Abstract
Shared molecular programs govern the formation of heart and head during mammalian embryogenesis. Development of both structures is disrupted in human chromosomal microdeletion of 22q11.2 (del22q11), which causes DiGeorge syndrome (DGS) and velo-cardio-facial syndrome (VCFS). Here, we have identified a genetic pathway involving the Six1/Eya1 transcription complex that regulates cardiovascular and craniofacial development. We demonstrate that murine mutation of both Six1 and Eya1 recapitulated most features of human del22q11 syndromes, including craniofacial, cardiac outflow tract, and aortic arch malformations. The mutant phenotypes were attributable in part to a reduction of fibroblast growth factor 8 (Fgf8), which was shown to be a direct downstream effector of Six1 and Eya1. Furthermore, we showed that Six1 and Eya1 genetically interacted with Fgf8 and the critical del22q11 gene T-box transcription factor 1 (Tbx1) in mice. Together, these findings reveal a Tbx1-Six1/Eya1-Fgf8 genetic pathway that is crucial for mammalian cardiocraniofacial morphogenesis and provide insights into the pathogenesis of human del22q11 syndromes.
Introduction
Progenitors of the cardiac and craniofacial skeletal muscles arise from subdomains of a contiguous field of cephalic mesoderm: the ventrolateral primary heart field (PHF) and the dorsomedial cranial paraxial mesoderm (CPM), respectively. A distinct population of mesodermal progenitors located in between the PHF and CPM, called the secondary heart field (SHF), forms the cardiac outflow tract (OFT) and the RV in mammals (1–5). Recent studies demonstrated that genes critical for SHF development are also required for formation of the head branchiomeric muscles, which control jaw movement, facial expression, and pharyngeal functions (6, 7), and that head muscles and heart may a share common lineage (8). However, the molecular mechanisms that couple development of heart and face are poorly understood.
In humans, deletions of chromosome 22q11.2 cause del22q11 syndrome, DiGeorge syndrome (DGS), and velo-cardio-facial syndrome (VCFS). Affected patients exhibit a wide spectrum of developmental defects, including craniofacial anomalies, dysmorphogenesis of cardiovascular structures, and hypoplasia of the thymus and parathyroid glands (9, 10). Additionally, more than 30% of del22q11 patients have renal defects; however, the underlying etiology remains to be elucidated. T-box transcription factor 1 (TBX1) is located in the 22q11.2 critical region, and TBX1 haploinsufficiency is a major contributor to human del22q11 phenotypes and to murine models of the syndrome (11, 12). Mouse Tbx1 is expressed in SHF cardiac progenitors and is required for their proliferation as well as for cranial skeletal muscle development (13–17). Similar to human hemizygous del22q11 phenotypes, mouse Tbx1 haploinsufficiency causes aortic arch defects, whereas Tbx1-null mutants exhibit more severe cardiac and craniofacial anomalies (18–20).
Although human fibroblast growth factor 8 (FGF8) does not map to the 22q11.2 region, FGF8 and FGF signaling pathways are critical for cardiovascular development and have been directly implicated in the etiology of del22q11 syndromes (12, 21). Autocrine function of murine Fgf8 controls proliferation and survival of SHF cardiac progenitors (22, 23), whereas paracrine Fgf8 signals from the pharyngeal ectoderm and endoderm epithelia control formation and remodeling of both craniofacial structures and aortic arch arteries and cardiac OFT (13, 24–26). Double-heterozygous Tbx1+/–Fgf8+/– mutant embryos exhibit increased incidence of pharyngeal arch (PA) artery remodeling anomalies (27, 28), demonstrating a genetic link between Tbx1 and Fgf8 during vascular morphogenesis. Fgf8 activity has to be precisely controlled, as too much or too little is detrimental to cardiovascular development (29). For example, mutations of components of the Notch signaling pathway cause congenital heart defects in humans (30), which depends in part on modulation of Fgf8 expression in the SHF (31). Fgf8 hypomorphic mice with reduced Fgf8 expression display a range of pharyngeal and cardiovascular phenotypes (32). Forced expression of Fgf8 in a Tbx1-null background could not fully rescue the cardiac defects of Tbx1-null mutants and even increased the incidence of cardiovascular defects (28, 33). Despite the critical role of Fgf8, little is known about the direct transcriptional mechanism responsible for its gene regulation during cardiovascular development (14, 27, 34).
We reported previously that the evolutionarily conserved homeodomain transcription factor Six1 and its canonical coactivator Eya1 synergistically control organ-specific progenitor cell proliferation and survival (35). Deletion of either Six1 or Eya1 phenocopies human branchio-oto-renal (BOR) syndrome, which is characterized by congenital preauricular pits, branchial fistulas, hearing loss, and renal agenesis (35–37). Human EYA1 mutations are also found in patients with congenital cardiac abnormalities (38, 39). Mutation of the EYA1 homolog EYA4 causes dilated cardiomyopathy (40). Therefore, the Six1/Eya1 complex may have yet unidentified roles during cardiovascular development.
Here, we demonstrate that double-null Six1–/–Eya1–/– mutant mice exhibited phenotypes similar to Tbx1 and Fgf8 mouse mutants and phenocopied human del22q11 syndromes, including the major craniofacial and cardiovascular phenotypes. We provide genetic and molecular evidence indicating that a Tbx1-Six1/Eya1-Fgf8 pathway is critical for morphogenesis of cardiocraniofacial structures and propose that mutations of SIX1 and EYA1 may contribute to the pathogenesis of both del22q11-like and BOR syndromes in humans.
Results
Six1 and Eya1 regulate cardiovascular and craniofacial morphogenesis.
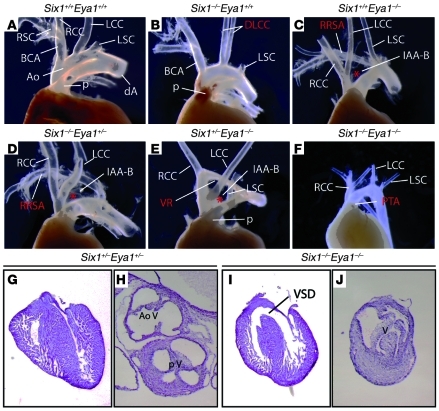
To determine a potential role of Six1 and Eya1 in cardiovascular development, we analyzed the cardiac phenotype of embryos with null mutation of 1 or both copies of Six1 and Eya1. While Six1+/–Eya1+/– embryos had normal morphology of the aorta and pulmonary arteries, further loss of function of either Six1 or Eya1 caused a spectrum of abnormalities of great artery patterning (Figure 1 and Table 1) consistent with abnormal development of the PA arteries and characteristic of del22q11 syndromes (9, 10). For instance, 78% of Eya1–/– mutants had aortic arch defects (n = 13), including 54% with interrupted aortic arch type B (IAA-B), in which the transverse arch is disrupted between the left common carotid and left subclavian arteries. Other vascular anomalies found in Eya1–/– mutants included retroesophageal right subclavian artery (23%) and vascular ring (16%). Six1–/–Eya1–/– mutant embryos had a higher incidence and greater severity of cardiovascular phenotypes (Table 1). Instead of the normal OFT pattern, in which the aorta and pulmonary arteries connect to the LV and RV, respectively, 100% of Six1–/–Eya1–/– embryos exhibited OFT defects (n = 7). We found 14% had double-outlet RV (DORV), in which both great vessels arise from the RV, and egress of blood from the LV is via a ventricular septal defect (Figure 1I). Such defects were present in 100% of compound mutants. Approximately 70% of Six1–/–Eya1–/– mutants had a single, unseptated outflow vessel, and the outflow valve contained 3 or 4 leaflets (Figure 1J). These SHF-related defects are found in the human del22q11 patient population: persistent truncus arteriosus (PTA; a single unseptated OFT and outflow vessel reflecting failure of the embryonic truncus arteriosus to septate into 2 ventricular outflows); pulmonary atresia; tetralogy of Fallot; and, rarely, DORV (41, 42). They are also seen in animal models, including mouse mutants of Tbx1 and Fgf8 (13, 18–20, 22, 23, 25, 26, 43), manipulation of avian Fgf8 expression (29), and SHF ablation in chicks (44).
Figure 1. Six1–/–Eya1–/– mutants phenocopy features of human del22q11 syndromes.
(A–F) Gross morphological defects of the OFT and great arteries of newborn mutants (B–F) versus WT control (A). (G–J) H&E histological staining of sectioned hearts revealed VSD and dysmorphology of the outflow valves in a Six1–/–Eya1–/– mutant (I and J) compared with Six1+/–Eya1+/– control (G and H). Ao, aortic artery; BCA, brachiocephalic artery; dA, descending aorta; DLCC, duplicated left common carotid artery; LCC, left common carotid artery; LSC, left subclavian artery; p, pulmonary artery; RCC, right common carotid artery; RRSA, retroesophageal right subclavian artery; RSC, right subclavian artery; V, ventricle; VR, vascular ring. Asterisks denote interrupted aortic arches. Original magnification, ×50 (G and I); ×80 (H and J).
Table 1 .
Six1 and Eya1 synergistically regulate cardiovascular development
In addition to cardiovascular defects, Six1/Eya1 compound mutants exhibited a spectrum of craniofacial phenotypes, including micrognathia (small jaw) and cleft palate phenotypes found in human del22q11 patients (Supplemental Figure 1 and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI44630DS1) (9, 10, 35, 36, 45). Six1–/– mutants had micrognathia, dysmorphogenesis of the midface, and deformation of nasal structures (Supplemental Figure 1, A–F). Eya1–/– mutants had milder facial defects. Compound mutants exhibited progressively worse phenotypes, with the most severe defects found in the Six1–/–Eya1–/– mutants. To determine whether Six1 and Eya1 are part of the genetic program required for formation of both cardiac and branchiomeric skeletal muscles (6, 7), we examined all cranial skeletal muscles using an antibody marker, MF20 (myosin heavy chain–specific antibody) (Supplemental Figure 1, G–H). Similar to the mouse Tbx1 mutant model of del22q11 syndromes (17), Six1 and Eya1 compound mutants had severely hypoplastic branchiomeric muscles. Six1–/–Eya1–/– mutants also had hypoplastic extraocular muscles, muscles of facial expression, and tongue. Thus, Six1 and Eya1 are required for both cardiovascular and craniofacial development.
Six1 is transiently expressed in a subset of cardiac progenitors.
The surprising SHF-related OFT septation and alignment defects suggest a direct involvement of Six1 and Eya1 in the function of cardiac progenitors in the SHF. We therefore reexamined the early expression pattern of these genes during cardiogenesis, initially using a gene-specific RNA in situ hybridization approach (Supplemental Figures 2 and 3 and refs. 46, 47). At the cranial level, Six1 and Eya1 exhibited a lateromedial mesoderm expression gradient at E8.5: strong lateral expression in the CPM, faint but detectable expression in the SHF/splanchnic mesoderm (SHF/SpM), and undetectable expression in the PHF (Supplemental Figures 2 and 3). These findings indicated that Six1 and Eya1 are indeed expressed in the cardiac progenitors in the SHF/SpM. However, their signal intensities, a reflection of both gene expression level and sensitivity of the gene-specific probe, were considerably less than those of the known SHF progenitor marker genes Tbx1 and islet LIM homeobox 1 (Isl1) (Supplemental Figures 2 and 3).
To better characterize the Six1 expression pattern and cell lineages, we generated a Six1Cre/hpAP knockin/knockout mouse allele that expresses the Cre recombinase and a sensitive reporter, human placental alkaline phosphatase (hpAP). The dicistronic Cre/hpAP gene replaces and inactivates Six1 using the same targeting strategy we reported previously (35). hpAP activity driven by this allele provided a surrogate for Six1 expression and allowed us to better define the location, number, and fate of Six1-positive (Six1+) progenitors. We detected hpAP activity in the cardiogenic mesoderm at E7.5 (Figure 2, B–D), but not in the primitive heart tube at E8.5 (Figure 2, E and F) or in the heart at subsequent stages (data not shown). Consistent with the RNA in situ analysis (Supplemental Figures 2 and 3), we also detected strong endoderm staining as early as E7.5 and ectoderm staining at E8.5 (Figure 2, C and F). The hpAP activity was maintained in skeletal muscle and thymus, but absent from mature cardiac and renal tissues at later developmental stages (data not shown).
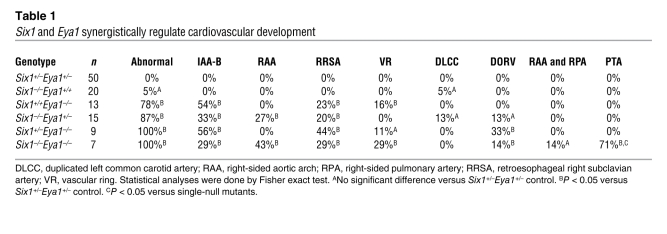
Figure 2. Six1 is expressed in cardiac progenitors.
(A–F) Six1 is expressed in cardiac/skeletal muscle progenitors and in cranial ectoderm and endoderm at E7.5 and E8.5. WT (A) and Six1Cre/hpAP (B–F) embryos were stained for hpAP activity. Cross-sections of B are shown in C and D; cross-section of E is shown in F. CC, cardiac crescent; Ect, ectoderm; End, endoderm; NE, neural epithelium; SM, somite. (G) Whole-mount dorsal-right view of the looped heart from an E9.0 Six1Cre/hpAPR26RmTmG embryo. eGFP+ cells (Rosa/eGFP) in the Six1 lineage were found in inflow tract (IFT), OFT, and developing apex. (H and I) eGFP+ cells in the Six1 lineage and Isl1 colabeled SHF cardiac progenitor cells (arrowheads) in E9.0 sagittal sectioned tissue. Boxed region in H is enlarged in I. (J–N) Coimmunostaining of eGFP+ cells in the Six1 lineage with TNNT2 (J) and ACTN2 (K) in the RV and with SM22α (L) and CNN1 (M and N, arrows) in the proximal region of the great arteries from E17.5 hearts. Boxed region in M is enlarged in N. Original magnification, ×115 (A and B); ×100 (C, D, and F); ×63 (E); ×200 (G–N). (O) Summary of Six1+ progenitor cell lineage, based on results from Figure 2 and Supplemental Figure 5 (WT1, RALDH2; epicardial progenitor data).
Six1+ progenitors contribute to multiple cardiac lineages.
To determine whether Six1+ progenitors contribute to mature cardiovascular tissues, we used double-heterozygous Six1Cre/hpAPR26RmTmG and Six1Cre/hpAPR26RlacZ embryos, in which Six1+ progenitors and their progeny are permanently labeled with membrane-localized eGFP and β-gal reporters, respectively (Figure 2 and Supplemental Figure 4). eGFP+ cells were detected in the inflow tract and OFT of the looped heart at E9.0 (Figure 2, G–I), consistent with Six1 expression in the SHF. This was further supported by colocalization of eGFP with the cardiac progenitor marker Isl1 (4, 48). Consistently, Six1+ progenitors differentiated into multiple cell types, including ventricular cardiomyocytes and smooth muscle cells of the proximal aortic and pulmonary arteries, based on coimmunostaining with the cardiac-specific TNNT2 and ACTN2 and the smooth muscle–specific SM22α and CNN1 antibodies (49), respectively (Figure 2, J–N). In addition, eGFP colocalized with WT1 and RALDH2 (Figure 2O and Supplemental Figure 4, C–J), which suggests that Six1 is expressed in epicardial progenitors (49–51). Thus, in addition to the previous reports of Six1 expression in the pharyngeal ectoderm and endoderm, and the cranial skeletal muscle progenitors (35, 36), our detailed expression and fate-mapping analyses demonstrated that Six1 was also expressed in cardiac progenitors in the SHF and proepicardium during cardiovascular development (Figure 2O). We posit that the combination of Six1 and Eya1 function in all these cells is required for normal cardiovascular and craniofacial morphogenesis.
Six1 and Eya1 are required for cell survival and proliferation.
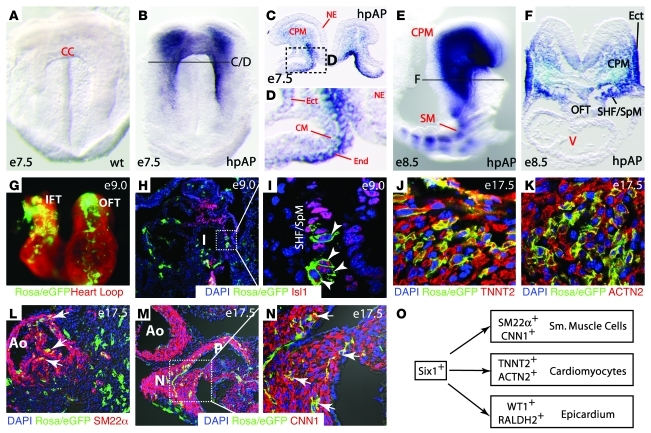
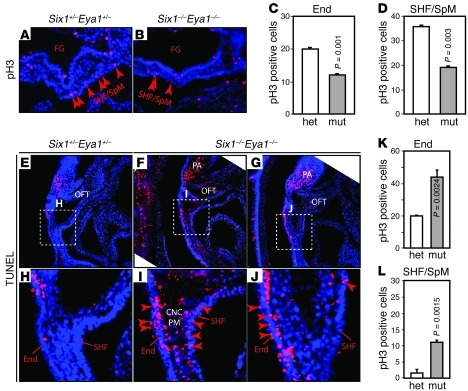
We have shown previously that the Six1/Eya1 transcription complex is essential for renal progenitor cell survival and proliferation at E11.5 and E12.5 (35, 36, 52). Thus, we analyzed early-stage (E9.5 and E10.5) Six1–/–Eya1–/– mutants to determine whether a similar mechanism contributes to cardiovascular morphogenesis (Figure 3). We detected an approximately 50% reduction in phospho–histone H3–labeled (pH3-labeled) cells in the PAs and OFT of whole-mount and sectioned Six1–/–Eya1–/– specimens (Figure 3 and data not shown). Cardiac progenitors in the SHF/SpM had an approximately 48% reduction in pH3 labeling, and those in the foregut endoderm had an approximately 40% reduction (Figure 3, A–D). More apoptotic cells were detected in the SHF pharyngeal ectoderm and endoderm of the Six1/Eya1 mutants relative to controls, based on both LysoTracker and TUNEL staining (Figure 3, E–L, and data not shown). Thus, reduced proliferation and survival of the SHF and the pharyngeal ectoderm and endoderm epithelia in Six1/Eya1 mutants likely contributes to the observed cardiovascular phenotypes, as has been reported in Tbx1 and Fgf8 mouse mutants with similar phenotypes.
Figure 3. Six1 and Eya1 are required for cell proliferation and survival in PA and OFT.
(A–D) Anti-pH3 immunostaining (red) of E9.5 transverse sections to label proliferating cells. Quantification of results are shown in C and D. FG, foregut; mut, Six1–/–Eya1–/– mutant; het, heterozygous control. n = 3. (E–L) TUNEL staining of sagittal sections revealed increased cell death in pharyngeal endoderm, SHF/SpM, and pharyngeal mesenchyme (PM) that contained CNCs and mesoderm. Boxed regions in E–G are enlarged in H–J. Original magnification, ×200 (A, B, and E–J). n = 3. P values were determined by Student’s t test.
We also observed a substantial increase in apoptosis in the pharyngeal mesenchyme cells derived from either mesoderm or neural crests (Figure 3I). Defects of a specialized population of cardiac neural crest cells (CNCs) result in cardiovascular phenotypes similar to those found in the Six1/Eya1 mutants (53–55). To analyze CNCs specifically, we examined the expression patterns of transcription factor AP-2α (AP2) and plexin A2 (Plxna2) (Supplemental Figure 5). These genes were detected in migrating CNCs in the mutant pharynx (AP2) and postmigratory CNCs in the mutant OFT (Plxna2) (Supplemental Figure 5). AP2 expression in the ectoderm was unaffected (Supplemental Figure 5H), but staining in the caudal pharynx was decreased (Supplemental Figure 5, D and G, arrow). Consistent with a potential increase of CNC apoptosis (Figure 3I), the intensity of Plxna2 staining in the prongs of CNCs invading the OFT appeared to be decreased (Supplemental Figure 5B), suggestive of fewer invading CNCs in the mutants. Together, these findings suggest that Six1 and Eya1 are required for CNC survival and that decreased CNC number and/or CNC dysfunction in the mutants may also contribute to the hypoplastic OFT cushions and septation failure.
Fgf8 expression depends on Six1 and Eya1 in vivo.
To identify Six1/Eya1-dependent genetic pathways that regulate cardiovascular morphogenesis, we examined expression of a panel of molecular markers known to be important in SHF survival and function, including Tbx1, Isl1, Fgf8, and Fgf10 (Figure 4 and Supplemental Figures 6–8). Of these genes, Fgf8 exhibited a drastic reduction in expression throughout the PAs and the SHF at E9.5 in Six1–/–Eya1–/– mutants; expression levels of Tbx1 and Isl1 were unaltered despite the severe hypoplastic caudal arch defect (Figure 4, A–D and Supplemental Figures 6–8). Consistent with the gross cardiocraniofacial phenotypes (Figure 1), we noted that Fgf8 expression in the caudal pharyngeal region and the SHF was more sensitive to the loss of Eya1 than of Six1, and the reverse was true in the first arch (Figure 4, B and C). Similar to Six1 and Eya1, Fgf8 is required for SHF progenitor cell proliferation and survival (22, 23, 25, 26), and Fgf8 mouse mutants display phenotypic features of del22q11 syndrome (32, 56). In contrast to Tbx1 mutants, in which both Fgf8 and Fgf10 are reduced, expression of Fgf10 in Six1–/–Eya1–/– mutants was slightly increased (Supplemental Figure 7). Baldini and colleagues have demonstrated that Tbx1 can directly regulate Fgf10 in vitro (16), but it is not yet clear whether Fgf8 is also a direct target of Tbx1 in some tissues (14). Our results suggest an alternate mechanism, in which Tbx1 may regulate Fgf8 expression through the Six1/Eya1 transcription complex.
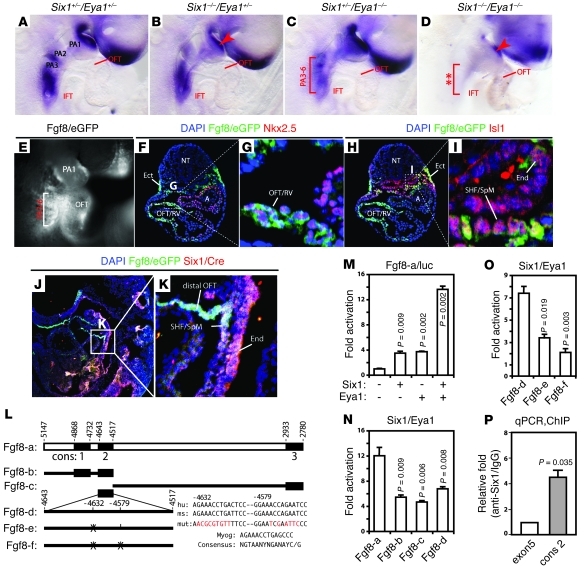
Figure 4. Six1 and Eya1 directly regulate Fgf8 expression.
(A–D) Fgf8 expression was dependent on Six1 and Eya1. Fgf8 was downregulated in the PA ectoderm (arrowhead) of E9.5 Six1–/–Eya1+/– (B) and Six1–/–Eya1–/– (D) mutants and other PA regions (bracket) of Six1+/–Eya1–/– (C) and Six1–/–Eya1–/– (D) mutants. (E–I) Six1hpAP/Cre efficiently turned on the Fgf8eGFP reporter allele in the pharynx (E, bracket) and OFT. eGFP+ (Fgf8/eGFP) cells were found in the pharyngeal ectoderm (F), endoderm (I), SHF/SpM (I), and OFT/RV (F and G). eGFP staining colocalized with Nkx2.5 in the OFT/RV (F and G) and Isl1 in the SHF/SpM (G and I). A, atrium; NT, neural tube. (J and K) Colabeling of Six1-expressing cells with Cre antibody (red) and Fgf8-expressing cells with eGFP antibody (green) of E9.5 Six1hpAP/CreFgf8eGFP embryos (sagittal section). Colabeling appeared white with blue DAPI counter staining. Boxed regions in, F, H, and J are enlarged in G, I, and K, respectively. Original magnification, ×100 (A–D); ×200 (E–K). (L) Potential murine Fgf8 enhancer. Dark boxes mark evolutionarily conserved regions (cons); putative Six1 binding sites and mutations (red) are listed. hu, human; ms, mouse. (M–O) Six1 and Eya1 synergistically regulated reporters containing WT Fgf8 enhancers (M and N), but not the putative Six1 binding site mutant enhancers (O), in transiently transfected HEK293 cells. Reporter constructs are as in L. (P) Quantitative PCR analyses of in vivo ChIP assays of E9.5 mouse PA/heart tissues. Six1 protein was selectively bound to the second conserved region of Fgf8 enhancers (cons2; ~4.4-fold relative to anti-Six1/IgG enrichment). Fgf8 coding region (exon5) served as a negative control.
The Six1/Eya1 transcription complex directly controls Fgf8 expression in vivo.
To test whether Six1 and Eya1 directly control Fgf8 gene expression, we first questioned whether they are expressed in the same cells. We used double-heterozygous Six1hpAP/Cre/+Fgf8eGFP/+ embryos to address this question. Expression of eGFP in these embryos depends on the endogenous promoter activity of both Fgf8 and Six1 and therefore can be used as a sensitive indicator of the same cell lineages (25). Six1hpAP/Cre/+Fgf8eGFP/+ embryos displayed strong Fgf8/eGFP expression in the pharyngeal region and the cardiac OFT (Figure 4E), which is similar to previously reported Fgf8/eGFP activity (25). These Fgf8/eGFP+ lineages included Nkx2.5+ cells in the bulbus cordis and the conotruncus (which contribute to the RV and OFT, respectively; Figure 4, F and G) and Isl1+ cells in the pharyngeal endoderm and ectoderm and the SHF/SpM (Figure 4, H and I). Furthermore, Fgf8/eGFP colocalized with Six1/Cre immunoreactivity in the SHF/SpM and pharyngeal endoderm (Figure 4, J and K), which suggests that Six1 and Fgf8 are coexpressed in multiple cell lineages.
To examine whether Six1/Eya1 can directly regulate Fgf8 expression, we analyzed a genomic fragment containing an Fgf8 enhancer sufficient to drive transgene reporter expression in pharyngeal ectoderm and endoderm and in cardiac OFT (14). The proximal Fgf8 promoter has 3 conserved regions in mice, rats, cows, and primates, and we identified 4 putative Six1 binding sites distributed among those 3 regions (Figure 4L). The second and third sites were clustered in the second conserved region (~130 bp). A luciferase reporter driven by a 2.4-kb mouse Fgf8 enhancer fragment had negligible activity after transient transfection in HEK293 cells. Cotransfection of either Six1 or Eya1 resulted in a 3-fold increase in reporter activity, and there was synergistic enhancement of the reporter activity to approximately 13-fold when both factors were present (Figure 4M). Because truncation analyses indicated that the second conserved region (Fgf8-d) had relatively higher activity than the first and third regions (Figure 4N), we focused on the 2 putative Six1-binding sites within the second conserved region. Mutation of these 2 sites significantly decreased reporter activity (Figure 4O). To confirm that Six1 binds directly to the endogenous Fgf8 enhancer, we purified Six1-associated chromatin from E9.5 mouse embryonic PAs and cardiac tissues. Real-time quantitative PCR analyses indicated that binding of Six1 to the endogenous Fgf8 enhancer was enriched more than 4-fold relative to the coding region of Fgf8 and IgG control (Figure 4P). Together, these findings strongly suggest that the Six1/Eya1 transcription complex directly regulates Fgf8 gene expression in vivo.
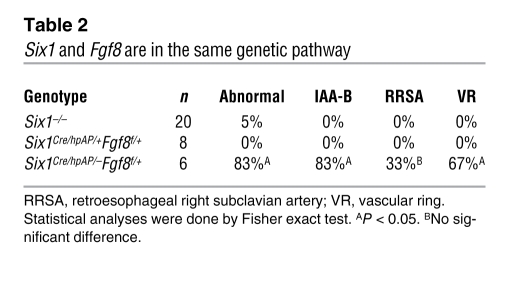
To further establish the functional relationship in vivo between Six1 and Fgf8, we analyzed compound Six1Cre/hpAP/–Fgf8f/+ mutants (ref. 32 and Supplemental Figure 9). In these mutants, a single allele of Fgf8 is inactivated specifically in Six1-expressing cells. Six1–/– and Six1Cre/hpAP/+Fgf8f/+ mutants had only rare great vessel defects (Table 2); however, 83% of Six1Cre/hpAP/–Fgf8f/+ mutants exhibited IAA-B or cervical aortic arch phenotypes. This functional interaction strongly suggests that Six1 and Fgf8 are in the same genetic pathway and synergistically regulate cardiovascular morphogenesis. Decreased expression of Fgf8 likely contributes to the observed del22q11-like phenotypes in Six1–/–Eya1–/– mutants.
Table 2 .
Six1 and Fgf8 are in the same genetic pathway
Exogenous Fgf8 rescues endothelial-to-mesenchymal transformation defects.
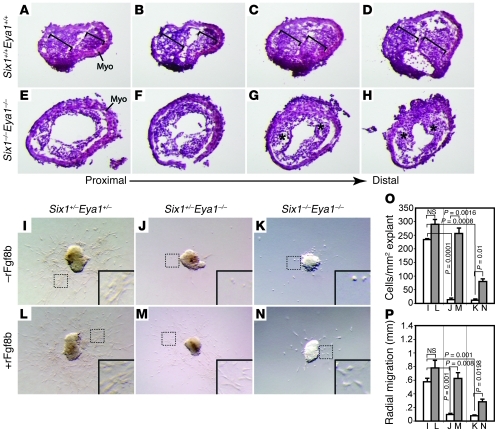
Fgf8 signaling is critical for endothelial-to-mesenchymal transformation (EMT) in the proximal OFT cushions and for CNC migration and invasion of both proximal and distal OFT cushions (26, 31). Consistently, we found that Six1–/–Eya1–/– mutants had severely hypoplastic proximal and distal OFT cushions compared with WT littermate controls (Figure 5, A–H). Using an established OFT explant culture assay (26, 31, 57), we found that both Six1+/–Eya1–/– and Six1–/–Eya1–/– mutants had a dramatic reduction in the number of mesenchymal cells that invaded the gel and migrated away from the beating myocardial explants (Figure 5, I–P). These results suggest that the Six1/Eya1 complex is required indirectly through Fgf8 for EMT of OFT endothelial cells during cushion remodeling.
Figure 5. Fgf8 rescues EMT defects in Six1/Eya1 mutant OFT explants.
(A–H) Six1–/–Eya1–/– mutants (E–H) had hypoplastic OFT cushions compared with WT littermate controls (A–D). Shown are serial histological sections of E11.5 outflow vessels proceeding from proximal (A and E) to distal (D and H). Brackets in A–D indicate WT OFT cushions, which were hypoplastic in the mutants (asterisks). Myo, myocardium. (I–K) The compound mutants (J and K) had fewer endothelial cells migrating from the OFT explant, undergoing EMT, and invading the gel than did WT controls (I) in cultured E9.5 OFTs. (L–N) The EMT defect was rescued by rFgf8b in the explant culture. Original magnification, ×230 (A–H); ×100 (I–N). (O and P) Quantification of results in I–N. White bars, without rFgf8b; gray bars, with rFgf8b. Student’s t test. n = 6 (control); 3 (Six1+/–Eya1–/– and Six1–/–Eya1–/–).
To determine whether Fgf8 is sufficient to rescue EMT in Six1+/–Eya1–/– mutant phenotypes, we treated WT and mutant explants with recombinant Fgf8 isoform b protein (rFgf8b) or BSA control and assayed EMT by counting the total number of mesenchymal cells that invaded the gel as well as the distance migrated from the explant after invasion. Whereas very few mesenchymal cells were present in untreated compound mutant explants and the few present had not migrated far (Figure 5J), rFgf8b treatment almost fully rescued EMT and migration defects of Six1+/–Eya1–/– mutants (Figure 5, I–P). The Six1–/–Eya1–/– mutant phenotypes were also rescued, but to a lesser extent, indicating, not surprisingly, that additional factors downstream of Six1/Eya1 contribute to defects in the Six1–/–Eya1–/– mutant.
Tbx1 is a genetic upstream regulator of Six1 and Eya1.
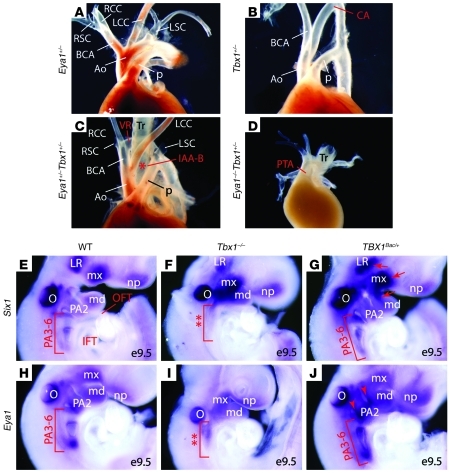
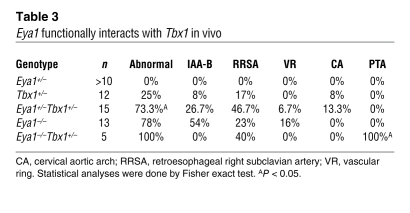
Although human SIX1 and EYA1 do not map to chromosome 22q11.2, the shared morphological defects and molecular phenotype of decreased Fgf8 expression in Six1/Eya1 and Tbx1 mutant mouse models suggests that human SIX1 and EYA1 may be genetic modifiers of the del22q11 syndrome. To test this possibility using mouse models, we examined the functional relationship between Six1 or Eya1 and Tbx1 by analyzing compound mutants of these genes in a C57BL6/CD-1 mixed genetic background (Figure 6, A–D, and Table 3). Whereas 25% of Tbx1+/– mutants (n = 12) had cardiovascular defects at E17.5, none of the Eya1+/– mutants did (n > 10). Penetrance of these defects increased to 73% in Eya1+/–Tbx1+/– mutants (n = 15). Remarkably, although approximately 80% of Eya1–/– mutants had cardiovascular phenotypes (n = 13), none of them had the severe defect of a single outflow vessel seen in Tbx1–/– mutants, in contrast to 100% of Eya1–/–Tbx1+/– mutants (n = 5; Figure 6D and Table 3). Similar to Eya1/Tbx1 compound mutants, 63% of Six1–/–Tbx1+/– mutants (n = 8) had cardiovascular phenotypes, in contrast to less than 5% of Six1–/– mutants (n = 20) and 33% of Tbx1+/– mutants (n = 9). Together, these findings demonstrated that Six1, Eya1, and Tbx1 are in a common genetic pathway controlling cardiovascular and craniofacial development.
Figure 6. Eya1 functionally interacts with Tbx1 in vivo.
(A–D) Gross views of cardiovascular structures of newborn Eya1/Tbx1 compound mutants. Asterisk in C denotes interrupted aortic arches. (E–J) Whole-mount RNA in situ hybridization revealed altered Six1 and Eya1 expression in Tbx1–/– (F and I) and TBX1Bac/+ gain-of-function mutants (G and J). Tbx1–/– mutants had hypoplastic and unsegmented caudal PAs (F and I, double asterisks). Brackets in G and J indicate increased expression of Six1 and Eya1 in the TBX1 gain-of-function mutants. Arrows in G denote Six1 upregulation. Arrowheads in J indicate enhanced Eya1 expression. CA, cervical aortic arch; LR, lateral rectus muscle; md, mandible; mx, maxillary; np, nasal placode; O, otic vesicle; Tr, trachea. Original magnification, ×63 (E–J).
Table 3 .
Eya1 functionally interacts with Tbx1 in vivo
Since Tbx1 expression in the pharyngeal epithelia appeared to be unaffected in Six1–/–Eya1–/– mutants (Supplemental Figures 7 and 8), we next examined the possibility that Six1 and Eya1 function downstream of Tbx1. Both Six1 and Eya1 were downregulated in the presumptive caudal arch tissue (PA3–PA6 region) of Tbx1 mutants (Figure 6, F and I). It is not clear whether the observed reduction of Six1 and Eya1 expression in Tbx1 mutants is attributable to arch hypoplasia and failed segmentation (i.e., fewer cells to express the genes), or rather reflects a direct effect of Tbx1 loss of function on expression of these genes (i.e., less expression in affected cells). We reasoned that if Six1 and Eya1 are genetically downstream of Tbx1, increased levels of Tbx1 should result in their upregulation. We therefore examined transgenic mice that overexpress human TBX1 using a BAC (20). As expected, 54% of the transgenic mice had vascular defects involving the aortic arch and/or great vessels in the face of relatively normal morphology of the pharyngeal apparatus (20, 58, 59). We noted enhanced expression of both Six1 and Eya1 in the PA3–PA6 region (Figure 6, G and J). Six1, but not Eya1, was also upregulated in pharyngeal mesoderm (Figure 6G, arrows), including primordium of the lateral rectus eye muscle and the first branchial arch core mesoderm. Eya1 expression in the pharyngeal epithelia, particularly in the pharyngeal clefts, was increased in TBX1Bac/+ transgenic mice (Figure 6J, arrowheads). Together, these loss-of-function and gain-of-function analyses revealed a functional relationship in which Tbx1 is genetically upstream of Six1 and Eya1 during cardiocraniofacial morphogenesis. It remains to be determined whether Tbx1 directly regulates Six1 and Eya1 gene expression in vivo.
Discussion
Here, we uncovered what we believe to be a novel function for the Six1/Eya1 transcription complex in cardiovascular development and delineated a Tbx1-Six1/Eya1-Fgf8 regulatory cascade that controls morphogenesis of mammalian heart and face. Our findings suggest that dysregulation of this molecular pathway is likely to be a critical mechanism underlying the pathogenesis of del22q11-like syndromes in humans.
The present study provides evidence that Six1 is transiently expressed in cardiac progenitors in the SHF and proepicardium and contributes directly to cardiovascular development. The restricted distribution pattern of Six1 progeny, which is reminiscent of the SHF in chicks (3), suggests that Six1 is expressed in a subpopulation of the SHF lineages marked by Isl1 or Tbx1 (4, 60). It is also possible, however, that the lineage tracing experiments may be limited by inefficient recombination rate as a result of weak enhancer activity of the endogenous Six1 locus in these cells. We showed that Six1 and Eya1 were broadly expressed in the pharyngeal ectoderm, endoderm, and mesoderm (including SHF). Consequently, deletion of both genes resulted in downregulation of Fgf8 in all 3 cell layers, all of which are likely to contribute to the observed morphological defects of cardiocraniofacial structures (21, 24, 25). Future conditional knockout experiments will be needed to define the tissue-specific roles of Six1/Eya1.
Six1–/–Eya1–/– mutants had a single unseptated OFT in addition to other typical vascular defects seen in del22q11 human patients (Figure 1). It has been proposed that the lesion diagnosed as PTA in some cases of human del22q11 and animal models of this syndrome may in fact be a severe form of pulmonary atresia in which the entire RV outflow fails to form, resulting in a single outflow vessel that is the aorta (43, 61). However, at this point, the morphological and molecular criteria for this important distinction remain to be established. Based on the variable and abnormal outflow valve morphology and the size and location of the outflow vessel in Six1–/–Eya1–/– mutants, we think the defect is likely PTA, but recognize the possibility that the spectrum of defects in these mutants may include pulmonary atresia.
To our knowledge, no direct in vivo transcriptional regulator of Fgf8 has previously been described. Loss of Tbx1 causes a significant reduction of both Fgf8 and Fgf10, which may have redundant roles during cardiovascular development (12, 14, 16, 21, 27, 62). Fgf10 in the SHF, but not Fgf8, is a direct downstream target of Tbx1 (1, 14, 16). Hu et al. demonstrated that Tbx1 is required for an orthologous region upstream of the human FGF8 locus to enhance reporter activity in the OFT of transgenic mice; expression of the enhancer in the PAs (including SHF) was not affected by loss of Tbx1 (14). Furthermore, mutation of 3 putative Tbx1 binding sites found in the FGF8 enhancer did not affect its activity in vivo. Thus, it is unclear whether Tbx1 directly activates Fgf8 in vivo. We found that Tbx1 expression was unaltered in the Six1–/–Eya1–/– mutants (Supplemental Figures 6–8), despite the severe morphological caudal arch defects. Both Six1 and Eya1 were reduced in Tbx1–/– loss-of-function mutants and enhanced in the Tbx1Bac/+ gain-of-function transgenics (Figure 6). Tbx1 may therefore be genetic upstream of Six1 and Eya1. Furthermore, we demonstrated that Six1 and Fgf8 were coexpressed in the pharyngeal ectoderm and endoderm and in some cardiac progenitors in the SHF. Fgf8, but not Fgf10, was severely reduced in all these cells in the Six1–/–Eya1–/– mutants (Figure 4 and Supplemental Figure 7). We showed that not only did Six1/Eya1 compound mutants share cardiovascular and craniofacial features with Fgf8 mutants, they synergistically interacted to regulate pharyngeal artery patterning, which strongly suggests that they are in the same genetic pathway. Taken together, our results indicate that the Six1/Eya1 transcription complex directly controls Fgf8 expression through an evolutionarily conserved enhancer element (Figure 4). Future studies will determine whether Tbx1 is a direct regulator of Six1 and Eya1.
Recent studies in Drosophila melanogaster demonstrate that the fly eya1 ortholog is a direct downstream target of tinman. Nkx2.5, the mammalian ortholog of tinman, is required for both PHF and SHF development (63, 64). In Drosophila, eya1 and tinman are initially coexpressed throughout the mesoderm, after which their localization becomes mutually exclusive, with eya restricted to ventral mesoderm and tinman present only dorsally. tinman is also required for Drosophila expression of six4 and differentiation of muscle along the dorsoventral axis (65). In addition, zebrafish six1 and eya1 were reported to function downstream of tbx1 during craniofacial myogenesis (66). Thus, an evolutionarily conserved network consisting of Tbx1, Nkx2.5, Six1, and Eya1 may be critical for specification and differentiation of cranial skeletal and cardiac muscle.
Haploinsufficiency of SIX1 and EYA1 in humans causes BOR syndrome (35, 36), which often exhibits kidney and ear anomalies but does not routinely feature cardiovascular defects. It is important to note that frequent and severe cardiovascular defects were found only in Six1–/–Eya1–/– mutant mice (Table 1). Renal defects are seen in nearly one-third of del22q11 patients (9, 10, 41, 42), but Tbx1-null mutants do not exhibit any apparent renal defects (18–20). Thus, TBX1 haploinsufficiency is unlikely to be the genetic basis for the renal defects in affected human del22q11 patients. In contrast, Six1 and Eya1 were critical regulators of kidney development (Supplemental Table 1 and refs. 35, 52). We hypothesize that mutations of human SIX1 and EYA1 likely contribute to the pathogenesis of del22q11 syndrome and that the pathogenesis of human BOR and del22q11-like syndromes may be linked.
Methods
Knockout mice and histological analyses.
Six1Cre/hpAP was generated using the same targeting strategy as previously described (35). Six1/Eya1 compound mutants were generated by crossing Six1+/– and Eya1+/– mice (36), and all mutants were confirmed by PCR. Fgf8eGFP, Fgf8f/f, R26RlacZ, and R26RmTmGCre reporter mice were reported previously (20, 25, 32, 67). All mice were maintained on C57BL6, 129svj, or CD-1 mixed genetic backgrounds. Heads, hearts, and aortic arch arteries were dissected, embedded in OCT, sectioned, and stained with H&E using standard procedures. All animal studies were performed according to protocols reviewed and approved by the Institutional Animal Care and Use Committee at the Children’s Hospital Boston.
Gene expression analyses.
Whole-mount RNA in situ hybridization with digoxigenin-UTP–labeled riboprobes and specific gene probes was performed as previously described (35, 67–69). For quantitative PCR analyses, relative gene expression levels were normalized to a GAPDH internal control from the E9.5 pharynx and heart, analyzed using the SYBR Green method on an ABI-7500 detector (Applied BioSystems).
Immunohistochemistry and TUNEL assay.
Staged embryos were dissected, fixed in 4% paraformaldehyde for 1–2 hours at 4°C, and then sectioned at 8–10 μm. Anti-PECAM1 (BD Biosciences), cardiac Troponin T (Thermo scientific), WT1, NKX2-5, GATA4 (Santa Cruz Biotech), ISLET1 (DSHB), GFP (Rockland), Calponin, SM22α (Abcam), and cardiac α-actin (Sigma-Aldrich) were all incubated at 1:100 in blocking solution, except for anti-pH3 (Upstate), which was incubated at 1:200. TUNEL assay was performed on cryostat sections according to the manufacturer’s instructions (Roche). The number of pH3+ or TUNEL+ cells was averaged from 3 sections per embryo. In total, 3 embryos per genotype were quantified.
Whole-mount and section β-galactosidase and hpAP staining.
As described previously (35, 68), we detected β-galactosidase activity and hpAP activity in whole-mount embryos using X-gal and BM purple substrate, respectively.
In vivo ChIP.
ChIP was performed as described previously (67, 70). PAs and cardiovascular tissues from 40 E9.5 embryos were used for each ChIP experiments. Normal serum (IgG) and nonspecific Fgf8 intragenic region were used as negative controls. Anti-Six1 antibody (Sigma-Aldrich) specific oligos for real-time PCR reactions were as follows: Fgf8 promoter (con2) forward, 5′-CTGTGAGGGGTGGGGACCAGTC-3′; Fgf8 promoter (con2) reverse, 5′-CGAGGTTGATTCCCAGGGGCTC-3′; Fgf8 negative control (exon5) forward, 5′-ACGGCGCTGCAGAACGCCAAG-3′; Fgf8 negative control (exon5) reverse, 5′-CAGGCTCTGCTCGGTGGTGTG-3′. 3 independent ChIPs with triplicate real-time PCR reactions were performed per experiment.
Luciferase reporter assays.
HEK293 cells were transfected with the designated plasmids (100 ng/plasmid/well, 24-well plate) using Lipofectamine 2000 (Invitrogen). Transfected cells were collected and lysed 48 hours after transfection with 50 mM Tris-MES, pH 7.8; 1 mM DTT; and 0.1% Triton X-100. The resulting cell lysates were measured using a Sirius Single Tube Luminometer (Berthold Detection Systems) according to the manufacturer’s instructions. Results represent 3 independent assays, each having 3 replicates.
OFT explant culture.
The OFT explants were cultured as previously described (26, 31, 57). OFTs of E9.5 embryos were microdissected and divided into halves along the longitudinal axis from proximal to distal. For the rescue experiments, 50 ng/ml mouse rFgf8b (R&D Systems) or vehicle solution (0.1% mg/ml BSA in PBS) was added. Cell migration distance from the edge of the explant was measured at 8 different locations every 45° and then averaged. The number of migrating cells was normalized to the explant area. Data represent 3 independent experiments.
Statistics.
Paired 2-tailed Student’s t test was used for data analysis between groups for qRT-PCR, pH3 and TUNEL staining, in vivo ChIP, reporter assay, and OFT explant culture. Data are presented as mean ± SEM (n = 3). Phenotypic data were analyzed with Fisher exact test. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We would like to thank M. Freeman, X. He, Z. He, and S. Wu for commenting on the manuscript; A.P. McMahon, J. Yu, M.G. Rosenfeld, S. Evans, Y. Chang, K.R. Chien, and T. Kume for RNA in situ probes; A. Retik for support; and I. Teng, K. Gehrig, T. Tang, R. Huo, Z.F. Chai, P. Gargollo, X. Shi, and X. Fang for technical assistance. We also thank M. Thompson from the Mouse Gene Manipulation Facility of the Children’s Hospital Intellectual and Developmental Disabilities Research Center (IDDRC) (NIHP30-HD 18655). Y. Sun is an American Urological Association Foundation Research Scholar. X.K. Li is a ChangJiang Scholar. X. Li is supported by March of Dimes Birth Defects Foundation (5-FY05-1228), Hood Foundation, Harvard Stem Cell Institute, and NIH/NIDCR (1R01DE019823).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(4):1585–1595. doi:10.1172/JCI44630.
Bin Zhou’s present address is: Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, and Graduate School of the Chinese Academy of Sciences, Shanghai, China.
References
- 1.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1(3):435–440. doi: 10.1016/S1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 2.Mjaatvedt CH, et al. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238(1):97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 3.Waldo KL, et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128(16):3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 4.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131(16):3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 6.Grifone R, Kelly RG. Heartening news for head muscle development. Trends Genet. 2007;23(8):365–369. doi: 10.1016/j.tig.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Tzahor E. Heart and craniofacial muscle development: a new developmental theme of distinct myogenic fields. Dev Biol. 2009;327(2):273–279. doi: 10.1016/j.ydbio.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137(19):3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- 9.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2(11):858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 10.Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370(9596):1443–1452. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- 11.Paylor R, Lindsay E. Mouse models of 22q11 deletion syndrome. Biol Psychiatry. 2006;59(12):1172–1179. doi: 10.1016/j.biopsych.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal VS, Morrow BE. Genetic modifiers of the physical malformations in velo-cardio-facial syndrome/DiGeorge syndrome. Dev Disabil Res Rev. 2008;14(1):19–25. doi: 10.1002/ddrr.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105(9):842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131(21):5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Huynh T, Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133(18):3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131(13):3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 17.Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13(22):2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- 18.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27(3):286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 20.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104(4):619–629. doi: 10.1016/S0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 21.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104(8):933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- 22.Park EJ, et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133(12):2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilagan R, et al. Fgf8 is required for anterior heart field development. Development. 2006;133(12):2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 24.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13(23):3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130(25):6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park EJ, et al. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135(21):3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002;129(19):4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 28.Vitelli F, Zhang Z, Huynh T, Sobotka A, Mupo A, Baldini A. Fgf8 expression in the Tbx1 domain causes skeletal abnormalities and modifies the aortic arch but not the outflow tract phenotype of Tbx1 mutants. Dev Biol. 2006;295(2):559–570. doi: 10.1016/j.ydbio.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutson MR, et al. Cardiac arterial pole alignment is sensitive to FGF8 signaling in the pharynx. Dev Biol. 2006;295(2):486–497. doi: 10.1016/j.ydbio.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 30.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9(1):49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 31.High FA, et al. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest. 2009;119(7):1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank DU, et al. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129(19):4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitelli F, Lania G, Huynh T, Baldini A. Partial rescue of the Tbx1 mutant heart phenotype by Fgf8: genetic evidence of impaired tissue response to Fgf8. J Mol Cell Cardiol. 2010;49(5):836–840. doi: 10.1016/j.yjmcc.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lania G, et al. Early thyroid development requires a Tbx1-Fgf8 pathway. Dev Biol. 2009;328(1):109–117. doi: 10.1016/j.ydbio.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426(6964):247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 36.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23(1):113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 37.Ruf RG, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101(21):8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimasaki N, Watanabe K, Hara M, Kosaki K. EYA1 mutation in a newborn female presenting with cardiofacial syndrome. Pediatr Cardiol. 2004;25(4):411–413. doi: 10.1007/s00246-003-0271-3. [DOI] [PubMed] [Google Scholar]
- 39.Taylor MH, Wilton NC. Bradycardia with sevoflurane in siblings with Branchio-oto-renal syndrome. Paediatr Anaesth. 2007;17(1):80–83. doi: 10.1111/j.1460-9592.2006.02024.x. [DOI] [PubMed] [Google Scholar]
- 40.Schonberger J, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37(4):418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 41.Derbent M, Yilmaz Z, Baltaci V, Saygili A, Varan B, Tokel K. Chromosome 22q11.2 deletion and phenotypic features in 30 patients with conotruncal heart defects. Am J Med Genet A. 2003;116A(2):129–135. doi: 10.1002/ajmg.a.10832. [DOI] [PubMed] [Google Scholar]
- 42.Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion 22q11.2. J Pediatr. 2005;147(1):90–96. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Theveniau-Ruissy M, et al. The del22q11.2 candidate gene Tbx1 controls regional outflow tract identity and coronary artery patterning. Circ Res. 2008;103(2):142–148. doi: 10.1161/CIRCRESAHA.108.172189. [DOI] [PubMed] [Google Scholar]
- 44.Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol. 2005;284(1):72–83. doi: 10.1016/j.ydbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006;293(2):499–512. doi: 10.1016/j.ydbio.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124(1):219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- 47.Oliver G, et al. Homeobox genes and connective tissue patterning. Development. 1995;121(3):693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 48.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 49.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454(7200):109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454(7200):104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458(7240):E8–E9. doi: 10.1038/nature07916. [DOI] [PubMed] [Google Scholar]
- 52.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130(14):3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutson MR, Kirby ML. Neural crest and cardiovascular development: a 20-year perspective. Birth Defects Res C Embryo Today. 2003;69(1):2–13. doi: 10.1002/bdrc.10002. [DOI] [PubMed] [Google Scholar]
- 54.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18(1):101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16(6):704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129(19):4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 57.Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8(8):850–855. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17(1):150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- 59.Liao J, et al. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13(15):1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 60.Huynh T, Chen L, Terrell P, Baldini A. A fate map of Tbx1 expressing cells reveals heterogeneity in the second cardiac field. Genesis. 2007;45(7):470–475. doi: 10.1002/dvg.20317. [DOI] [PubMed] [Google Scholar]
- 61.Kirby ML. Pulmonary atresia or persistent truncus arteriosus: is it important to make the distinction and how do we do it? Circ Res. 2008;103(4):337–339. doi: 10.1161/CIRCRESAHA.108.174862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe Y, Miyagawa-Tomita S, Vincent SD, Kelly RG, Moon AM, Buckingham ME. Role of mesodermal FGF8 and FGF10 overlaps in the development of the arterial pole of the heart and pharyngeal arch arteries. Circ Res. 2010;106(3):495–503. doi: 10.1161/CIRCRESAHA.109.201665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu YH, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EE. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell. 2009;16(2):280–291. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Prall OW, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark IB, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev Biol. 2006;294(1):220–231. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 66.Lin CY, Chen WT, Lee HC, Yang PH, Yang HJ, Tsai HJ. The transcription factor Six1a plays an essential role in the craniofacial myogenesis of zebrafish. Dev Biol. 2009;331(2):152–166. doi: 10.1016/j.ydbio.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 67.Olson LE, et al. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125(3):593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 68.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297(5584):1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 69.Zhou W, et al. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat Genet. 2007;39(10):1225–1234. doi: 10.1038/ng2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc Natl Acad Sci U S A. 2007;104(27):11310–11315. doi: 10.1073/pnas.0611299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.