Abstract
Epithelial tumor cells transit to a mesenchymal state in response to extracellular cues, in a process known as epithelial-to-mesenchymal transition (EMT). The precise nature of these cues has not been fully defined, an important issue given that EMT is an early event in tumor metastasis. Here, we have found that a population of metastasis-prone mouse lung adenocarcinoma cells expresses Notch and Notch ligands and that the Notch ligand Jagged2 promotes metastasis. Mechanistically, Jagged2 was found to promote metastasis by increasing the expression of GATA-binding (Gata) factors, which suppressed expression of the microRNA-200 (miR-200) family of microRNAs that target the transcriptional repressors that drive EMT and thereby induced EMT. Reciprocally, miR-200 inhibited expression of Gata3, which reversed EMT and abrogated metastasis, suggesting that Gata3 and miR-200 are mutually inhibitory and have opposing effects on EMT and metastasis. Consistent with this, high levels of Gata3 expression correlated with EMT in primary tumors from 2 cohorts of lung adenocarcinoma patients. These findings reveal what we believe to be a novel Jagged2/miR-200–dependent pathway that mediates lung adenocarcinoma EMT and metastasis in mice and may have implications for the treatment of human epithelial tumors.
Introduction
Lung cancer is the foremost cause of cancer-related death in Western countries, and metastasis is the leading cause of death in patients with lung cancer. Improving clinical outcomes will require a better understanding of the biological processes that initiate metastasis. Toward that goal, mouse models have been generated that develop lung adenocarcinomas with high or low propensities for invasion and metastasis. Mice that express K-rasG12D alleles inducibly, conditionally, or somatically develop lung adenocarcinomas with low invasive and metastatic potential (1–5), whereas mice that express K-rasG12D and p53R172H alleles develop lung adenocarcinomas that metastasize widely (6–9). Thus, K-ras–driven mouse models of lung cancer acquire metastatic potential with the addition of a second mutation commonly found in lung cancer.
Investigators have used mouse models of cancer to study the biological basis of metastasis. In one working hypothesis, epithelial tumor cells acquire the ability to invade and disseminate by undergoing epithelial-to-mesenchymal transition (EMT), which is characterized by a loss of cell-cell attachments and apical-basal polarization and gain of mesenchymal and invasive properties (10–19). The process of EMT is regulated by several transcriptional suppressor families, including the zinc-finger proteins Snail1 and Snail2, the 2-handed zinc-finger δEF1 family factors (δEF1/Zeb1 and SIP1/Zeb2), and the basic helix-loop-helix factors Twist and E12/E47 (17, 20–22). Zeb1 and Zeb2 expression are regulated by the microRNA-200 family, which includes 5 members clustered in 2 genomic loci (200b-200a-429 and 200c-141; collectively referred to here as miR-200), and several groups have demonstrated that miR-200 targets Zeb1 and Zeb2 (23–25). We have reported that metastasis-prone tumor cells derived from KrasLA1/+p53R172HΔG/+ mice transit reversibly between epithelial and mesenchymal states, forming highly polarized epithelial spheres in 3-dimensional culture that undergo EMT after treatment with TGF-β or subcutaneous injection into syngeneic mice, indicating that the metastatic capacity of these tumor cells is not limited to the lung microenvironment (7). Furthermore, this EMT is associated with profound suppression of miR-200 levels, and forced expression of miR-200 locks these tumor cells into an epithelial state and abrogates their metastatic capacity in syngeneic mice (7). Consistent with these findings, low miR-200b levels in early-stage lung cancer specimens predicts disease recurrence (26). Collectively, these findings suggest that miR-200 is a central regulator of tumor metastasis and demonstrate a critical need to understand the extracellular cues that regulate miR-200 expression.
Environmental signals regulate EMT through cell-cell contacts mediated by families of transmembrane receptors and ligands expressed on adjacent cells (27). A notable example of this is the Notch axis, which includes 4 Notch family members (Notch1–Notch4) and 5 Notch ligands — 3 Delta-like ligands (Dll1, Dll3, and Dll4) and 2 serrate-like ligands (Jagged1 and Jagged2) (28, 29). Jagged1-induced Notch activation promotes EMT of breast epithelial cells through Snail2 (30), but the mechanism by which Notch increases Snail2 expression has not been elucidated. Ligand binding induces Notch cleavage by a transmembrane protease of the a disintegrin and metalloproteinase (ADAM) family and a membranous protein complex containing presenilin, which has γ-secretase activity (28). The intracellular portion of Notch is thereby released and translocates to the nucleus in which it acts as a cofactor for the transcription factor CBF-1/suppressor of hairless/Lag1 (CSL). One of the transcriptional targets of CSL is the transcription factor GATA3, a key regulator of embryonic development and adult progenitor cell maturation (31, 32). The Notch pathway is constitutively activated in cancer cells originating from the lung and other tissues, due to gain-of-function mutations in Notch genes and overexpression of Notch ligands (33, 34). However, GATA3 reportedly restrains tumorigenesis in breast tumor models by suppressing INK4C transcription (35, 36), suggesting that the net effect of Notch and GATA3 on tumorigenesis is context dependent.
In this study, we sought to discover extracellular cues that regulate metastasis by using mice that develop widely metastatic lung adenocarcinomas, owing to expression of p53R172H and a latent, somatically activated K-rasG12D allele (9), and share transcriptional features of primary tumors from lung cancer patients with poor prognosis (6). Using a syngeneic tumor model derived from these mice (7), we isolated metastasis-prone tumor cells by sorting tumors on the basis of CD133 (also known as Prominin-1), a putative marker of tumor-initiating cells and metastasis-prone tumor cells in some human tumors and mouse models of human cancer (37, 38). We show that tumor cell EMT and metastasis are dependent upon Jagged2, which promotes EMT by decreasing miR-200 expression.
Results
Isolation of metastasis-prone tumor cells.
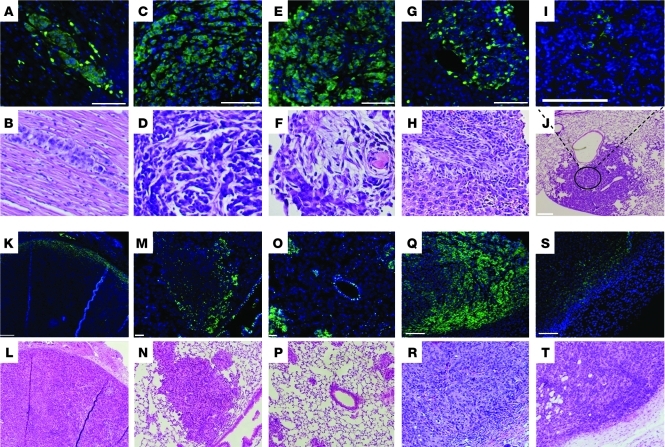
We examined CD133 expression in primary and metastatic tumor deposits isolated from KrasLA1/+p53R172HΔG/+ mice and in lung adenocarcinoma cell lines derived from these mice that metastasize with defined frequency (high or low) in syngeneic mice (7). In KrasLA1/+p53R172HΔG/+ mice, CD133-expressing tumor cells formed large clusters in metastatic deposits (Figure 1, A–H), whereas they were found as solitary cells or in small clusters in primary lung tumors (Figure 1, I and J). A metastasis-prone lung adenocarcinoma cell line derived from KrasLA1/+p53R172HΔG/+ mice (344SQ) expressed CD133 in culture (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI42579DS1) and after subcutaneous injection into syngeneic mice, in which it was detected on the periphery of primary tumors (Figure 1, K and L); diffusely throughout small, nascent metastatic deposits (Figure 1, O and P); and on the periphery of large metastases (Figure 1, M and N). The percentage of tumor cells that expressed CD133 per metastasis was estimated by counting CD133pos cells and total DAPI-stained tumor cells within metastatic deposits (Figure 1, A–H). On average, 87% of the tumor cells expressed CD133. Collectively, these findings raised the possibility that CD133 marks a population of metastasis-prone tumor cells.
Figure 1. CD133 expression in primary and metastatic tumors in KrasLA1/+p53R172HΔG/+ mice and in syngeneic mice injected with 344SQ cells.
CD133 staining (green) in metastases to (A and B) heart, (C and D) diaphragm, (E and F) chest wall, and (G and H) liver and (I and J) a primary lung tumor from KrasLA1/+p53R172HΔG/+ mice, (K and L) in a primary subcutaneous tumor and (M and N) large and (O and P) small lung metastases from syngeneic mice injected with 344SQ cells, and in a primary subcutaneous tumor in mice injected with (Q and R) CD133hi and (S and T) CD133lo 344SQ tumor cells. Sections were costained with DAPI (blue). Under each CD133/DAPI-stained panel is a panel with hematoxylin and eosin staining of an adjacent tissue section. Original magnification, ×20 (A–I and M–T); ×4 (J–L). Scale bars: 100 μm (A–I, L, N, and P–T); 200 μm (J and K); 50 μm (M and O).
To examine this possibility, 344SQ syngeneic tumors were sorted into CD133hi and CD133lo fractions by flow cytometry (Supplemental Figure 1B), and these fractions were injected into the flanks of syngeneic mice (10,000 cells per mouse, n = 14 mice per cohort) (Supplemental Table 1). Immunofluorescent staining revealed CD133 expression in the majority of tumor cells arising from the CD133hi fraction and in rare tumor cells arising from the CD133lo fraction (Figure 1, Q–T). Although the 2 fractions exhibited similar tumorigenicity, generating subcutaneous tumors in 13 out of 14 mice injected with CD133hi cells and 11 out of 14 mice injected with CD133lo cells, lung metastases arose 4-fold more frequently in mice injected with the CD133hi fraction than in those injected with the CD133lo fraction (8 out of 13 mice bearing CD133hi fraction tumors vs. 2 out of 11 mice bearing CD133lo fraction tumors, respectively; P = 0.047, 2-sided Fisher’s exact test; Supplemental Table 1), indicating that the CD133hi fraction was enriched in metastasis-prone tumor cells.
Transcriptional profiling of CD133hi and CD133lo 344SQ cells.
To identify transcriptional features of the CD133hi tumor cells that regulate metastasis, RNA samples were prepared from 344SQ CD133hi and CD133lo tumor fractions (n = 3 for each) and subjected to Affymetrix gene expression profiling, which revealed 681 unique differentially expressed genes (nominal P < 0.01, fold change >1.5), of which 254 were increased and 427 were decreased in the CD133hi fraction (Figure 2A; lists of genes are available via Gene Expression Omnibus accession number GSE15587; http://www.ncbi.nlm.nih.gov/geo/). Using all 20,959 genes on the array as a reference, enrichment analysis using Gene Ontology terms demonstrated highly significant enrichment of the increased-expression gene set in “Notch-binding” (3 out of 8 genes; 1-sided Fisher’s exact test, P = 9.42 × 10–5), “calcium ion binding” (21 out of 781 genes; P = 5.7 × 10–4), “EMT” (2 out of 6 genes; P = 2.1 × 10–3), “extracellular space” (38 out of 1,910 genes; P = 1.6 × 10–3), “integral to membrane” (60 out of 3,618 genes; P = 5.7 × 10–3), and “tight junctions” (5 out of 67 genes; P = 1.3 × 10–3). Such broad differences in gene expression are remarkable given that these fractions were derived from a uniform population of tumor cells that differed on the basis of a single cell surface marker.
Figure 2. Transcriptional profiling of CD133hi and CD133lo fractions of 344SQ tumors.
(A) Heat map of 681 genes that were differentially expressed in CD133hi and CD133lo fractions of 344SQ tumors (n = 3 for each). Notch ligands (top) and Gene Ontology (GO) terms (right) are indicated. (B) Quantitative RT-PCR analysis performed on paired triplicate RNA samples was normalized on the basis of L32 mRNA, and CD133hi values were expressed as mean values (± SD) relative to those of CD133lo values, which were set at 1.0; P values are from paired t test. (C) Western blotting of CD133, Notch ligands, Notch isoforms, and Actin in CD133lo and CD133hi fractions of a 344SQ tumor. Actin indicates relative protein loading. Protein band density was quantified by densitometry, normalized to that of CD133lo cells (which was set at 1.0), and is indicated below each blot.
From the standpoint of extracellular signals that might regulate metastasis, “Notch binding” was a term of particular interest, because Notch ligands mediate cell-cell interactions. We confirmed that the CD133hi cell fraction had significantly higher expression of Jagged1 (Jag1), Jagged2 (Jag2), Dll4, Notch1, and Notch2 (Figure 2B). Furthermore, the CD133hi cell fraction had increased activated (cleaved) intracellular forms of all 3 Notch family members (Figure 2C) and Hes1 mRNA, a Notch1 transcriptional target (ref. 28 and Figure 2B). Interestingly, Dll3 was significantly more abundant in CD133lo tumor cells (Figure 2, B and C), which leads one to speculate that Dll3 plays an opposite role to that of other Notch ligands in this tumor model. Tumor tissues were immunostained to determine whether Notch ligand expression is restricted to those tumor cells that express CD133 on the basis of signal colocalization using antibodies against CD133 and Jagged1 (optimal Jagged2-specific antibodies were not available for these studies). Dual immunofluorescence staining of a 344SQ subcutaneous tumor revealed that Jagged1 was detected solely in CD133-expressing tumor cells (Supplemental Figure 2, top and middle rows), but a large fraction of CD133-expressing tumor cells had no detectable Jagged1 (Supplemental Figure 2, bottom row), indicating the potential for differential Notch signaling within the pool of CD133-expressing tumor cells.
Metastatic activity is Jagged2 dependent.
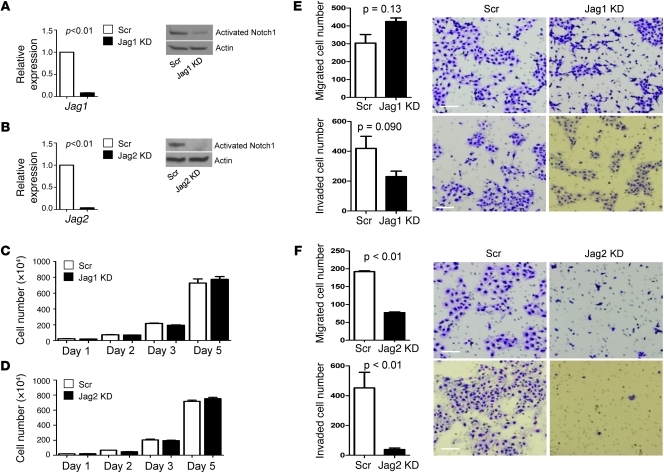
To examine the role of these Notch ligands in metastasis, 344SQ cells were stably transfected with retroviral vectors expressing shRNA against Jagged1 (Figure 3A) or Jagged2 (Figure 3B), which decreased the levels of Jagged mRNA and cleaved (activated) Notch1 protein (Figure 3, A and B), reflecting a reduction in Notch activity. We examined the ability of the transfectants to proliferate, migrate, and invade in vitro and to form tumors and metastasize in syngeneic mice. Although the proliferation of 344SQ cells in monolayer cultures was not affected by knockdown of either Notch ligand (Figure 3, C and D), the migratory and invasive capacities of 344SQ cells were profoundly reduced by knockdown of Jagged2 but not Jagged1 (Figure 3, E and F). Tumor incidence after subcutaneous injection into syngeneic mice was reduced relative to that of matched control transfectants for Jagged2-depleted 344SQ cells (15 out of 15 mice bearing control tumors vs. 10 out of 15 mice bearing Jagged2-depleted tumors, respectively; P = 0.021) but not Jagged1-depleted 344SQ cells (8 out of 8 mice bearing control tumors vs. 6 out of 8 mice bearing Jagged1-depleted tumors, respectively; P = 0.23) (Supplemental Table 2). Furthermore, the incidence of lung metastases in mice bearing visible subcutaneous tumors was reduced relative to that of control transfectants for Jagged2-depleted 344SQ cells (14 out of 15 mice bearing control tumors vs. 1 out of 10 mice bearing Jagged2-depleted tumors, respectively; P < 0.01) but not Jagged1-depleted 344SQ cells (8 out of 8 mice bearing control tumors vs. 3 out of 6 mice bearing Jagged1-depleted tumors, respectively; P = 0.055) (Supplemental Table 2). Jagged2 knockdown in 344P, a second lung adenocarcinoma cell line derived from KrasLA1/+p53R172HΔG/+ mice that has mesenchymal features and is metastasis-prone (7), abrogated metastasis but not tumorigenicity (Supplemental Table 2).
Figure 3. Characterization of Jagged-depleted 344SQ cells.
(A and B) Jagged depletion inactivates Notch. Quantification of (A) Jag1 and (B) Jag2 mRNA by quantitative PCR (bar graph) and Notch activation by Western blotting of cleaved Notch1 (gels) in Jagged1-depleted (Jag1 KD), Jagged2-depleted (Jag2 KD), and control (Scr) 344SQ transfectants. Quantitative PCR values were normalized based on L32 mRNA and expressed as the mean values of replicate (triplicate) samples relative to that of controls, which were set at 1.0. Actin indicates relative protein loading. (C and D) Jagged depletion has no effect on 344SQ cell proliferation. Quantification of (C) Jagged1-depleted and (D) Jagged2-depleted 344SQ cells grown in monolayer, counted at the indicated time points and expressed as the mean values (± SD) of replicate (triplicate) wells. (E and F) Inhibition of 344SQ cell migration and invasion by depletion of Jagged2 but not Jagged1. Images of migrated (top rows) and invaded (bottom rows) (E) Jagged1-depleted, (F) Jagged2-depleted, and (E and F) control 344SQ cells, which were counted and expressed as the mean values (± SD) of replicate (triplicate) wells (bar graphs), with P values from Welch’s t test using log-transformed data. Scale bars: 100 μm.
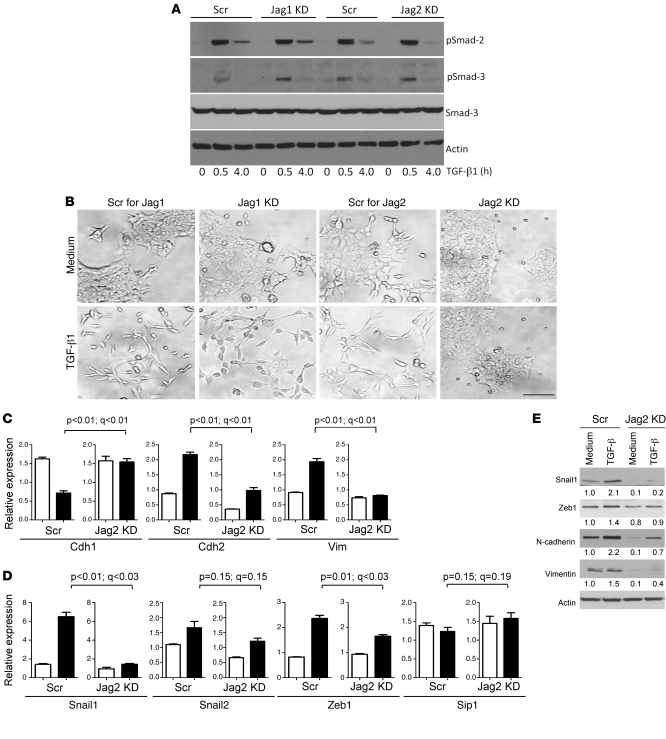
To explore the biologic basis for metastasis inhibition in Jagged2-depleted cells, we examined whether Jagged2 depletion abrogated the propensity to undergo EMT, a prominent feature of 344SQ cells treated with TGF-β (7). Despite having no detectable effect on Smad2 or Smad3 phosphorylation (Figure 4A), Jagged2 depletion abrogated TGF-β–induced changes in cellular morphology (Figure 4B) and biochemical evidence of EMT, including expression of epithelial (Cdh1) and mesenchymal (Cdh2 and Vim) markers (Figure 4, C and E) and the transcription factors that regulate them (Zeb1 and Snail1) (Figure 4, D and E). We conclude that Jagged2 is required for TGF-β–induced EMT.
Figure 4. Abrogation of TGF-β–induced EMT by depletion of Jagged2 but not Jagged1.
(A) Jagged depletion has no effect on Smad phosphorylation. Western blotting of total and phosphorylated (p) Smads in Jagged-depleted and control transfectants treated for the indicated time points with TGF-β. Actin indicates relative protein loading. (B) Loss of TGF-β–induced morphologic changes in Jagged2-depleted but not Jagged1-depleted or control 344SQ cells. Jagged2-depleted cells retain epithelial features after treatment for 48 hours with TGF-β (1 ng/ml) (bottom right panel). Original magnification, ×20. Scale bar: 100 μm. (C and D) Loss of TGF-β–induced EMT by Jagged2 depletion. Quantitative PCR analysis of (C) epithelial/mesenchymal markers and (D) transcriptional regulators of EMT in Jagged2-depleted and control shRNA transfectants treated for 48 hours with (black bars) or without (white bars) TGF-β (1 ng/ml). Values represent the mean (± SD) of replicate (triplicate) samples. P values are based on comparison of TGF-β–treated samples (control vs. Jagged2 depleted). Q values estimate the fraction of comparisons with the given nominally significant P value that may arise from multiple testing. (E) Western blotting of Snail1, Zeb1, Cdh2, Vim, and Actin in control and Jagged2-depleted cells treated with medium or TGF-β (1 ng/ml) for 48 hours. Actin indicates relative protein loading. Protein band density was quantified by densitometry, normalized to that of medium-treated control cells (which is set at 1.0), and is indicated below each blot.
Jagged2 transcriptionally suppresses miR-200.
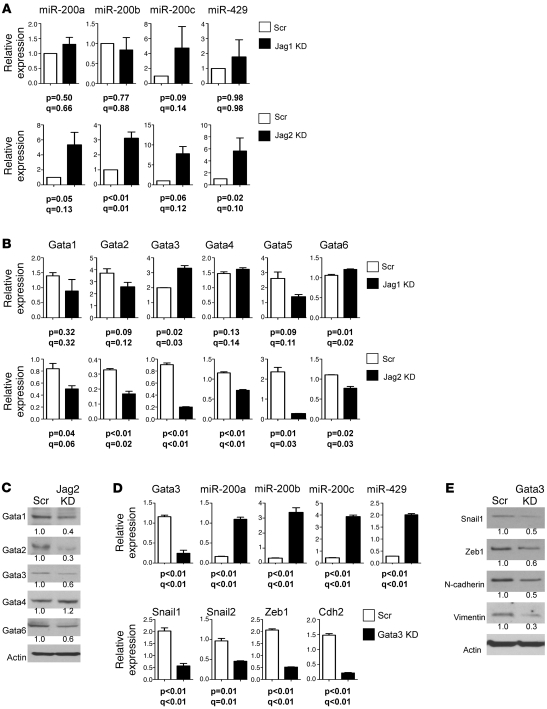
Metastasis-prone tumor cells from KrasLA1/+p53R172HΔG/+ mice have a marked plasticity, undergoing EMT and the reverse process (MET) in a manner that is entirely dependent upon miR-200 (7), but the extracellular cues that regulate miR-200 expression are not clear. On the basis of the above evidence that Jagged2 is required for invasion and metastasis, we postulated that Jagged2 is one of those extracellular cues. Analysis of miR-200 family members revealed that Jagged2 knockdown increased the levels of miR-200a (5 fold), miR-200b (3 fold), miR-200c (7.8 fold), and miR-429 (5.5 fold), whereas Jagged1 depletion did not have this effect (Figure 5A). To query the transcriptional basis for miR-200 regulation by Jagged2, we examined the genomic sequences 5ι of the miR-200b cluster. Of note were multiple binding sites for Gata transcription factors, which are direct transcriptional targets of Notch (31, 32), including 1 binding site for Gata1/2/3 from –79 to –71 bp upstream of the transcription start site, between the pair of E-boxes to which Zeb-1 binds (39). 344SQ cells expressed all 6 Gata family members. After Jagged2 depletion, all Gata family members except Gata1 decreased at the mRNA level (Figure 5B), and all but Gata4 decreased at the protein level (Figure 5C). Thus, the RNA and protein data were corroborative for all of the Gata factors except Gata1 and Gata4, and Jagged2 shRNA coordinately decreased the expression of multiple Gata factors. Given reports of cross-regulation between Gata family members (40), we posited that the multiplicity of Gata factors regulated by Jagged2 is attributable to Gata factor cross-regulation. In fact, knockdown of Gata3, which plays a central role in breast cancer models (35, 36), decreased the levels of Gata1, Gata2, and Gata5 (Supplemental Figure 3A), and add back of Gata3 to the Gata3 shRNA transfectants restored expression of Gata1, Gata2, and Gata5 (Supplemental Figure 3B). Moreover, introduction of Gata3 shRNA into 344SQ cells sharply increased miR-200a (6 fold), miR-200b (10 fold), miR-200c (9 fold), and miR-429 (7 fold) (Figure 5D) and decreased the mesenchymal markers Snail1, Zeb1, N-cadherin, and Vimentin (Figure 5E). Similarly, Jagged2 knockdown in 344P cells decreased Gata3 mRNA levels by 72% and increased miR-200 family members by 6 to 7 fold; Gata3 knockdown in 344P cells increased miR-200 family members by 4 to 7 fold (Supplemental Figure 4); and Jagged2 knockdown in HCC15 human lung cancer cells increased miR-200 family members by 1.8 to 8 fold (Supplemental Figure 5). Collectively, these findings suggest that Gata3 functions as an intermediate between Jagged2 and miR-200, but the relatively modest reductions in Gata3 mRNA (Supplemental Figure 5) and protein (Figure 5C) induced by Jagged2 depletion leave open the possibility that Jagged2 regulates miR-200 levels through a Gata3-independent mechanism.
Figure 5. Jagged2 promotes EMT by inhibiting miR-200.
(A and B) Quantitative PCR analysis of (A) miR-200 and (B) Gata family members in Jagged1-depleted, Jagged2-depleted, and control shRNA transfectants. Values represent the mean (± SD) of replicate (triplicate) samples. Q values estimate the fraction of comparisons with the given nominally significant P value (2-tailed welch’s t test) that may arise from multiple testing. (C) Western blotting of GATA family members in Jagged2-depleted and control 344SQ transfectants. Densitometric quantification of bands indicated under each gel is expressed relative to that of control, which was set at 1.0. (D) Quantitative PCR analysis of Gata3, miR-200 family members, and EMT-related genes in Gata3-depleted and control shRNA transfectants. Values represent the mean (± SD) of replicate (triplicate) samples. (E) Western blotting of Snail1, Zeb1, Cdh2, Vim, and Actin in control and Gata3-depleted cells. Actin indicates relative protein loading. (C and E) Protein band density was quantified by densitometry, normalized to that of control cells (which is set at 1.0), and is indicated below each blot.
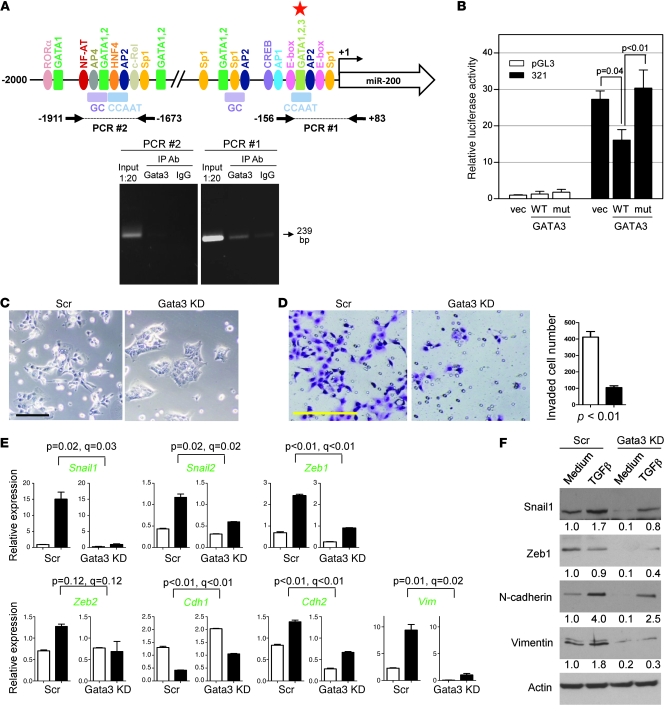
To further examine the role of Gata3 as a transcriptional regulator of miR-200, we analyzed the promoter region of the miR-200b cluster and identified multiple putative Gata binding sites, including 1 Gata1/2/3 binding site adjacent to the E-box to which Zeb binds (Figure 6A). We examined whether Gata3 binds directly to the promoter region of the miR-200b-200a-429 cluster in human H322 lung cancer cells by performing ChIP assays using primers to the proximal E-box–containing region and to an upstream region, which revealed that Gata3 bound to the proximal but not the upstream promoter region (Figure 6A). Furthermore, cotransfection of a Gata3 expression vector into human MCF-7 breast cancer cells suppressed the activity of a luciferase reporter containing the proximal promoter region of interest (–321 to +19 bp from transcription start site) (Figure 6B), and repression by Gata3 required the putative Gata3 binding site (–79 to –71 bp from transcription start site) but not the 2 E-boxes to which Zeb-1 binds (–101 to –95 bp and –57 to –51 bp, respectively, from transcription start site) (Supplemental Figure 6), suggesting that Gata3-induced miR-200b repression does not require Zeb1. Collectively, these findings suggest that Gata3 binds directly to the miR-200b promoter and suppresses its activity.
Figure 6. Gata3 directly suppresses miR-200 transcription and induces EMT.
(A) ChIP assays examined Gata3 binding to the miR-200b promoter in human H322 lung cancer cells. A 239-bp PCR product (arrow) was generated from the Gata3 IP samples (Gata3), using primers for a proximal promoter segment (PCR #1), but not from upstream regulatory sequences (PCR #2), which are illustrated graphically above the gel. Putative binding sites for GATA3 (red star) and other transcription factors (colored ovals and rectangles) predicted by TRANSFAC (BIOBASE Biological Databases). (B) MCF-7 cells cotransfected with reporters containing the proximal miR-200 promoter (–321 to +15 bp from the transcription start site) or empty luciferase vector (pGL3) and vectors containing Gata3 (WT), inactive Gata3 mutant (mut), or empty vector (vec). Values were expressed as the mean (± SD) of triplicate wells. The differences between groups were estimated by 1-way ANOVA (P < 0.01). (C) Images of Gata3-depleted or control 344SQ transfectants. Scale bar: 100 μm. (D) Images of invaded Gata3-depleted (black bar) and control (white bar) 344SQ transfectants, which were counted and expressed as the mean values (± SD) of replicate (triplicate) wells (bar graph). Scale bar: 100 μm. (E) Quantitative RT-PCR analysis of the indicated genes (green) in Gata3-depleted and control shRNA transfectants treated for 48 hours with (black bars) or without (white bars) TGF-β (1 ng/ml). Values represent the mean (± SD) of triplicate samples. (F) Western blotting of control and Gata3-depleted cells treated with medium or TGF-β (1 ng/ml) for 48 hours. Band density was quantified by densitometry, normalized to that of medium-treated control cells (which are set at 1.0), and is indicated below each blot.
On the basis of the above data, we posited that Gata3 is required for lung adenocarcinoma cells to undergo EMT, form tumors, and metastasize. We first explored the expression of Gata3 in metastases. Gata3 was abundantly expressed in distant metastases but not in lung tumors, based on immunofluorescent staining of tissues isolated from KrasLA1/+p53R172HΔG/+ mice (Supplemental Figure 7A). To determine whether Gata3 plays a role in Jagged2-mediated metastasis, we first examined whether Gata3 is detectable and coexpressed with Jagged2 in early metastases. Lungs were isolated from mice 3 weeks after subcutaneous injection of GFP-tagged 344SQ cells, when microscopic lung metastases are present. Lung tissues were dispersed and sorted by flow cytometry (Supplemental Figure 7B) to isolate GFPpos cells. Gata3 was coexpressed with Jagged2 in GFPpos cells, and these levels were similar to those of the CD133hi fraction from a 344SQ subcutaneous tumor (Supplemental Figure 7C). We next examined whether Gata3 deficiency abrogates tumor cell EMT and invasion in vitro and tumor growth and metastasis in syngeneic mice. 344SQ cells transfected with a Gata3 shRNA exhibited an epithelial morphology (small, round cells that grew in clusters) (Figure 6C), reduced invasive activity in Matrigel (Figure 6D), and resistance to EMT induction by TGF-β (Figure 6, E and F). Although Gata3-depleted and control transfectants were similarly tumorigenic in syngeneic mice (tumors developed in 5 out of 5 mice injected with either transfectant), the incidence of lung metastases in mice bearing visible subcutaneous tumors was reduced in Gata3 shRNA transfectants relative to that of control transfectants (1 out of 5 mice bearing Gata3-depleted tumors vs. 5 out of 5 mice bearing control tumors; P = 0.02) (Supplemental Table 3). Similarly, 5 out of 8 mice bearing control 344P tumors developed lung metastases, whereas only 1 out of 9 mice bearing Gata3-depleted 344P tumors developed lung metastases (P = 0.043), but 344P tumorigenicity was not affected by Gata3 knockdown (Supplemental Table 3). We conclude that Gata factors are required for lung adenocarcinoma cells to undergo EMT, invade, and metastasize.
The relevance of these findings to human lung cancer was examined by mining the transcriptional profiles of resected primary lung tumors from 2 lung adenocarcinoma patient cohorts (n = 193 and n = 117 patients, respectively) (41, 42). Tumors were stratified dichotomously into CD133hi or CD133lo groups on the basis of having PROM1 expression above or below the median value, respectively. For CD133hi tumors, GATA3 correlated modestly with low CDH1 (r = –0.21, P = 0.04; r = –0.31, P = 0.02 for the 2 cohorts, respectively, 1-sided Spearman’s rank test), high ZEB1 (r = 0.46, P < 0.001; r = 0.31, P = 0.02, respectively), and high VIM (r = 0.36, P < 0.001; r = 0.42, P = 0.001, respectively) (Supplemental Table 4). For CD133lo tumors, GATA3 correlated weakly with CDH1 (r = –0.29, P = 0.004; r = –0.24, P = 0.06, respectively), weakly with ZEB1 (r = 0.20, P = 0.05; r = 0.23, P = 0.07, respectively), and did not at all with VIM (r = 0.03, P = 0.74; r = 0.25, P = 0.06, respectively) (Supplemental Table 4). Collectively, these findings suggest that GATA3 correlates with EMT in human lung cancers and may do so more strongly in CD133hi tumors than it does in CD133lo tumors, but none of these genes (PROM1, GATA3, CDH1, ZEB1, and VIM) correlated with survival duration in either patient cohort.
miR-200 inhibits GATA3 expression.
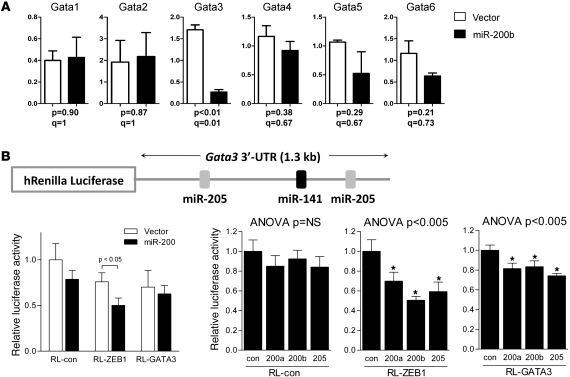
The MET features of the Gata3-deficient cells phenocopy the effects of forced expression of miR-200 in 344SQ cells (7). Therefore, we postulated that Gata3 is a downstream mediator of miR-200. Comparison of Gata levels in 344SQ cells stably transfected with miR-200b or a control lentiviral expression vector revealed that miR-200b suppressed Gata3 levels by 82% (P < 0.01) but did not significantly change other Gata family members (Figure 7A). Examination of Gata3 genomic sequences revealed putative binding sites for miR-205 and miR-141 in the 3′-untranslated region (3′-UTR) of the Gata3 mRNA (TargetScan; www.targetscan.org). To determine whether Gata3 is a direct gene target of miR-200, the miR-200b stable transfectants, which express increased levels of all the miR-200 family members and miR-205 (7), were transiently transfected with a reporter containing luciferase fused to 3′-UTR sequences from GATA3 or ZEB1 (as positive control). Transfection of the miR-200b expression vector suppressed ZEB1 but not GATA3 3′-UTR reporter activity (Figure 7B). As a second approach, 344SQ cells were transiently cotransfected with the same reporters and synthetic miR-200 precursors (200a, 200b, or 205). Relative to control oligomers, the synthetic miR-200a, miR-200b, and miR-205 precursors suppressed GATA3 3′-UTR reporter activity by 19%, 17%, and 26%, respectively. Although this decrease in GATA3 3′-UTR reporter activity was significant, it was considerably less than the decrease in ZEB1 3′-UTR reporter activity (Figure 7B). Collectively, these findings suggest that miR-200 inhibits GATA3 expression without directly targeting the GATA3 3′-UTR.
Figure 7. miR-200 inhibits Gata3 expression.
(A) Forced miR-200 expression inhibits Gata3. Quantitative PCR analysis of 344SQ cells subjected to forced miR-200b expression or empty lentiviral vector transfection (vector). Values represent the mean (± SD) of replicate (triplicate) samples. P values are from 2-tailed Welch’s t test. Q values estimate the fraction of comparisons with the given nominally significant P value that may arise from multiple testing. (B) Gata3 is not a direct miR-200 gene target. Reporter assays were performed using reporters fused to 3′-UTR sequences from Gata3 (RL-GATA3); ZEB1 (RL-Zeb1), which was included as a positive control; or nothing (RL-con). The Gata3 3′-UTR reporter construct (with positions of putative miR-200 binding sites) is illustrated graphically. These reporters were transiently transfected into miR-200b (miR200) or control 344SQ stable transfectants or were transiently cotransfected with synthetic miR-200 precursors (200a, 200b, or 205) or control oligomers (con) into 344SQ cells. Values were normalized based on renilla luciferase and expressed as the mean values (± SD) of replicate (triplicate) wells relative to those of controls cotransfected with empty reporter and empty expression vector or scrambled precursors, which were set at 1.0. Asterisks indicate F-test contrast P < 0.005 versus control. Values of 1-way ANOVA analysis are indicated.
Discussion
A growing body of evidence suggests that metastases emanate from a population of “migrating cancer stem cells” located at the interface between tumor and surrounding stroma (7, 15, 20, 21). Here, we show in a mouse model of lung adenocarcinoma that CD133 marks a population of metastasis-prone tumor cells that localize on the periphery of tumors. CD133 was detected in the majority of tumor cells in nascent distant metastases and in only a paucity of cells in primary lung tumors, implying a positive selection for CD133-expressing cells during metastasis. Direct examination of the metastatic potential of tumor cells sorted on the basis of CD133 expression indicated that CD133hi tumor cells were prone to metastasis. One interpretation of the peripheral location of these tumor cells is that metastasis competence requires physical interactions with cells in the surrounding stroma. However, several findings presented here support the conclusion that metastasis is driven by homotypic, rather than heterotypic, cell-cell interactions. CD133hi tumor cells expressed both Notch and Notch ligands, supporting their capacity for homotypic, Notch-dependent interactions, and Jagged2 depletion from tumor cells abrogated EMT and metastasis, which would not have occurred if the cell-cell interactions driving these processes were heterotypic.
The findings presented here demonstrate that metastasis is a regulable property of tumor cells that is governed by Gata3 and miR-200, which are mutually inhibitory and have opposing effects on EMT and metastasis. Jagged2 perturbs this equilibrium, shifting the balance toward high levels of Gata3, low levels of miR-200, EMT, and metastasis. These findings suggest that Gata3 functions as an intermediate between Jagged2 and miR-200, but the relatively modest reductions in Gata3 mRNA and protein induced by Jagged2 depletion leave open the possibility that Jagged2 regulates miR-200 levels through a Gata3-independent mechanism. The complexity of this pathway was made more apparent by the finding that Jagged2 regulates the expression of all 6 Gata factors and Gata3 regulates the expression of Gata1, Gata2, and Gata5, which supports the existence of an intricate regulatory network among the Gata factors, as reported previously in Gata1-null erythroid cells (43), and raises the possibility that the phenotypic changes in Gata3-deficient tumor cells are mediated in part by other Gata factors. Moreover, the plasticity of tumor cell metastatic potential in this model and complexity of the regulatory apparatus that controls it challenge the paradigm that metastasis is a constitutive trait owing to somatic mutations that accumulate in tumor cells over time (44).
The finding that Gata3 promotes metastasis contrasts strikingly with previous reports on the role of Gata3 in breast cancer. GATA3 is expressed in tumors from a subset of human breast cancer patients, in which it has emerged as a strong predictor of breast tumor differentiation, estrogen receptor status, and clinical outcome (45). In mouse models of breast cancer, Gata3 expression is lost with malignant progression, and restoration of Gata3 expression in late breast carcinomas induces tumor differentiation and suppresses metastasis, an effect mediated in part through suppression of the Gata3 target gene INK4c, which constrains luminal progenitor cell expansion and suppresses tumorigenesis (35, 36). Possibly contributing to its divergent roles in mouse models of breast and lung cancer, Gata3 varies its transcriptional output through dimerization with other Gata family members. Gata factors form homodimeric and heterodimeric Gata complexes that have distinct DNA-binding specificities and regulate unique sets of target genes (46–48) that include, as shown here, each other. Thus, tissue-specific differences in Gata factor expression may contribute to the distinct biologic roles of Gata3 in cancers of the lung and breast.
The phenotypic differences between the Jagged1- and Jagged2-deficient tumor cells were unexpected, given that Jagged1 and Jagged2 reportedly bind to Notch family members with similar specificities (49, 50). These phenotypic differences may be related to the unique ability of Jagged1 to transmit both forward and retrograde signals, the former through interactions with Notch and the latter through PDZ-binding motifs located at the extreme C terminus (29). An alternative explanation is that Jagged1 and Jagged2 activate distinct Notch-dependent intracellular signaling pathways, which is supported by the finding that Gata family members were more prominently diminished by knockdown of Jagged2 than they were by knockdown of Jagged1.
Strategies to detect metastasis-prone tumor cells and to inhibit their spread are of critical importance to the improvement of clinical outcome for lung cancer patients. Of note, human lung adenocarcinoma cells express CD133 and Jagged2 (37, 51, 52), which tempts speculation that Notch inhibitors will prevent metastasis in lung adenocarcinoma patients with tumors that express high levels of Jagged2 and low levels of miR-200. Although such an approach would not lead to disease eradication, it may prolong recurrence-free survival, a beneficial outcome made more germane by the report that CD133-expressing lung cancer cells are resistant to cytotoxic chemotherapy (37). However, human epithelial malignancies are genetically heterogenous, and the biologic basis for plasticity in human tumors will undoubtedly differ between patients. Regardless, these findings provide a basis for efforts to determine whether metastases in cancer patients emanate from a pool of highly plastic tumor cells and, if so, whether metastasis is preventable by targeting those cues that regulate tumor cell EMT.
Methods
Animal husbandry.
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center. For syngeneic tumor experiments, 10- to 16-week-old 129/Sv mice were injected with the indicated numbers of tumor cells into the posterior flanks and sacrificed at the first signs of morbidity.
Antibodies.
We purchased rabbit anti-CD133 (Abcam, ab19898), anti-Notch1 (Abcam, ab27526), anti-Notch2 (Millipore, 07-1233), anti-Notch3 (Santa Cruz Biotechnology Inc., sc-5593), APC-conjugated anti-CD133 (eBioscience, 17-1331-81), goat anti-Jagged1 (Santa Cruz Biotechnology Inc., sc-6011), anti-actin (Santa Cruz Biotechnology Inc., sc-1616), goat anti-Delta (Santa Cruz Biotechnology Inc., sc-8155), rabbit anti-Delta3 (Santa Cruz Biotechnology Inc., sc-67269), rat anti-GATA1 (Santa Cruz Biotechnology Inc., sc-265), mouse anti-GATA2 (Santa Cruz Biotechnology Inc., sc-267), rabbit anti-GATA3 (Santa Cruz Biotechnology Inc., sc-9009), mouse anti-GATA4 (Santa Cruz Biotechnology Inc., sc-25310), rabbit anti-GATA6 (Santa Cruz Biotechnology Inc., sc-9055), goat anti-Zeb1 (Santa Cruz Biotechnology Inc., sc-10572), rabbit anti-Snai1 (Santa Cruz Biotechnology Inc., sc-28199), rabbit anti–N-cadherin (BD Transduction Laboratory, 610921), goat anti-Vimentin (Santa Cruz Biotechnology Inc., sc-5565), rabbit anti-phospho-SMAD2 (Cell Signaling Technology, 3108), anti-phospho-SMAD3 (Cell Signaling Technology, 9520), anti-total SMAD3 (Cell Signaling Technology, 9513), biotinylated anti-CD45 (BD Pharmingen, 553771), anti-CD31 (BD Pharmingen, 558737), and streptavidin-PE (BD Pharmingen, 554061) antibodies.
Antigen detection in tissues.
Four-μm paraffin-embedded sections were baked at 60°C for 30 minutes and then cooled to ambient temperature. Sections were sequentially incubated in Xylene (5 minutes twice), 100% ethyl alcohol (5 minutes twice), 95% ethyl alcohol (5 minutes twice), and 80% ethyl alcohol (5 minutes). After washing with water, the sections were antigen-retrieved using citrate buffer (pH 6.0; DAKO) in a steamer for 20 minutes and cooled to ambient temperature. Sections were then washed with Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBS-T) and quenched with 3% hydrogen peroxide in TBS for 10 minutes, blocked for avidin/biotin activity, blocked with serum-free blocking reagent, and incubated with primary antibody as follows: for CD133 staining, sections were incubated with antibody for 1 hour at ambient temperature or overnight at 4°C; for Jagged1 and Gata3 staining, sections were incubated with antibody overnight at 4°C. Immunohistochemical staining was developed using the DAB substrate system (DAKO), and immunofluorescence staining was developed using TSA Kits (Invitrogen). For CD133 staining of 393P and 344SQ cell cultures, cells were grown on 24-well plates to 70% confluence, washed with ice-cold PBS, and then fixed in methanol for 15 minutes at –20°C and washed with PBS. Cells were blocked with serum-free blocking reagent and incubated with antibody overnight at 4°C. The staining was visualized with Alexa Fluor 488 anti-rabbit antibody (Invitrogen).
Tissue dissociation and flow cytometric analysis.
Freshly isolated murine lung tissues were washed with ice-cold PBS and digested in PBS supplemented with 3 mg/ml dispase II (Roche) and 2 mg/ml collagenase I (Worthington) at 37°C for 45 minutes. Single cell suspension was achieved by sequentially filtering through 100-μm (Falcon) and 40-μm (Falcon, 352340) cell strainers. Red blood cells were removed by red blood cell lysis buffer. Cells were resuspended in PBS supplemented with 0.5% FBS and stained with appropriate antibodies. Cell sorting was performed on a 3-laser, 13-color FACSAria fluorescence-activated cell sorter (BD Biosciences) that is capable of sorting 4 subpopulations at a low pressure at a maximum rate of 20 million cells per hour.
Gene expression profiling.
CD133hi and CD133lo cell fractions (at least 1,000,000 cells of each) were sorted from 344SQ subcutaneous tumors by using flow cytometry. Cells from 3 independent experiments were lysed in buffer RLT (Qiagen) separately and snap-frozen in liquid nitrogen. Lysates (3 for each cell type and 6 in total) were shipped on dry ice to Asuragen for processing and analysis on Affymetrix Mouse Expression Array 430A 2.0 chips. Array data have been deposited in the Gene Expression Omnibus (accession number GSE15587; http://www.ncbi.nlm.nih.gov/geo/). Data processing, determination of differentially expressed genes, and Gene Ontology term enrichment analysis were carried out essentially as described previously (53). Briefly, the data were analyzed with dChip version 2006 ( www.dchip.org), using Affymetrix default analysis settings and invariant set as normalization method. Two-sided homoscedastic t tests (using log-transformed data) and fold changes (ratio of averages of 2 groups, unlogged) were used to determine differentially expressed genes. We used the method of Storey et al. (54) to estimate the false discovery rate (FDR) from multiple hypothesis testing; of the approximately 45,000 Affymetrix probe sets in the entire data set, 1,127 were nominally significant with a nominal P value of less than 0.01 (no fold criteria), which yielded an FDR of 40%, which was considered acceptable for use of the array data as an initial screen to identify gene candidates for validation (e.g., using RT-PCR).
RNA extraction and quantitative RT-PCR.
Cells were grown to 80% confluence in RPMI 1640 supplemented with 10% FBS on 24-well plates, washed with ice-cold PBS, and subjected to RNA extraction using 1 ml of TRIzol (Invitrogen). mRNA was reverse transcribed using the SuperScript First-Strand Synthesis System (Invitrogen). For quantitative PCR reactions, 1:10 dilutions of cDNA products were amplified using SYBR Green PCR Master Mix (Applied Biosystems) and analyzed by using ABI Prism 7500 Fast System (Applied Biosystems). For the analysis of miR-200 family members, TaqMan assays (Applied Biosystems) were performed using 10 ng of total RNA as described previously (7). mRNA and miR expression values were normalized on the basis of L32 mRNA and miR-16 levels, respectively.
Generation of shRNA transfectants.
The shRNA retroviral constructs were purchased (Jagged1 [GeneCopoeia], Jagged2 [Origene], and Gata3 [SABiosciences]). Purified plasmids (1 μg of each) were transfected into 344SQ cells by using LipofectAmine and PLUS (Invitrogen). After 48 hours, transfectants were replated in RPMI 1640 medium containing 10% FBS and 15 μg/ml puromycin for selection and passed serially for 4 weeks to generate stable transfectants.
Protein extraction and immunoblotting.
Cells or tissues were lysed in lysis buffer containing 10 mM Tris (pH 7.4), 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7, 10 H2O, 1 mM PMSF, and protease inhibitors cocktail (Sigma-Aldrich, P8340). The lysates were quantified for protein concentrations with BCA protein assay reagents (Pierce). Proteins (30 μg per sample) were electrophoresed in SDS-polyacrylamide gel and transferred onto nitrocellulose membranes (Amersham, Hybond ECL). The membranes were blocked with 5% skim milk in TBS-T and subsequently incubated with primary antibodies overnight at 4°C. After washing with TBS-T for 30 minutes at room temperature, the membrane was further incubated with horseradish peroxidase–conjugated secondary antibodies for 1.5 hours, followed by 30 minutes of washing with TBS-T. Protein bands were visualized with Pierce ECL substrates (Pierce).
Cell migration and invasion assays.
Migration and invasion assays were performed by using Transwell plates (BD Biosciences) as instructed by the manufacturer. Briefly, cells (50,000 cells/well for the migration assay and 100,000 cells/well for the invasion assay) were plated in triplicate in serum-free RPMI in the upper chambers and in RPMI 1640 with 10% FBS placed in the bottom wells, at 37°C for 24 hours. The migrated or invaded cells were visualized by Diff-Quik Stain (Dade Behring). Three microscopic fields (×10) were photographed and counted per chamber, and results were expressed as the mean ± SD of results from the replicate wells.
Cell proliferation assays.
Cells (200,000/well) were plated in triplicate in RPMI 1640 supplemented with 10% FBS on 60-mm cell culture dishes for 1–5 days as indicated. Cell proliferation was quantified by counting cells under a light microscope (×10) using a hemacytometer.
ChIP assays.
Briefly, H322 cells were cross-linked with 1% formaldehyde and then incubated in lysis buffer (50 mM Tris-HCl [pH 8.1], 1% SDS, 10 mM EDTA, and protease inhibitors cocktail) on ice for 10 minutes. After sonication at 4°C (Cole-Parmer GEX-130 Sonicator; at 50% power for 20 cycles of pulse-on for 10 seconds and pulse-off for 10 seconds), samples were immunoprecipitated with anti-GATA3 antibody or anti-rabbit IgG (Santa Cruz Biotechnology Inc.). DNA was eluted and purified with a PCR Purification Kit (Qiagen), and then PCR was carried out using PCR #1 (5′-CTGCGTCACCGTCACTGG-3′ and 5′-ACAACTCGCCCGTCTCTG-3′) and PCR #2 (5′-CAGCAGGTTTTCCACCACAG-3′ and 5′-AAGCTGCTCTTTCTCCAAGG-3′) primer sets.
Luciferase assays.
MCF7 cells were seeded on 24-well plates (1 × 105 cells/well) 1 day before transfection. Cells were cotransfected with 500 ng pGL3-basic (Promega) or miR-200 promoter (construct containing –321 to +17 bp from transcription start site; ref. 39) and 250 ng Gata3 wild-type (Addgene ID 1332) or inactive mutant (Addgene ID 1295) plasmids. pRL-TK (50 ng, Promega) was cotransfected as an internal control. After 48 hours of transfection, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
GATA3 3′-UTR reporter assays.
The GATA3 3′-UTR (1.3 kb) was isolated from mouse BAC clone (BACPAC Resource Center at Children’s Hospital Oakland Research Institute, RP24-155K15) by PCR and ligated into the pCI-neo-hRL vector. The ZEB1 3′-UTR (1.9 kb) subcloned into the same vector was used as a control. These reporters (500 ng) were transiently transfected into 344SQ cells stably transfected with miR-200b or control expression vectors (1 × 105 cells/well). Alternatively, these reporters were cotransfected with synthetic miR-200 precursors (5 nM, Ambion) into 344SQ cells. After 48 hours, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega).
Statistics.
For cell line experiments, comparisons between 2 groups were performed using the 2-tailed Welch’s t test, unless otherwise indicated. One-way ANOVA was performed to compare multiple experimental groups. Where multiple genes were each individually tested in a given experiment, the FDR was estimated as V/R, where V is the expected number of false positives (nominal P value × number of comparisons made) and R is the number of both true and false positives. The FDR q value is the fraction of comparisons yielding the given nominal P value that are expected to represent false positives (54). P values that are less than 0.05 are considered statistically significant.
Supplementary Material
Acknowledgments
We would like to thank all the members of the Kurie and Goodall laboratories for assistance and comments on the work; Dror Berel for assistance with the mRNA expression data analysis; and Waun Ki Hong for financial support. We acknowledgement support from NIH grant 5 T32 CA009666 (to D.L. Gibbons and W. Lin); NIH grants P50 CA70907 (Lung Cancer SPORE), R01 CA132608, and R01 CA117965, David M. Sather Memorial Fund, and Armour Family Lung Cancer Research Fund (to J.M. Kurie); and NIH grant P30 CA125123 and Dan L. Duncan Cancer Center at Baylor College of Medicine (to C.J. Creighton). D.L. Gibbons was also supported by a Young Investigator Award from ASCO Cancer Foundation and an International Association for the Study of Lung Cancer Fellow Grant. J.M. Kurie is the Elza A. and Ina Shackelford Freeman Endowed Professor in Lung Cancer.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(4):1373–1385. doi:10.1172/JCI42579.
References
- 1.Fisher GH, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15(24):3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4(2):111–120. doi: 10.1016/S1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 3.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25(14):2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- 5.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410(6832):1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons DL, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PloS ONE. 2009;4(4):e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65(22):10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 9.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26(48):6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 10.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67(5):1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 11.Comijn J, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–1278. doi: 10.1016/S1097-2765(01)00260-X. [DOI] [PubMed] [Google Scholar]
- 12.Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci U S A. 2006;103(50):18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17(5):548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4(7):487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 17.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 18.Savagner P, et al. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J Cell Physiol. 2005;202(3):858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Aigner K, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eger A, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14):2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 22.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 23.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 25.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. . Cancer Res. 2010;70(1):36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 27.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 28.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194(3):459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 29.D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27(38):5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong KG, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. . J Exp Med. 2007;204(12):2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amsen D, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27(1):89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5(2):168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Westhoff B, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106(52):22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouros-Mehr H, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13(2):141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei XH, et al. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell. 2009;15(5):389–401. doi: 10.1016/j.ccr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertolini G, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaBarge MA, Bissell MJ. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest. 2008;118(6):2021–2024. doi: 10.1172/JCI36046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. . Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko H, Shimizu R, Yamamoto M. GATA factor switching during erythroid differentiation. Curr Opin Hematol. 2010;17(3):163–168. doi: 10.1097/MOH.0b013e32833800b8. [DOI] [PubMed] [Google Scholar]
- 41.Chitale D, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28(31):2773–2283. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomida S, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27(17):2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 43.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8(10):1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 44.Chiang AC, Massague J. Molecular basis of metastasis. New Eng J Med. 2008;359(26):2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouros-Mehr H, Kim JW, Bechis SK, Werb Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol. 2008;20(2):164–170. doi: 10.1016/j.ceb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beuling E, et al. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008;322(1):179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyronlahti A, et al. GATA-4 regulates Bcl-2 expression in ovarian granulosa cell tumors. Endocrinology. 2008;149(11):5635–5642. doi: 10.1210/en.2008-0148. [DOI] [PubMed] [Google Scholar]
- 48.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domain. Mol Cell Biol. 1995;15(5):2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu K, et al. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol Cell Biol. 2000;20(18):6913–6922. doi: 10.1128/MCB.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu K, et al. Mouse Jagged1 physically interacts with Notch2 and other Notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274(46):32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 51.Choi K, et al. Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. J Biol Chem. 2009;284(26):17766–17774. doi: 10.1074/jbc.M109.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 53.Creighton CJ, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26(25):4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.