.8% of HIV-infected patients (py = .28; OR, 1.71 [95% CI, 0.69 - –4.22) (Table 1 and Figure 1). Final analyses evaluating allele and genotype frequencies for Bw4-80I and Bw4-80T subgroups did not show any signficant difference as is shown in Supplementary Table 1.
DISCUSSION
We compared the distribution of KIRs and their HLA ligands in Italian HESN individuals and HIV-infected individuals to verify whether the preferential presence of specific KIR genes and their HLA class I ligands could contribute to resistance to HIV infection. Our results show that HESN status is negatively associated with the inhibitory Bw4/KIR3DL1 functional complex; the data also suggest the presence of a possible association between HESN status and the activatory KIR3DS1 allele and the Bw4+/3DL1–/3DS1+ complex. An increased proportion of KIR3DS1 was previously reported in a group of Canadian HESN individuals [12]; our results expand these data by showing that the activatory Bw4+/3DL1–/3DS1+ complex is significantly more frequent in HESN individuals in association with a decreased frequency of the inhibitory Bw4/KIR3DL1 structure. Notably, these differences are observed in Bw4 carriers alone.
NK cells play a pivotal role in the first line of immune defense against viruses [4]. These cells express a panel of stimulatory and inhibitory molecules [2]; the activation or lack thereof of NK cells depends on the reciprocal balance of such functionally different molecules [2, 3]. KIRD3S1 is one of the main activatory molecules; its action is counterbalanced by KIR3DL1, an inhibitory receptor binding to HLA Bw4 [7]. Previous studies have revealed that the coexpression of KIRD3S1 and a particular subset of Bw4 (Bw4-180) is associated with slower progression of HIV infection [5], indicating a role for NK cells in modulating the clinical history of HIV disease [8]. Functional analyses of NK cells in HESN individuals have also shown that these cells are more activated in such individuals, which suggests that NK activity may be involved in protection against HIV [12]. Our results include both individuals who express Bw4*80I alleles and individuals who express Bw4*80T alleles and were not confirmed in subanalyses performed on HLA Bw4*80I-positive individuals alone, probably because of the lower number of HLA Bw4*80I carriers among study participants.
Our results suggest that in HESN individuals a reduced quantity of the KIR3DL1 (inhibitory) allele, associated with an increased amount of the KIR3DS1 (activatory) allele, would reduce KIR3DL1-mediated inhibition of NK cells within the context of an augmented interaction between the activatory KIR3DS1 and Bw4. The net result would be increased NK cell activity. The reciprocal equilibrium between inhibitory and activatory NK receptors and their ligands could thus be skewed in HESN individuals, in whom an “NK activatory milieu” would be present. The early accumulation of highly activated NK cells may provide a potent first-line defense allowing for the initial control of acute HIV replication while adaptive immune responses are still developing.
Resistance to HIV infection is an extremely complex phenomenon associated with a variety of both genetic and immune correlates; our results, although obtained in a relative small cohort of individuals, confirm that NK cells play a role in this phenomenon and suggest a possible explanation for the increased NK activity seen in HESN individuals.
Supplementary Data
Supplementary data are available at http://jid.oxfordjournals.org online.
Funding
This work was supported in part by grants from Istituto Superiore di Sanità “Programma Nazionale di Ricerca sull’AIDS,” the EMPRO and AVIP EC WP6 Projects, the nGIN EC WP7 Project, the Japan Health Science Foundation, 2008 Ricerca Finalizzata (Italian Ministry of Health), 2008 Ricerca Corrente (Italian Ministry of Health), Progetto FIRB RETI: Rete Italiana Chimica Farmaceutica CHEM - PROFARMA - NET (RBPR05NWWC), and Fondazione CARIPLO. No funds were received for this work from any of the following organizations: National Institutes of Health (NIH), Wellcome Trust, Howard Hughes Medical Institute (HHMI).
References
- 1.Kosmrlj A, Read EL, Qi Y, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–4. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 4.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–9. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 5.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 6.Fusco C, Guerini FR, Nocera G, et al. KIRs and their HLA-ligands in remitting-relapsing multiple sclerosis. J Neuroimmunol. 2010;229:232–7. doi: 10.1016/j.jneuroim.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alter G, Martin MP, Teigen N, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulet S, Sharafi S, Simic N, et al. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS. 2008;22:595–9. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]
- 10.Miyazawa M, Lopalco L, Mazzotta F, et al. The “immunologic advantage” of HIV-exposed seronegative individuals. AIDS. 2009;23:161–76. doi: 10.1097/QAD.0b013e3283196a80. [DOI] [PubMed] [Google Scholar]
- 11.Piacentini L, Fenizia C, Naddeo V, Clerici M. Not just sheer luck! Immune correlates of protection against HIV-1 infection. Vaccine. 2008;26:3002–7. doi: 10.1016/j.vaccine.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 12.Ravet S, Scott-Algara D, Bonnet E, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 13.Mazzoli S, Trabattoni D, Caputo SL, et al. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat Med. 1997;3:1250–7. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- 14.Castro KG, Ward JW, Slutsker L, et al. 1993 Revised Classification System for HIV Infection and Expanded Surveillance Case Definition for AIDS Among Adolescents and Adults. Morb Mortal Wkly Rep. 1992;41:RR-17. [PubMed] [Google Scholar]
- 15.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol. 2005;14:135–42. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]