Abstract
Helicobacter pylori is a Gram-negative bacteria that infects the human stomach of half of the world’s population. Colonization is followed by infiltration of the gastric mucosa by lymphocytes and myeloid cells. These cells are activated by various bacterial factors, causing them to release immune/inflammatory mediators, including reactive nitrogen species and polyamines that contribute to cellular damage and the pathogenesis of H. pylori-associated gastric cancer. In vitro experiments have revealed that H. pylori induces macrophage polyamine production by upregulation of the arginase 2/ornithine decarboxylase (ODC) metabolic pathway and enhances hydrogen peroxide synthesis through the activity of spermidine oxidase (SMO). In this chapter, we present a survey of the methods used to analyze the induction and the role of the enzymes related to polyamine metabolism, i.e. arginase, ODC, and SMO in H. pylori-infected macrophages.
Keywords: Macrophage, arginase, ornithine decarboxylase, spermine oxidase, Helicobacter pylori
1. Introduction
Worldwide human infection with Helicobacter pylori is the main cause of chronic gastritis, peptic ulcer and gastric cancer (1). Importantly, gastric cancer is the second-leading cause of cancer death worldwide. The persistence of this pathogen in the stomach despite a strong induction of a mucosal immune response is a critical hallmark of the infection (2). It is therefore of importance to determine how H. pylori modulates the innate immune response of the cells with which it interacts, such as gastric epithelial cells and macrophages.
Enzymes that metabolize l-arginine are essential for macrophage function. First, inducible nitric oxide (NO) synthase (iNOS) converts l-arginine into citrulline and NO. The free radical NO possesses numerous antimicrobial (3) and proinflammatory properties (4). In addition, l-arginine is also a substrate for both arginase 1 and arginase 2. Therefore, through consumption of l-arginine, arginase is a natural competitor of iNOS by decreasing substrate availability. Arginase synthesizes l-ornithine, which is metabolized by ornithine decarboxylase (ODC) into the three polyamines putrescine, spermidine, and spermine. Intracellular polyamine catabolism then occurs through two distinct enzymatic pathways: i) spermidine/spermine N1-acetyltransferase (SSAT) acetylates both spermine and spermidine, providing acetylated polyamines as substrates for further back-conversion to putrescine by N1-acetylpolyamine oxidase (APAO); and ii) spermine oxidase (SMO), also termed polyamine oxidase (PAO1), directly converts spermine to spermidine. Of importance, both metabolic pathways generate hydrogen peroxide.
We have shown that H. pylori i) induces macrophage apoptosis through the arginase 2/ODC/SMO pathway (5,6,7) and ii) regulates iNOS translation in macrophages by increasing ODC expression and endogenous polyamine synthesis (8). Furthermore, the induction of ODC and its biological consequences is dependent on c-Myc binding to the ODC promoter (7). Together these data suggest that there is a strong interaction between H. pylori and the inducible metabolism of l-arginine in macrophages (Fig. 1).
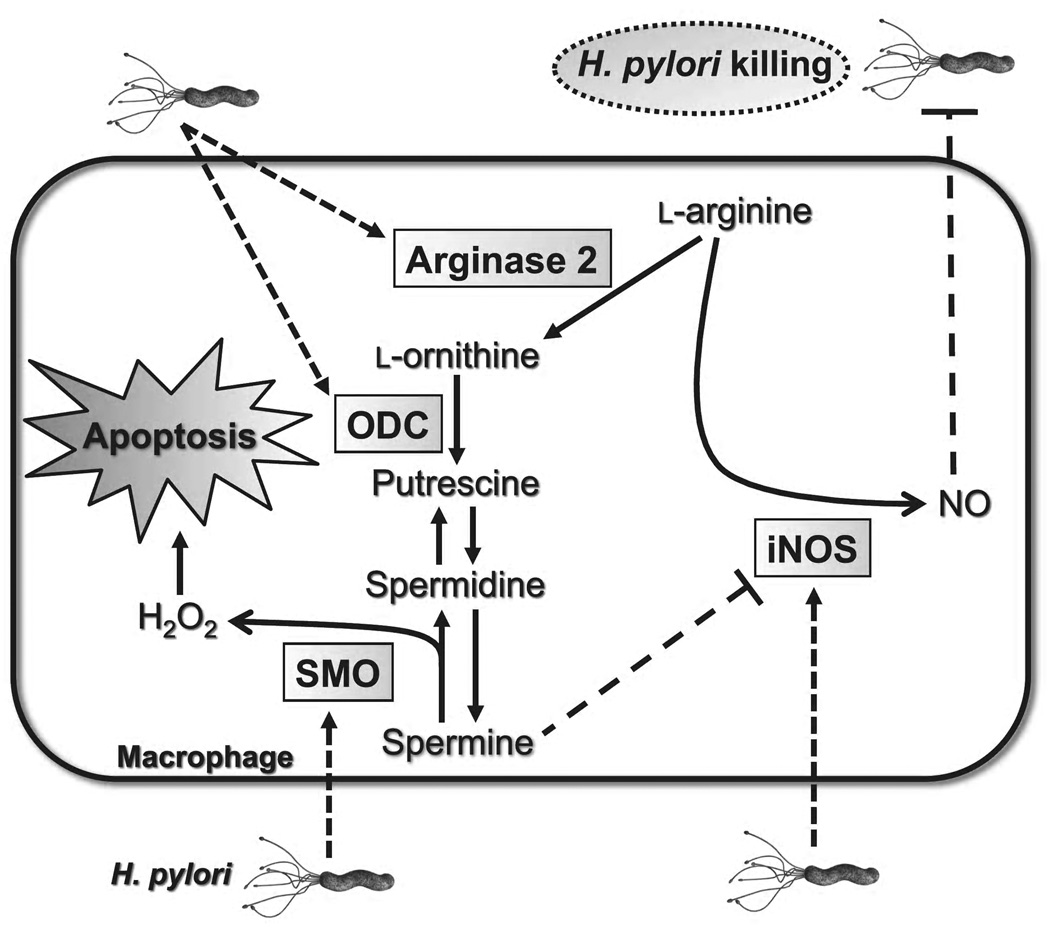
Fig. 1.
H. pylori stimulated synthesis and metabolism of polyamines in macrophages. H. pylori stimulates expression of iNOS, arginase 2, ODC, and SMO. High output iNOS-derived NO kills H. pylori. Polyamines are synthesized through the arginase 2/ODC metabolic pathway. Spermine inhibits iNOS translation. The back-conversion of spermine to spermidine by SMO releases H2O2 that induces macrophage apoptosis.
The modulation of the induction of arginase 2, ODC, and SMO in macrophages by H. pylori may occur at both the transcriptional and translational levels. Below we will describe straightforward assay protocols for understanding of polyamine metabolism regulation and role in H. pylori-infected macrophages. These include: transient transfection of macrophages with small interfering RNA (siRNA) or reporter plasmids, analysis of mRNA expression by reverse transcription (RT)-polymerase chain reaction (PCR) and real-time PCR, detection of proteins by Western blotting, and determination of relevant enzymes activities involved in polyamine metabolism.
It should be noted that our laboratory also works extensively on the response to H. pylori in other experimental systems. Space does not permit description of these experimental protocols in the current review, but we would like to refer the reader to our published protocols for isolation of mouse peritoneal macrophages (5,8) and mouse gastric macrophages (9) and infection of mice with H. pylori (5). Additionally, further information about the adaptation of the macrophage protocols described below to the gastric epithelial cell model has also been described by our laboratory (10).
2. Materials
2.1. Cell Culture
Complete RAW 264.7 cell culture medium: Dulbecco’s Modified Eagle’s Medium with glutamine (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% decomplemented (heat-inactivated) fetal bovine serum (FBS; Invitrogen), 1 % sodium pyruvate, 10 mM HEPES, 100 U/ml penicillin, and 100 µg/ml streptomycin.
2.2. Transfections
SMO siRNA sequence: 5′-GGACGUGGUUGAGGAAUUC-3′ and 5′-CCUGCACCAACUCCUUAAG-3′; ODC siRNA sequence 5′-CUCAUGAAACAGAUCCAGA-3′ and 5′-UCUGGAUCUGUUUCAUGAG-3′.
Prepare 20 µM stock solutions of the above siRNA duplexes dissolved in 10 mM Tris-HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA) buffer.
The pcDNA3.1-SMO plasmid contains the gene encoding SMO under the control of the constitutive SV40 promoter; this plasmid is provided by RA Casero, Jr. (Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD).
The pSV-β-galactosidase plasmid is obtained from Promega (Madison, WI).
2.3. Reagents for RNA Extraction and PCR
Add 1 ml of diethylpyrocarbonate (DEPC) to 1 l of distilled water, treat from 6 h to overnight, and autoclave for 20 min at 120°C.
Prepare a solution of 75% ethanol in DEPC water
A 5 × RT buffer is composed of 50 mM Tris-HCl (pH 9), 250 mM KCl, and 0.5% Triton X-100, all obtained from Sigma (St. Louis, MO).
RT mastermix, for each sample: 4 µl of 5× RT buffer, 1 µl of 10 mM deoxyribonucleotide triphosphate (dNTP), 2 µl of 0.1 M dithiothreitol (DTT), 1 µl of 40 U/µl RnaseOUT, and 0.5 µl of 100 U/ml SuperScript II Reverse Transcriptase®, all obtained from Invitrogen.
A 10 × PCR buffer contains 500 mM KCl, 100 mM Tris-HCl (pH 8.3), and 15 mM MgCl2.
Sense and antisense primer sequences and PCR product sizes are as follows: murine iNOS, 5'- GCCTCGCTCTGGAAAGA-3' and 5'-TCCATGCAGACAACCTT-3', 499 bp or 5'- CACCTTGGAGTTCACCCAGT-3' and 5'-ACCACTCGTACTTGGGATGC-3', 170 bp; murine arginase 1, 5'-AAGAAAAGGCCGATTCACCT-3' and 5'-CACCTCCTCTGCTGTCTTCC-3', 201 bp; murine arginase 2, 5'-ACAGGGTTGCTGTCAGCTCT-3' and 5'-TGATCCAGACAGCCATTTCA-3', 298 bp; murine ODC, 5'-CAGCAGGCTTCTCTTGGAAC6 3' and 5'-CATGCATTTCAGGCAGGTTA-3', 602 bp; murine SMO, 5′- CACGTGATTGTGACCGTTTC-3’ and 5′-TGGGTAGGTGAGGGTACAGTC-3, 222 bp; murine APAO, 5′-CTTTTCCAGGGGAGACCTTC-3′ and 5′-CACACCACCTGGATGAACTG-3′, 250 bp; murine SSAT, 5′-GACCCCTGAAGGACATAGCA-3’ and 5′-CCGAAGCACCTCTTCTTTTG-3′, 248 bp; and murine/human β-actin, 5'-CCAGAGCAAGAGAGGTATCC-3' and 5'-CTGTGGTGGTGAAGCTGTAG-3', 436 bp. The stock concentration of each primer is 15 pmoles/µl, and they are stored at −20°C.
PCR mastermix, for each sample: 2.5 µl of 10 × PCR buffer, 1 µl of 50 mM MgCl2, 0.5 µl of 10 mM dNTP, 0.5 µl of 25 U/µl Platinum Taq DNA polymerase (Invitrogen). Add i) 0.5 µl each of the of 5' and 3' murine iNOS, arginase 1 or 2, ODC, SMO, APAO, or SSAT primers, and ii) 0.1 µl each of the of 5' and 3' murine β-actin primers. Complete with 18.9 µl of DEPC H2O.
10× Tris-acetate-EDTA (TAE): 48.4 g/l Tris base, 10.9 g/l glacial acetic acid, and 2.92 g/l EDTA, pH 8.0.
2.4. Reporter Gene for ODC Promoter
Primer sequence for ODC promoter region: sense, 5′-GGGGTACCTGTCCGACACGAG-3′; antisense, 5′-GGAAGATCTTTAGCCAAGAACTC-3′, 2.8 kbp.
The ODC-CAT plasmid is provided by JL Cleveland (St. Jude Children's Research Hospital, Memphis, TN).
The Luciferase Reporter pGL3-Enhancer vector (Promega) contains an SV40 enhancer located downstream of the luciferase gene (luc+) and the poly(A) signal, and an ampicillin resistance gene.
2.5. Western blot
-
1
Stock solution of 100 mM phenylmethanesulphonylfluoride (PMSF): 0.174 g PMSF in 10 ml ethanol; keep at −20°C.
-
1
Radioimmunoprecipitation (RIPA) buffer: Tris-HCl 50 mM, pH 7.4, sodium deoxycholate 0.25%, NaCl 150 mM, EDTA 1 mM, Na3VO4 1 mM, NaF 1 mM. Before use, add 1 mM PMSF, 1% NP-40, and an anti-protease cocktail (Sigma).
-
2
The following 6× protein loading buffer can be prepared in advance and stored at −20°C: Tris HCl 375 mM, pH 6.8, 3 ml glycerol, 1 g SDS, 0.93 g DTT, and 1.2 mg bromophenol blue, in a final volume of 10 ml.
-
3
A 5× solution of electrophoresis running buffer consists of Tris 125 mM, glycine 960 mM and SDS 0.5%, and stocked at 4°C.
-
4
A 5× solution of transfer buffer is composed of 125 mM Tris and 950 mM glycine. The working TB buffer is: 20% 5× TB buffer, 20% methanol and 60% H2O.
-
5
Tris-buffered saline with Tween (TBS-T): Prepare 10× stock with 1.37 M NaCl, 27 mM KCl, 250 mM Tris-HCl, pH 7.4, and 1% Tween-20.
-
6
Blocking buffer: 5% nonfat dry milk in TBS-T.
-
7
Antibody dilution buffer: TBS-T supplemented with 2% bovine serum albumin.
-
8
Stripping buffer: Tris HCl 62.5 mM, pH 6.8, SDS 2%, and 0.8% β-mercaptoethanol. Warm this buffer to 50°C before use.
2.6 Arginase Activity Assay
Arginase activity lysis buffer: 0.1% Triton ×100, 0.01% pepstatin, 0.01% aprotinin, and 0.01% antipain in distilled H2O. Make fresh as required.
9% α-isonitrosopropiophenone (Sigma) dissolved in ethanol.
2.7. ODC Activity Assay
ODC lysis buffer: 1 mM Tris-HCl (pH 7.4), 1 mM EDTA, 0.05 mM pyridoxal-5'-phosphate, and 5 mM DTT.
2.8. SMO and APAO Activity assay
Borate buffer: 4.76 g boric acid and 2.54 g sodium tetraborate in 1 l of distilled H2O. Equilibrate to pH 8.4 with NaOH.
Assay reaction mixture for SMO activity: 5.7 U/ml of horseradish peroxidase (HRP) and 50 µM spermine (6,10) in 80 mM borate buffer, pH 9.0.
Assay reaction mixture for APAO activity: 5.7 U/ml of HRP and 50 µM N1-acetylspermine (Fluka Chemie, Buchs, Switzerland) in 80 mM borate buffer, pH 9.0.
Luminol: Prepare fresh just before addition to assay reaction mixture as 100 nM solution in H20.
2.9. SSAT Activity assay
The SSAT assay reaction mixture is composed of 3 mM spermidine and 12.7 µm l-[14C]acetyl-CoA (specific activity, 63 mCi/mmol).
2.10. Griess Reagents
Griess A: sulfanilamide 1% in HCl 1.2 N.
Griess B: naphtylethylenediamine 0.3% in distilled H2O.
Keep solutions at 4°C and protect from light.
3. Methods
3.1. Cell Culture and Infection
H. pylori is cultured on trypticase soy agar blood agar plates (BD Biosciences, Sparks, MD) at 37°C under 5% CO2. The day before the infection, bacteria are grown overnight on plates and are then harvested using a cotton tip in sterile phosphate buffered saline (PBS) solution. Bacteria are washed once and resuspended in complete DMEM without antibiotics. The absorbance (A) of the bacterial suspension is then measured at a wavelength of 600 nm using a spectrophotometer; we have reported that a solution of 109 bacteria/ml corresponds to an A600 nm of 1 (11). These bacteria are used to infect the macrophages.
The murine macrophage cell line RAW 264.7 (ATCC # TIB-71) is maintained in complete DMEM in T75 flasks. The day of the experiment, adherent cells are washed twice with 10 ml PBS and detached from the culture flask using a scraper. Cells are harvested in 10 ml PBS, and centrifuged at 300×g for 10 min. Pellets are resuspended with 1 ml of complete medium without antibiotics (see Note 1); then 5 ml of this same medium is added. Cells are counted using a hemocytometer and are plated on 6-well plates (2–4 × 106 cells/well in 2 ml) or on 24-well plates (0.5–1 × 106 cells/well in 1 ml) for 2 h. Medium is removed and fresh complete medium without antibiotics is added to the wells. Bacteria are added to the cells at a multiplicity of infection (MOI; number of bacteria per cells) of 1–100 together with pharmacological inhibitors of iNOS, arginase, ODC, or SMO if required; inhibitors of signal transduction or of transcriptional factors are added 30 min prior to the infection. At the end of the infection (see Note 2), cocultures are washed with PBS and RNA or proteins are extracted from cells. In addition to live bacteria directly cocultured with the macrophages, the reader is referred to other studies documenting methods for coculture of H. pylori separated from macrophages by Transwell filter supports (5), and other preparations of H. pylori that can be used, including lysates prepared in a French Pressure Cell (11) and water extracts (12).
3.2. Transient Transfection of Macrophages
Transfection of siRNA for ODC or SMO in macrophages: To knock-down H. pylori-induced expression of SMO or ODC, we developed an siRNA-based approach. These strategies are particularly useful because the SMO inhibitor, MDL 72527 also blocks APAO, and the ODC inhibitor, α-difluoromethylornithine does not prevent spermine accumulation in short-term culture. For these studies, plate 2 × 106 cells in 6-well plates and grow them in complete culture medium overnight. The next day, discard the medium and add 1 ml of optiMEM medium (Invitrogen). For each well, mix 20 µl of the 20 µM stock solution of either SMO, ODC, or control scrambled (Qiagen, Valencia, CA) siRNA with 100 µl of optiMEM medium, and incubate for 15 min at room temperature. Add this mixture to a solution containing 5 µl of LipofectAMINE 2000 (Invitrogen) in 100 µl of optiMEM and incubate for 15 min at room temperature. Gently overlay this combination of siRNA, lipid transfection reagent, and medium on RAW 264.7 cells, and incubate for 18 h. Wash the cells, add complete medium without antibiotics, and proceed with H. pylori infection. To confirm knockdown of SMO in macrophages, analyze SMO mRNA expression by RT-PCR/real-time PCR after an infection of 6 h by H. pylori, as described in paragraph 3.3. ODC knockdown can be confirmed by RTPCR/real-time PCR or immunobloting (see paragraph 3.5.)
Transfection of plasmid expressing SMO in macrophages: To overexpress SMO activity in macrophages, RAW 264.7 cells are transfected with the pcDNA3.1-SMO plasmid; the pSV-β-galactosidase plasmid is transfected concomitantly to determine the efficacy of transfection. Plate 5 × 106 cells in 6-well plates and grow them in complete culture medium overnight. The next day, discard the medium and add 800 µl of optiMEM medium (Invitrogen). For each well, mix 400 ng of pSV-β-galactosidase and 200 ng of pcDNA3.1-SMO plasmids in a final volume of 100 µl of optiMEM. Separately mix 20 µl LipofectAMINE Plus (Invitrogen) with 100 µl of optiMEM medium. Incubate both tubes for 15 minute at room temperature. Mix both solutions and incubate the mixture for 15 min at room temperature before adding it to cells. Incubate the RAW 264.7 cells for 18 h, change the medium to complete DMEM, and activate cells with H. pylori. Measure transfection efficiency by measuring β-galactosidase activity in transfected cells using the β-galactosidase Enzyme Assay System with Reporter Lysis Buffer kit (Promega). SMO enzyme activity is measured as described in paragraph 3.8.
3.3. mRNA Analysis
RNA extraction: Wells containing RAW 264.7 cells are washed twice with PBS and liquid completely removed. TRIzol (1 ml; Invitrogen) is directly added to the cells for 10 min. The plates are then gently vortexed and the lysates are harvested in RNase-free 1.5 ml Eppendorf tubes (see Note 3 and Note 4). Add 0.2 ml chloroform and mix vigorously by manual shaking for 30 seconds; do not use a vortex. Leave the tubes at room temperature for 10 min. Centrifuge at 12000×g at 4°C for 15 min. Carefully collect the upper aqueous phase and transfer to new 1.5 ml Eppendorf tubes and add 0.5 ml isopropanol. Gently mix the contents by repeatedly inverting and let tubes stand at room temperature for 10 min. Centrifuge at 12000×g at 4°C for 15 min. Aspirate the supernatant using a Pasteur pipette and a vacuum line, and resuspend the pellet in ml of 75% ethanol in DEPC water. Centrifuge at 8000×g at 4°C for 10 min, and again remove the supernatant with a Pasteur pipette. Dry the pellet (see Note 5) and add 20 µl DEPC H2O. RNA is then solubilized by incubation of the tubes at 56°C for 20 min.
Removing of residual DNA. During RNA extraction, DNA contamination can occur. This DNA could be amplified by PCR together with complementary DNA (cDNA). Each RNA sample is treated with DNase I obtained from Promega according to the manufacturer’s protocol (see Note 6).
-
RNA concentration is determined by a spectrophotometric approach. RNA suspension is diluted 50–100 times in DEPC water and A260 and A280 are determined to measure nucleic acid and protein concentration, respectively (see Note 7). Ideally, the A260/A280 ratio should be close to 2 for high purity nucleic acid. RNA concentration is determined by the following formula:
- [RNA] (µg /µl) = (A260 × 40 × dilution factor)/1000.
RNA is stored at −80°C or in liquid nitrogen.
Reverse transcription: This step corresponds to the synthesis of cDNA from messenger RNA (mRNA). All the steps described below are performed on ice. Add 1 µl of Oligo dT (500 µg/ml) to 10.5 µl of a solution of DEPC water containing 2 µg total RNA. Incubate at 65°C for 5 min and chill quickly on ice. Centrifuge the tubes briefly and add 8.5 µl of RT mastermix to each sample. Incubate 50 min at 42°C, and 15 min at 70°C to stop the reaction. cDNA samples are stored at −20°C. Amplification of cDNA is then performed by PCR and/or real-time PCR.
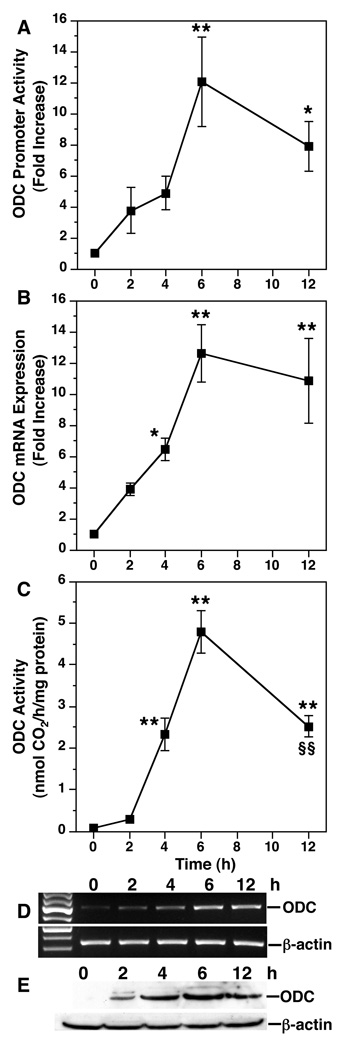
PCR: Prepare one PCR mastermix for all the samples; dispense 24 µl in 0.2 ml Eppendorf tubes and add 1 µl of each cDNA in each tube. PCR is conducted in an iCycler apparatus (Bio-Rad, Hercules, CA). One PCR cycle consists of the following: 94°C for 2 min; (94°C for 30 s, 59°C for 30 s, and 72°C for 60 s) for a total cycle number of 30–35; 72°C for 5 min. Harvest 10 µl of PCR product and add 2 µl of Loading Buffer (Invitrogen); perform the electrophoresis on 1.5% agarose gel in 1× TAE buffer containing 0.4 µg/ml ethidium bromide (see Note 8). Stained bands are visualized under UV light and photographed. An example of H. pylori-stimulated ODC mRNA expression is shown in Fig. 2D.
Real-time PCR: cDNA (1 µl) is amplified by the SYBR® Green SuperMix for iQ kit (Bio-Rad) containing 0.12 pmol/µl each of sense and antisense primers, in a Bio-Rad iQ5 real-time PCR machine. One PCR cycle consists of the following: 94°C for 2 min; (94°C for 30 s, 59°C for 30 s, and 72°C for 30 s) for a total cycle number of 45; 72°C for 2 min. For each sample, the cycle threshold (Ct; number of PCR cycles needed to get a fluorescent signal) is provided by the software of the individual real-time PCR machine. Variations in crossing point reflect different DNA quantities between samples. Results are calculated using the comparative Ct method in which the amount of target gene mRNA is normalized to the internal control β-actin and in which mRNA expression in H. pylori infected cells (Hp) is compared to uninfected cells (Ctrl).
Fig. 2.
Time course of induction of ODC in H. pylori-stimulated macrophages. RAW 264.7 cells were exposed to H. pylori for the times indicated. A, promoter activity determined by luciferase reporter assay using a −264-bp functional ODC promoter. B, mRNA levels determined by real-time PCR. C, ODC activity determined by radiochemical assay. D, RT-PCR for ODC. E, Western blot for ODC (53 kDa) and β-actin (42 kDa). *, p < 0.05; **, p < 0.01 versus time 0; in A–C, each experiment was performed a minimum of three times, each in duplicate. In D and E, the data are representative of three experiments. (Reproduced from ref. 7 with permission from The American Society for Biochemistry and Molecular Biology.)
The following formula is used:
Gene expression (fold increase/Ctrl) = 2[Ct β-actin(Ctrl) − Ct gene(Ctrl)] − [(Ct β-actin(Hp) − Ct gene(Hp)].
Additionally, specificity is checked by analysis of the melting curves in which the rate of change in fluorescence with changing temperature is monitored.
Representative published data for H. pylori-stimulated ODC mRNA expression is shown in Fig. 2B.
3.4. Luciferase Assay for ODC Promoter
Construction of the gene reporter: To generate desired promoter constructs, perform PCR using 2 µg ODC-CAT plasmid as a template with 0.5 pmol/l each of ODC promoter region primers and the Phusion™ High-Fidelity PCR Master Mix (New England Biolabs, Inc, Ipswich, MA), in a final volume of 50 µl. Use the following conditions for PCR: 95°C for 3 min; 30 cycles as follows: 98°C for 1 min, 50°C for 1 min, and 72°C for 2.5 min; 10 min at 72°C. Run PCR products on a 0.7% agarose gel in 1× TAE buffer containing 0.4 µg/ml ethidium bromide (see Note 8). Excise the 2.8 kbp band from the gel with a razor blade, and purify the PCR product using QIA Quick Gel Extraction kit from Qiagen. Measure the concentration of the DNA, using spectrophotometry, and adjust the concentration of the PCR product to 0.2 µg/µl. To 20 µl of purified PCR product, add 1µl of 0.5 mM dNTP and 5 units of the DNA polymerase I Klenow fragment (New England Biolabs, Inc) and incubate at 30°C for 15 min. Stop the reaction by heating to 75°C for 10 min. In parallel, the pGL3-Enhancer vector plasmid is linearized with 20 units of the restriction enzyme KpnI (New England Biolabs, Inc) overnight at 24°C. This plasmid is then isolated by electrophoresesis on an 0.7% agarose gel, treated with the Klenow fragment, and purified using the QIA Quick Gel Extraction kit (Qiagen), as described above. Combine 50 ng of pGL3-Enhancer vector with 1 µg of the insert corresponding to the ODC promoter region. Adjust volume to 10 µl with distilled H2O, add 10 µl of 2× Quick Ligation Buffer (New England Biolabs, Inc.) and 1µl of Quick T4 DNA Ligase (New England Biolabs, Inc.); mix thoroughly, centrifuge briefly, and incubate at 25°C for 5 min. Chill on ice and store this pGL3-ODC minimal functional promoter plasmid at −20°C.
The reporter gene is introduced into the competent Escherichia coli strain DH5α and selected on Luria Bertani (LB) agar plates containing 25 µg/ml ampicillin.
Selection of the clones containing pGL3-ODC plasmid: Transformed E. coli are then spread on LB plates. Isolated colonies are harvested and grown on 10 ml LB broth overnight. Half of the culture is frozen in 10% glycerol, and half is used to isolate the plasmid using the QIAprep Spin Miniprep Kit (Qiagen). Digest the plasmid with 20 units of the restriction enzyme XbaI (New England Biolabs, Inc.) overnight at 24°C and run on a 0.7% agarose gel. Check for 5.0 kpb band and keep the colonies that have been transformed.
Transient transfection and luciferase assay: RAW 264.7 cells are transfected with the purified pGL3-ODC and the pSV-β-galactosidase plasmids as described in paragraph 3.2. Transfection efficiency is evaluated by measuring β-galactosidase activity in transfected cells using the β-galactosidase Enzyme Assay System with Reporter Lysis Buffer kit (Promega). After infection with H. pylori, luciferase activity is measured using the Luciferase Assay System (Promega) following the manufacturer’s instructions. Representative data are shown in Fig. 2A.
3.5. Immunobloting for ODC
Scrape the RAW 264.7 cells in 1 ml PBS and harvest them in 1.5 ml Eppendorf tubes. Spin at 300×g for 10 min and discard the supernatant.
On ice, suspend each pellet with 100 µl of RIPA buffer; incubate for 20 min.
Centrifuge the lysate at 12,000×g for 20 min at 4°C. Transfer the supernatants to ice-chilled Eppendorf tubes and measure protein concentration.
Dilute 30 µg of each protein sample in a final volume of 20 µl of RIPA buffer, add 4 µl of loading buffer, and boil the samples for 5 min. Chill on ice.
Load samples and the colored molecular weight markers in 12.5% Tris-HCl polyacrylamide Criterion Precast Gels (Bio-Rad).
Electrophoresis is performed in 1× running buffer at 100 V in a Criterion™ Cell (Bio-Rad), until the dye front reaches the bottom of the gel. The samples that have been separated by SDS-page are then transferred to nitrocellulose membrane.
Cut a nitrocellulose membrane and two pieces of GB005 Whatman filter paper to the same size as the gel, and incubate them in the transfer buffer at 4°C for 15 min. The gel unit is disconnected from the power supply and disassembled. On the base of the Transblot SD Semidry Transfer Cell apparatus (Bio-Rad), place one wet filter paper, the gel, the nitrocellulose membrane (see Note 9), and then the second filter paper. Gently roll a pipette on this block to remove bubbles present between the gel and the membrane. Transfer is performed at 400 mA for 1 h. The colored molecular weight markers should be clearly visible on the membrane.
The nitrocellulose is then incubated in 20 ml blocking buffer for 2 h at room temperature on a rocking platform.
Wash the membrane 2 times 5 min with 20 ml TBS-T.
Incubate the membrane with the rabbit polyclonal anti-ODC antibody (Santa Cruz Biotechnology, Santa Cruz, CA; 1:1000 dilution) overnight at 4°C and wash membrane three times with TBS-T for 15 min.
Incubate the membrane with a goat anti-rabbit IgG secondary antibody conjugated with HRP (Sigma; 1:5000 dilution) for 1 h at room temperature, and wash three times with TBS-T for 15 min.
After the final wash, mix 2 ml each of both ECL reagents (Invitrogen) according to the manufacturer’s instructions, and incubate membrane in this solution for 5 min. Remove membrane, gently blot on Kim wipes, and place it in between plastic sheets. In a dark room, expose the blot to Bio-Max ML film (Kodak, Rochester, NY) for 30 s – 5 min in an X-ray film cassette. Develop exposed film in developer. Films can be digitized using a flatbed scanner. Quantification of data may be done by analyzing densitometry of the bands. A representative example of H. pylori-stimulated ODC protein expression as detected by Western blotting is shown in Fig. 2E.
Once a satisfactory exposure is obtained, incubate the blot in stripping buffer for 30 min at 50°C. Wash membrane 3 times for 10 min each with 100 ml of TBS-T and incubate again in blocking buffer.
The membrane is then ready to be reprobed with mouse anti-β-actin (Sigma; 1:10,000 dilution); washes, secondary antibody addition, and ECL detection are performed as described above. A blot of β-actin after a strip of the membrane is depicted in Fig. 2E.
3.6. Determination of Arginase Activity
Arginase activity is measured in RAW 264.7 macrophages using the micromethod described by Corraliza et al. (13).
Macrophages are harvested at the end of the experiment, counted, and 2 × 105 cells are isolated in a 1.5 ml Eppendorf tube.
After washing with PBS, the pellet is resuspended in 50 µl of arginase activity lysis buffer. Tubes are vortexed and incubated 15 min on ice.
In parallel, serial dilutions of recombinant bovine arginase (Sigma) are prepared as a standard curve.
5 µl of a solution of 100 mM MnCl2 in PBS is added to each sample and to the standards for 10 min at 55°C. Then, 50 µl of a solution of 0.5 M l-arginine in PBS is added to each sample for 1 h at 37°C. The reaction is stopped by the addition of 400 µl of an acid mixture of H2SO4/H3PO4/H2O (1:3:7).
The concentration of urea synthesized by arginase metabolism is determined by addition of 25 µl of 9% α-isonitrosopropiophenone for 45 min at 100°C. The colored product is quantified by A540 nm in a spectrophotometer.
3.7. Assay for ODC Activity
ODC activity is determined by a radiometric analysis in which the amount of [14C]O2 liberated from l-[14C]ornithine is measured.
Macrophages (2 × 105) are lysed in 500 µl of ODC lysis buffer. Cells are frozen and thawed three times, then sonicated.
Prepare 2.5 cm diameter discs of Whatman filter paper impregnated with 20 µl of 2 N NaOH. These are left to dry at room temperature.
After centrifugation at 12,000×g for 15 min, 300 µl of the macrophage lysate supernatants are incubated at 37°C with 10 nmol L-[14C]ornithine (specific activity, 47.7 mCi/mmol; NEN, Boston, MA) for 15 min in a 15 ml glass tube fitted with a rubber stopper and a center well assembly containing the Whatman filter paper; the [14C]O2 liberated by the activity of the ODC is trapped on the filter paper. The reaction is stopped with 300 µl of 10%trichloroacetic acid. The rest of each lysate is used for measurement of protein concentration.
[14C]O2 present on the filter paper is analyzed by liquid scintillation counting.
- From the cpm value measured, the amount of [14C]O2 present in each sample is calculated using the following formula:
- [14C]O2 (fmoles) = cpm / [2.109 × specific activity (Ci/mmol)]
ODC activity is expressed as nmol CO2/h/mg protein. Representative data are shown in Fig. 2C.
3.8. Measurement of SMO and APAO Activity
The same assay is used to determine PAO1 and APAO activity. Only the reaction mixture is different between the assays.
Harvest 5 × 106 macrophages in 500 µl borate buffer (for 6-well plates) on ice, and homogenize with an Ultra-Turrax (IKA Works, Wilmington, NC). Centrifuge at 12000×g for 10 min and collect the supernatant.
Measure protein concentration using BCA protein estimation kit (Pierce/Thermo Scientific, Rockford, IL) as described in the manufacturer’s instructions.
On ice, add 100 µl of cell lysate to 100 µl of assay reaction mixture and incubate for 2 min at 37 °C (see Note 10).
For the standard curve, prepare solutions of H2O2 (0 to 160 nmol) in 100 µl of cell lysis buffer and 100 µl of assay buffer in 12 × 75 mm glass tubes.
Transfer the unknown and standard tubes to a luminometer. Add 10 µl of 100 nM luminol using an injector, and integrate the resulting chemiluminescence for 20 s.
Express activities as nmol of H2O2/min/mg of protein.
3.9. Determination of SSAT Activity
-
1
Scrape 5 × 106 RAW 264.7 cells in 250 µl borate buffer and homogenize with an Ultra-Turrax (IKA Works, Wilmington, NC) and measure protein concentration with BCA protein kit.
-
2
Add 100 µl of RAW 264.7 lysates to 100 µl SSAT assay reaction mixture and incubate for 10 min at room temperature.
-
3
Stop reaction by adding 10 µl of 1 M hydroxylamine, and boil for 3 min.
-
4
Spot the resulting samples onto P-81 phosphocellulose discs and wash the discs twice with 10 ml of 100% methanol.
-
6
Transfer discs in 5 ml of biodegradable scintillation fluid in counting vials and measure radioactivity in a scintillation counter.
-
7From the cpm value measured, the amount of 14C]acetyl-CoA incorporated into spermidine in each sample is calculated, using the following formula:
- l-[14C]acetylspermidine (fmoles)=cpm/[2.109 × specific activity (Ci/mmol)].
-
8
Express enzyme activity as nmol of l-[14C]acetylspermidine formed/min/mg of protein.
3.10. Measurement of NO2− Concentration
NO2− and NO3− concentration in a cell supernatant is a reliable indicator of NO production by activated macrophages. Here we present the Griess reaction that allows spectrophotometric detection of NO2− (see Note 11).
Place 50 µl of each cell supernatant in wells of 96-well plates in duplicate; add 60 µl of Griess A and then 60 µl of Griess B.
To ensure accurate NO2− quantitation, prepare a reference curve with NaNO2 for each assay (from 0 to 100 µM), using the same cell culture medium used for experimental samples.
Incubate at room temperature for 5–10 min, protected from light. A purple/magenta color will begin to form immediately. Measure A540 nm in a plate reader.
3.11. Measurement of Apoptosis
Plate 5 × 105 RAW 264.7 cells and stimulate with H. pylori.
Scrape the cells and collect them in the cell culture supernatant. Centrifuge at 1000×g at 4°C for 10 min.
Discard the supernatant and use the Annexin V:FITC Apoptosis Detection kit I from BD Biosciences to analyze the apoptosis by flow cytometry.
Acknowledgements
This work was supported by R01 DK053620, R01 AT004821, P01 CA116087, P01 CA028842, P30 DK058404 (Vanderbilt Digestive Disease Center), and a Merit Review Grant from the Office of Medical Research, Department of Veterans Affairs. APG is also supported by a grant from Philippe Foundation.
Footnotes
The cells have to be first dissolved in 1 ml medium with a pipetman to ensure a proper dissolution of the pellet before adding more medium.
The time of infection varies according to the type of experiment. Usually, mRNA expression and protein analysis are performed 3–9 hours and 6–24 hours after the beginning of the infection, respectively. NO2− concentration and apoptosis are analyzed 24–48 h post-infection. We recommend performing a kinetic analysis of earlier time points as appropriate.
Samples can be stored frozen at −20°C or −80°C in TRIzol for 3 and 12 months, respectively.
The material will be very viscous at this stage owing to release of DNA, and thus hard to pipette. The viscosity will decrease if samples are frozen at −20°C or −80°C.
The use of a vacuum chamber is advised.
DNase treatment is a recommended step. Contaminating DNA fragments will remain in the cDNA samples if not removed. As such, this DNA will be amplified by PCR along with the cDNA, and results could be compromised. To check for proper removal of contaminating DNA, it is suggested to perform PCR using 0.2 µg RNA as a template instead of 1 µl cDNA; no amplification should be observed if there is no DNA contamination.
We use a 96-well quartz microplate for absorbency determinations in the ultraviolet region of the light spectrum. Additionally, different disposable microplates with no background absorbance characteristics in the UV range are commercially available.
Add agarose to TAE buffer in an Erlenmeyer flask and cover it with plastic wrap and pierce a hole in the wrap for ventilation. Weigh the beaker and solution. Microwave the agarose solution for periods of 1 min. Between each series, take out the flask from the microwave using high heat gloves and swirl gently the solution to re-suspend any remaining agarose particles; any microwaved solution may be superheated and could foam over the container's rim if agitated. It is important to check the boiling of this solution in the microwave to avoid an overflow. Agarose is dissolved when a clear solution is obtained. Add enough hot distilled water to return the contents to the original weight; mix continuously. Then, let the temperature cool down to 55°C. Add ethidium bromide under a fume hood, mix thoroughly, and pour into gel casting apparatus.
One corner of the gel and the membrane are cut to allow orientation to be tracked. Also, wet transfer of SDS-PAGE gels to membranes can be used.
We have found that the standard curve is linear for values between 5 and 80 nmol of H2O2. Therefore, samples should be diluted to fit in this linear range.
When produced by activated cells, NO reacts with itself and water in the presence of oxygen to form NO2−. Nonetheless, other reactive nitrogen species, including NO3−, can be synthesized. To measure NO3− concentration, NO3− should be converted into NO2− by a NO3− reductase prior to performing the Griess assay. Numerous kits are commercially available to perform this experiment.
References
- 1.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson KT, Crabtree JE. Immunology of Helicobacter pylori: insights into the failure of the immune response and perspectives on vaccine studies. Gastroenterology. 2007;133:288–308. doi: 10.1053/j.gastro.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gobert AP, Cheng Y, Akhtar M, Mersey BD, Blumberg DR, Cross RK, Chaturvedi R, Drachenberg CB, Boucher JL, Hacker A, Casero RA, Jr, Wilson KT. Protective role of arginase in a mouse model of colitis. J. Immunol. 2004;173:2109–2117. doi: 10.4049/jimmunol.173.3.2109. [DOI] [PubMed] [Google Scholar]
- 5.Gobert AP, Cheng Y, Wang JY, Boucher JL, Iyer RK, Cederbaum SD, Casero RA, Jr, Newton JC, Wilson KT. Helicobacter pylori induces macrophage apoptosis by activation of arginase II. J. Immunol. 2002;168:4692–4700. doi: 10.4049/jimmunol.168.9.4692. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi R, Cheng Y, Asim M, Bussiere FI, Xu H, Gobert AP, Hacker A, Casero RA, Jr, Wilson KT. Induction of polyamine oxidase 1 by Helicobacter pylori causes macrophage apoptosis by hydrogen peroxide release and mitochondrial membrane depolarization. J. Biol. Chem. 2004;279:40161–40173. doi: 10.1074/jbc.M401370200. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y, Chaturvedi R, Asim M, Bussiere FI, Scholz A, Xu H, Casero RA, Jr, Wilson KT. Helicobacter pylori-induced macrophage apoptosis requires activation of ornithine decarboxylase by c-Myc. J. Biol. Chem. 2005;280:22492–22496. doi: 10.1074/jbc.C500122200. [DOI] [PubMed] [Google Scholar]
- 8.Bussiere FI, Chaturvedi R, Cheng Y, Gobert AP, Asim M, Blumberg DR, Xu H, Kim PY, Hacker A, Casero RA, Jr, Wilson KT. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem. 2005;280:2409–2412. doi: 10.1074/jbc.C400498200. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi R, Asim M, Lewis ND, Algood HM, Cover TL, Kim PY, Wilson KT. L-arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect. Immun. 2007;75:4305–4315. doi: 10.1128/IAI.00578-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, Chaturvedi R, Cheng Y, Bussiere FI, Asim M, Yao MD, Potosky D, Meltzer SJ, Rhee JG, Kim SS, Moss SF, Hacker A, Wang Y, Casero RA, Jr, Wilson KT. Spermine oxidation induced by Helicobacter pylori results in apoptosis and DNA damage: implications for gastric carcinogenesis. Cancer Res. 2004;64:8521–8525. doi: 10.1158/0008-5472.CAN-04-3511. [DOI] [PubMed] [Google Scholar]
- 11.Wilson KT, Ramanujam KS, Mobley HL, Musselman RF, James SP, Meltzer SJ. Helicobacter pylori stimulates inducible nitric oxide synthase expression and activity in a murine macrophage cell line. Gastroenterology. 1996;111:1524–1533. doi: 10.1016/s0016-5085(96)70014-8. [DOI] [PubMed] [Google Scholar]
- 12.Gobert AP, Mersey BD, Cheng Y, Blumberg DR, Newton JC, Wilson KT. Cutting edge: urease release by Helicobacter pylori stimulates macrophage inducible nitric oxide synthase. J. Immunol. 2002;168:6002–6006. doi: 10.4049/jimmunol.168.12.6002. [DOI] [PubMed] [Google Scholar]
- 13.Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. J. Immunol. Methods. 1994;174:231–235. doi: 10.1016/0022-1759(94)90027-2. [DOI] [PubMed] [Google Scholar]