Abstract
Systemic iron homeostasis is mainly controlled by the liver through synthesis of the peptide hormone hepcidin (encoded by Hamp), the key regulator of duodenal iron absorption and macrophage iron release. Here we show that the liver-specific microRNA miR-122 is important for regulating Hamp mRNA expression and tissue iron levels. Efficient and specific depletion of miR-122 by injection of a locked-nucleic-acid–modified (LNA-modified) anti-miR into WT mice caused systemic iron deficiency, characterized by reduced plasma and liver iron levels, mildly impaired hematopoiesis, and increased extramedullary erythropoiesis in the spleen. Moreover, miR-122 inhibition increased the amount of mRNA transcribed by genes that control systemic iron levels, such as hemochromatosis (Hfe), hemojuvelin (Hjv), bone morphogenetic protein receptor type 1A (Bmpr1a), and Hamp. Importantly, miR-122 directly targeted the 3′ untranslated region of 2 mRNAs that encode activators of hepcidin expression, Hfe and Hjv. These data help to explain the increased Hamp mRNA levels and subsequent iron deficiency in mice with reduced miR-122 levels and establish a direct mechanistic link between miR-122 and the regulation of systemic iron metabolism.
Introduction
The liver is the major body iron storage site and the endocrine organ responsible for the regulation of systemic iron homeostasis. The homeostatic system controls plasma iron availability in order to supply iron to cells and tissues and to prevent toxic iron excess. It reacts to the demand of the erythron, which requires most of the systemically available iron for erythroid heme synthesis and transmits signals to duodenal enterocytes (which absorb iron from the diet), macrophages (which release iron recycled from senescent erythrocytes), and hepatocytes (the major iron reserve; ref. 1). The liver orchestrates iron fluxes by synthesizing the small peptide hormone hepcidin (encoded by Hamp; also known as LEAP1; refs. 2, 3), which controls the amount of iron available in the circulation by interacting with the iron exporter ferroportin. Ferroportin is expressed on the surface of duodenal enterocytes, macrophages, and hepatocytes and upon hepcidin binding is internalized and degraded (4). Disturbance of the hepcidin/ferroportin regulatory system unbalances systemic iron levels and causes important disorders, such as iron deficiency anemia (e.g., iron refractory iron deficiency anemia; ref. 5) or iron overload disorders (e.g., the frequent genetic disorder hereditary hemochromatosis [HH]; ref. 6), or contributes to the iron overload of iron-loading anemia, such as the thalassemias (7). The liver senses systemic iron availability through genes mutated in HH — hemochromatosis (Hfe; refs. 8, 9), hemojuvelin (Hjv; refs. 10–12), and transferrin receptor 2 (Tfr2; refs. 13, 14) — and through the bone morphogenetic protein 6 (15, 16) and Smad4 proteins (17, 18), which serve to regulate hepcidin transcription. In addition, inflammatory cytokines secreted from macrophages stimulate hepcidin synthesis, resulting in iron retention in macrophages (e.g., hepatic Kupffer cells) and reduced intestinal iron absorption. Persistent induction of hepcidin by macrophage-secreted inflammatory cytokines results in the anemia of inflammation, a common disorder of patients with chronic inflammatory conditions (19).
MicroRNAs (miRNAs) are an abundant class of short, noncoding RNAs that regulate gene expression at the posttranscriptional level. miRNA activity has been associated with the control of a wide range of processes, such as development, differentiation, and metabolism (20). Mature miRNAs interact with the RNA-induced silencing complex to base pair with partially homologous sequences in the 3′–untranslated region (3′-UTR) of target mRNAs. Mechanistically, miRNAs prevent mRNA translation (21) and/or mediate target mRNA degradation (22). Computational analyses suggest that miRNAs may regulate up to 30% of the human protein-coding genes (23); however, thus far only a small number of target genes have been experimentally confirmed (TarBase database of validated miRNA targets; ref. 24).
miR-122 is an abundant, liver-specific miRNA (25, 26) whose expression is decreased in advanced liver diseases, such as cirrhosis (27) and hepatocellular carcinoma (28, 29). Several independent studies demonstrate that in vivo inhibition of miR-122 reduces systemic cholesterol levels by as-yet unidentified molecular mechanisms (30, 31). We noticed that these publications report increased mRNA levels of the HH-associated proteins Hfe and Hjv in response to miR-122 inhibition (31, 32). Moreover, miR-122 expression is critical for HCV infection, replication, and response to IFN therapy (33–35). Taken together, these data indicate that miR-122 plays a fundamental role in regulating hepatic functions.
In this study, we addressed the question of whether miR-122 is involved in maintaining systemic iron homeostasis. We showed that expression of miR-122 was decreased in a mouse model of HH and in liver biopsies of HH patients. Efficient inhibition of miR-122 in WT mice caused systemic iron deficiency, hallmarked by reduced plasma and liver iron levels, impaired hematopoiesis, and increased extramedullary erythropoiesis. Moreover, miR-122 depletion directly increased mRNA expression of activators of hepcidin transcription, such as Hfe, Hjv, and bone morphogenetic protein receptor type 1A (Bmpr1a), in liver tissue and murine primary hepatocytes. Elevated hepcidin levels help to explain the systemic iron deficiency in mice with reduced miR-122 levels.
Results
The liver-specific miR-122 is downregulated in Hfe–/– mice.
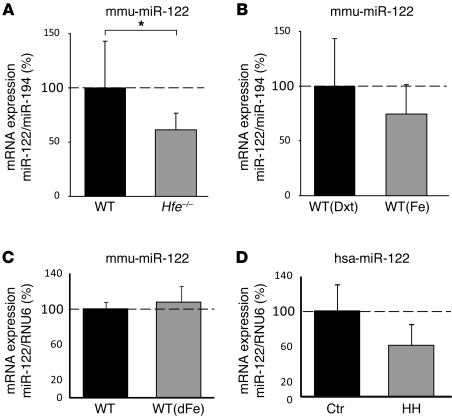
To investigate whether miRNA expression is altered in response to hepatic iron overload, we analyzed total liver RNA from mice with primary iron overload as a consequence of Hfe deficiency (36) or WT mice with iron overload caused by iron-dextran injection (8). miRNA expression profiling was performed using the miChip microarray platform (37–39). We found that miR-122, a miRNA abundantly and selectively expressed in the liver, was downregulated in Hfe–/– mice (data not shown). Subsequent real-time quantitative PCR (qPCR) analysis revealed significantly reduced hepatic miR-122 expression in untreated Hfe–/– mice compared with WT controls (1.63-fold; P = 0.039; Figure 1A), but not in WT mice injected with iron-dextran (P = 0.187; Figure 1B) or subjected to dietary iron overload (P = 0.471; Figure 1C). Similar to the data obtained in Hfe–/– mice, we observed reduced miR-122 levels in liver biopsies from HH patients with homozygous C282Y mutations compared with control subjects without Hfe mutations or iron overload (1.68-fold; P = 0.068; Figure 1D). However, statistical significance was not reached. Interpretation of the reduced miR-122 levels in HH patients is hampered by the fact that in addition to Hfe deficiency and iron overload, HH patients experience varying degrees of hepatic fibrosis and steatosis, as well as therapeutic interventions that may affect miR-122 expression (refs. 35, 40, and Supplemental Table 5; supplemental material available online with this article; doi: 10.1172/JCI44883DS1). In contrast, Hfe–/– mice do not demonstrate hepatic fibrosis or fat accumulation (41), which suggests that the lack of Hfe specifically causes decreased miR-122 levels. These data, together with previous observations that Hfe and Hjv mRNA expression increased in the livers of miR-122–depleted mice (31, 32), led us to hypothesized that miR-122 could be involved in maintaining iron homeostasis.
Figure 1. miR-122 levels are decreased in Hfe–/– mice and patients with HH.
(A) qPCR analysis of miR-122 expression in liver total RNA of WT (n = 8) and Hfe–/– (n = 11) mice (P = 0.039). mmu-miR-122, Mus musculus miR-122 stem-loop. (B) Analysis of miR-122 expression in the liver of WT mice injected with dextran (Dxt; n = 5) and iron-dextran (Fe; n = 6) (P = 0.187) and (C) WT mice on a regular (n = 4) or iron-supplemented diet (dFe; n = 4) (P = 0.471). (D) miR-122 levels were reduced in liver biopsies of HH patients (n = 6) compared with the control group without iron overload (Ctr; n = 4) (P = 0.068). hsa-miR-122, Homo sapiens miR-122 stem-loop. Data were normalized to the appropriate reference genes: miR-194 (A and B), mouse RNU6 (C), and human RNU6 (D). Data are mean ± SD, and values from WT mouse (A–C) and control patient (D) groups were set to 100%. *P < 0.05, 2-tailed Student’s t test.
Efficient and specific antagonism of miR-122 in murine liver.
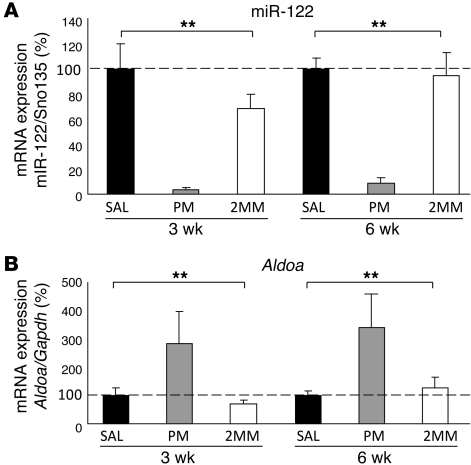
To functionally investigate a possible link between miR-122 and iron metabolism, we inhibited miR-122 by a single i.p. injection of locked nucleic acid–modified (LNA-modified) anti-miR oligonucleotides (31) into age- and sex-matched C57BL/6 WT mice. To inhibit miR-122 specifically, we injected an anti-miR compound with perfect complementarity to miR-122 (perfect match; referred to herein as PM_anti–miR-122). As negative controls, mice were injected either with an LNA control compound with 2 mismatches (referred to herein as 2MM_antimiR-122) or saline vehicle control (0.9% NaCl). Mice were sacrificed 3 and 6 weeks after injection. Independent of treatment, mice were viable and exhibited no overt physical or behavioral abnormalities. To assess the efficiency of miR-122 inhibition, hepatic miR-122 levels were measured by qPCR (Figure 2A). The amount of detectable miR-122 was reduced compared with saline-injected mice by 28- and 11-fold at 3 and 6 weeks, respectively, after injection with PM_anti–miR-122. Injection of the 2MM_anti–miR-122 control did not significantly reduce miR-122 detectability. Expression of the miR-122 primary transcript was not altered under the experimental conditions (Supplemental Figure 1A). To exclude that PM_anti–miR-122 administration disturbs the expression of other miRNAs we analyzed miRNA expression profiles in the livers, hearts and spleens of the same mice (Supplemental Figure 2). Our data show specific and exclusive inhibition of miR-122 in the liver of PM_anti–miR-122 treated mice. In the spleen, expression of 3 miRNAs was increased in PM_anti–miR-122–treated mice, which may be a consequence of increased extramedullary hematopoiesis (see below). Alteration of the miRNA profile was not detected in the heart, where miR-122 is not expressed (26).
Figure 2. miR-122 depletion is functional.
(A) miR-122 detectability was decreased in the liver of PM_anti–miR-122–injected mice. Mice were injected i.p. with a single dose of 25 μg/g PM_anti–miR-122 (PM), 2MM_anti–miR-122 (2MM), or saline (SAL) and sacrificed 3 or 6 weeks after injection. Hepatic miR-122 expression was analyzed by qPCR, and data were normalized to the expression of small nucleolar RNA Sno135. Detectability of miR-122 was reduced 3 (P = 0.0001) and 6 (P = 0.0001) weeks after PM_anti–miR-122 administration. (B) Aldoa was increased in the liver of PM_anti–miR-122–injected mice 3 (P = 0.0001) and 6 (P = 0.0001) weeks after treatment. Values were normalized to mRNA expression of reference gene Gapdh. Data are mean ± SD (n = 8), and the saline-treated group was set to 100%. **P < 0.01, 1-way ANOVA.
Furthermore, miR-122 inhibition by PM_anti–miR-122 increased hepatic mRNA levels of the miR-122 target gene aldolase A (Aldoa; ref. 31) at both 3 weeks (2.8-fold vs. saline; 4.0-fold vs. 2MM_anti–miR-122) and 6 weeks after treatment (3.4-fold vs. saline; 2.7-fold vs. 2MM_anti–miR-122; Figure 2B). Likewise, serum cholesterol levels were decreased (30, 31) 3 weeks after injection of PM_anti–miR-122 (Supplemental Figure 1B). Taken together, these data suggest that PM_anti–miR-122 injection specifically and efficiently antagonizes miR-122 and that miR-122 inhibition is functional.
PM_anti–miR-122–treated mice show altered hematological parameters and reduced plasma and liver iron levels.
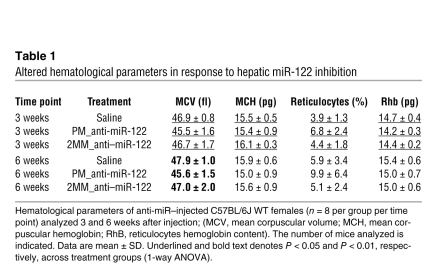
To assess potential consequences of miR-122 inhibition on systemic iron metabolism, we analyzed hematological parameters in PM_anti–miR-122–injected mice (Table 1 and Supplemental Table 1). The mean corpuscular volume (MCV) of erythrocytes was significantly decreased 3 and 6 weeks after PM_anti–miR-122 administration compared with 2MM_anti–miR-122– or saline-injected mice. In addition, reticulocyte counts were significantly increased at the 3-week time point. Reticulocyte hemoglobin (Rhb) content, a useful diagnostic parameter for iron deficiency (42), was significantly reduced in PM_anti–miR-122–treated mice 3 weeks after injection (Table 1). Whereas elevated reticulocyte numbers suggest ongoing hemolysis in humans, reticulocytosis in mice arises as a consequence of dietary iron deficiency (43). Taken together, the hematological analyses of PM_anti–miR-122–treated mice showed that miR-122 inhibition hinders hematopoiesis, possibly through inducing mild iron deficiency.
Table 1 .
Altered hematological parameters in response to hepatic miR-122 inhibition
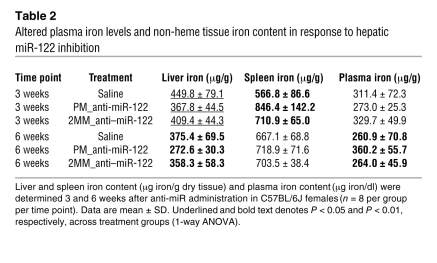
We next analyzed the iron content of the liver (site of iron storage), spleen (site of iron recycling), and plasma (site of iron transport) in miR-122–depleted mice (Table 2). Interestingly, the non-heme iron content was markedly decreased in the liver of PM_anti–miR-122–treated mice both 3 and 6 weeks after injection (3 weeks, 18% vs. saline and 10% vs. 2MM_anti–miR-122; 6 weeks, 27% vs. saline and 24% vs. 2MM_anti–miR-122). In contrast, a small but significant increase in iron content was observed in the spleen of PM_anti–miR-122–treated mice at the 3-week time point compared with both controls (49% vs. saline and 19% vs. 2MM_anti–miR-122); this was normalized 6 weeks after injection. Consistent with iron-deficient hematopoiesis, we observed a decrease in plasma iron levels (approximately 12.5% vs. saline and 17% vs. 2MM_anti–miR-122) and transferrin iron-binding capacity (Supplemental Table 1) at the 3-week time point. Interestingly, plasma iron levels started to increase in PM_anti–miR-122–treated mice 6 weeks after treatment (Table 2), which suggests that the slight recovery in miR-122 activity at the 6-week time point may be sufficient to trigger this effect. Alteration of both hematological parameters and tissue iron levels that occur in response to in vivo miR-122 depletion are supported by data from independent experiments that were conducted under modified experimental conditions or by using WT mice on a different genetic background (Supplemental Table 2). Taken together, these results indicate that in vivo inhibition of miR-122 decreases systemic iron and that an inadequate iron supply to the erythron mildly impairs hematopoiesis.
Table 2 .
Altered plasma iron levels and non-heme tissue iron content in response to hepatic miR-122 inhibition
miR-122 inhibition increases mRNA expression of Hfe, Hjv, Hamp, and Bmpr1a.
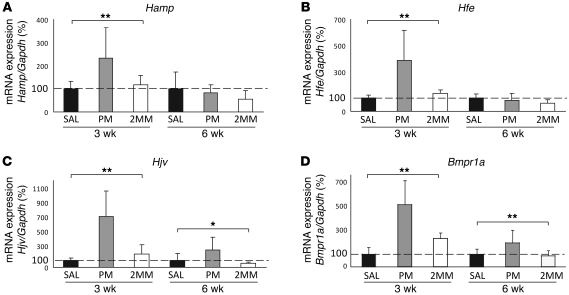
Systemic iron levels are sensed by genes mutated in hereditary hemochromatosis (i.e., Hfe, Hjv, and Tfr2), and the signal is transmitted, at least in part, via the Bmp/Smad signaling pathway to regulate transcription of hepcidin (44–47). To investigate whether mRNA expression of genes in this pathway is affected by in vivo miR-122 inhibition, we analyzed total RNA from the liver of PM_anti–miR-122–treated mice. mRNA expression of Hamp was significantly increased in PM_anti–miR-122–treated mice 3 weeks after injection compared with 2MM_anti–miR-122– and saline-injected controls (1.96- and 2.3-fold, respectively; Figure 3A). In addition, mRNA expression of 3 positive hepcidin regulators, Hfe (3.9-fold vs. saline; 2.9-fold vs. 2MM_anti–miR-122), Hjv (7.1-fold vs. saline; 3.8-fold vs. 2MM_anti–miR-122), and Bmpr1a (4.9-fold versus saline; 2.8-fold vs. 2MM_anti–miR-122), was significantly increased in PM_anti–miR-122–injected mice. In contrast, we did not detect any PM_anti–miR-122–specific change in the mRNA expression of Tfr2 or other hepcidin effectors (e.g., Smad7 and Smad4; Supplemental Figure 3). Consistent with the partial relief of miR-122 inhibition (Figure 2) and increased plasma iron levels (Table 2) at the 6-week time point, Hamp and Hfe mRNA expression returned to normal, and the changes in mRNA levels of Hjv (2.4-fold vs. saline and 4.1-fold vs. 2MM_anti–miR-122) and Bmpr1a (1.9-fold vs. saline and 2.2-fold vs. 2MM_anti–miR-122) diminished at 6 compared with 3 weeks.
Figure 3. Liver mRNA expression of Hamp, Hfe, Hjv, and Bmpr1a is increased in PM_anti–miR-122–treated mice.
Analysis of hepatic (A) Hamp (3 weeks, P = 0.0005; 6 weeks, P = 0.27), (B) Hfe (3 weeks, P = 0.0005; 6 weeks, P = 0.158), (C) Hjv (3 weeks, P = 0.0001; 6 weeks, P = 0.0225), and (D) Bmpr1a (3 weeks, P = 0.0001; 6 weeks, P = 0.011) mRNA expression by qPCR 3 and 6 weeks after anti-miR administration. Values were normalized to mRNA expression of reference gene Gapdh. Data are mean ± SD (n = 8), and the saline-treated group was set to 100%. *P < 0.05, **P < 0.01, 1-way ANOVA.
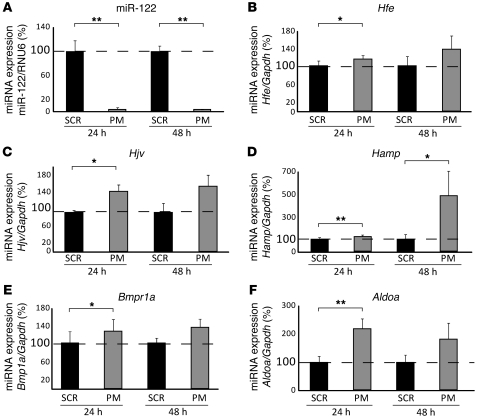
To investigate whether increased mRNA expression of Hfe, Hjv, Hamp, and Bmpr1a in the liver of PM_anti–miR-122–treated mice is a consequence of hepatocyte-specific miR-122 depletion, we transfected murine primary hepatocytes with either PM_anti–miR-122 or scrambled control oligos. Consistent with the observations in PM_anti–miR-122–treated mice, we found significantly increased mRNA expression of Hfe (24 hours, P = 0.026; 48 hours, P = 0.117), Hjv (24 hours, P = 0.014; 48 hours, P = 0.11), Hamp (24 hours, P = 0.005; 48 hours, P = 0.044), Bmpr1a (24 hours, P = 0.034; 48 hours, P = 0.175), and Aldoa (24 hours, P = 0.003; 48 hours, P = 0.119) in primary hepatocytes (Figure 4).
Figure 4. miR-122 inhibition in murine primary hepatocytes.
Primary hepatocytes were transfected with PM_anti–miR-122 or scrambled control oligos (SCR). (A) miR-122 expression was analyzed 24 (P = 0.01) and 48 (P = 0.01) hours after transfection. (B–F) Expression of (B) Hfe (24 hours, P = 0.026; 48 hours, P = 0.117), (C) Hjv (24 hours, P = 0.014; 48 hours, P = 0.11), (D) Hamp (24 hours, P = 0.005; 48 hours, P = 0.044), (E) Bmpr1a (24 hours, P = 0.034; 48 hours, P = 0.175), and (F) Aldoa (24 hours, P = 0.003; 48 hours, P = 0.119) mRNA 24 and 48 hours after transfection of PM_anti–miR-122 or scrambled control oligos. Data are mean ± SD (n = 4), and the value for scrambled control–transfected cells was set to 100%. Data were normalized to mRNA expression of the appropriate reference gene. *P < 0.05, **P < 0.01, 2-tailed Student’s t test.
Anti–miR-122 administration does not induce an IL-6–mediated inflammatory response.
In principle, LNA oligonucleotide–mediated activation of proinflammatory cytokines, such as IL-6, could stimulate Hamp mRNA expression via the Jak/Stat signaling pathway (19). Therefore, we analyzed mRNA expression of the inflammatory marker Il1b as well as Il6 and its downstream effector C-reactive protein (Crp) in PM_anti–miR-122–injected mice (Supplemental Figure 4A). Consistent with the lack of increased Il6 mRNA expression in PM_anti–miR-122–injected mice, we did not observe increased phospho-Stat3 levels in liver protein extracts (Supplemental Figure 4B), which suggests that the Jak/Stat signaling pathway is not activated in these mice. Moreover, phosphorylation of Smad1/5/8 and mRNA expression of DNA binding protein 1 (Id1), a downstream target gene of the Bmp/Smad signaling pathway (48), remained unchanged in the liver of PM_anti–miR-122–injected mice (Supplemental Figures 3 and 6). Analyses of liver morphology, as well as assessment of T cell and macrophage numbers by immunohistochemistry, did not detect any significant differences between PM_anti–miR-122–treated mice and respective controls (Supplemental Figure 4C). Taken together, these data show that hepcidin stimulation in PM_anti–miR-122–injected mice is unlikely the result of an inflammatory response mediated by IL-6 or IL-1β, which indicates that miR-122 directly or indirectly controls expression of one or more of the activators of Hamp levels (i.e., Hfe, Hjv, or Bmpr1a) or of Hamp itself.
Identification of predicted miR-122 binding sites within the 3′-UTRs of Hfe, Hjv, and Hamp mRNAs.
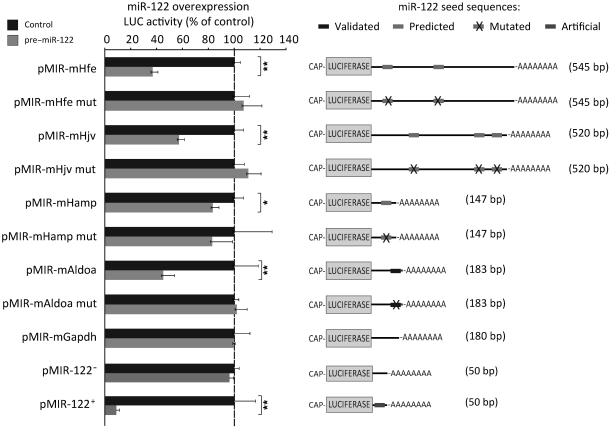
To investigate whether murine Hfe, Hjv, Bmpr1a, or Hamp are targets for miR-122, we queried several miRNA target prediction databases: miRanda (49), PicTar (50), miRTar ( http://mirtar.mbc.nctu.edu.tw/human/), TargetScanS (51), and TarBase (24). Applying this strategy, we identified 3 putative miR-122 binding sites in the 3′-UTR of Hjv and a single putative miR-122 target sequence in the 3′-UTR of Hamp mRNA (Supplemental Table 3). Application of the RNA22 algorithm (52) predicted 2 miR-122 binding sites in the 3′-UTR of the Hfe gene (Supplemental Table 3). No miR-122 seed sequences were identified within the Bmpr1a 3′-UTR; therefore, Bmpr1a was not further analyzed. We generated luciferase reporter genes bearing the full-length 3′-UTRs of mouse Hamp, Hfe, and Hjv (referred to herein as pMIR-mHamp, pMIR-mHfe, and pMIR-mHjv, respectively). As a positive control, we used the 3′-UTR of the mouse Aldoa gene (pMIR-mAldoa), a validated miR-122 target (31). As a negative control, the entire 3′-UTR of mouse Gapdh (pMIR-mGapdh) was inserted. To assess the specificity of putative miR-122 regulation, we also generated constructs where the seed sequences of the miR-122 binding sites were mutated (Supplemental Table 3; bold sequences). To further monitor expression control by miR-122 mimics (Ambion), we generated artificial positive and negative control vectors containing sequence with perfect complementarity to miR-122 and to the miR-122 sequences directly (pMIR-122+ and pMIR-122–, respectively).
Hfe and Hjv are miR-122 target genes.
To investigate whether the predicted 3′-UTRs of the Hfe, Hjv, and Hamp mRNAs convey miR-122–dependent regulation, we cotransfected miR-122 mimics (referred to herein as pre–miR-122) and the luciferase reporter plasmids into Hepa1-6 mouse hepatocarcinoma cells. As expected, luciferase activity from cells transfected with the positive control vector pMIR-122+ was strongly reduced upon miR-122 overexpression (7- to 10-fold, Figure 5). Conversely, pMIR-122––transfected cells were unaffected by miR-122 overexpression (Figure 5), which suggests that miR-122 overexpression is efficient and specific. Likewise, transfection of the miR-122 mimic significantly reduced luciferase counts from the positive control pMIR-mAldoa (2.23-fold; P = 0.003), whereas luciferase counts from pMIR-mAldoa mut, which contains mutated miR-122 seed sequences (Supplemental Table 3), were unaffected. Importantly, overexpression of the miR-122 mimic significantly reduced expression from pMIR-mHfe (2.72-fold; P = 0.00002) and pMIR-mHjv (1.74-fold; P = 0.0001), but not from constructs in which the predicted miR-122 binding sites are mutated (pMIR-mHfe mut and pMIR-mHjv mut) or from pMIR-mGapdh, which does not contain a predicted miR-122 target site (Figure 5). The suppressive effect of miR-122 on pMIR-mHfe and pMIR-mHjv was maintained for at least 72 hours after transfection (Supplemental Figure 5). We further observed a minor reduction of luciferase counts from pMIR-mHamp upon miR-122 overexpression. However, this effect appears not to be specific, because a similar level of reduction was obtained when the predicted miR-122 target site was mutated. Taken together, these data show that the 3′-UTRs of Hfe and Hjv are direct targets of miR-122.
Figure 5. Validation of Hfe and Hjv as miR-122 target genes.
Hepa1-6 cells were transfected with pMIR-mHfe, pMIR-mHjv, pMIR-mHamp, pMIR-mAldoa, or pMIR-mGapdh (see Supplemental Table 3 for localization of predicted miR-122 binding sites), miR-122 complementary sequence (pMIR-122+), or miR-122 antisense sequence (pMIR-122–), either with (pre–miR-122) or without (control) miR-122 mimics. Luciferase activity was measured 24 hours later. As a specificity control, we transfected luciferase reporter vectors linked to the 3′-UTRs of the genes of interest that contain mutations within the predicted miR-122 seed sequences (mut; see Supplemental Table 3). Experiments were performed at least in triplicate, and results of the luciferase reporter assay are presented as fold change ± SD of the pre–miR-122– and control-transfected cells. *P < 0.05, **P < 0.01, 2-tailed Student’s t test.
miR-122 inhibition induces extramedullary hematopoiesis.
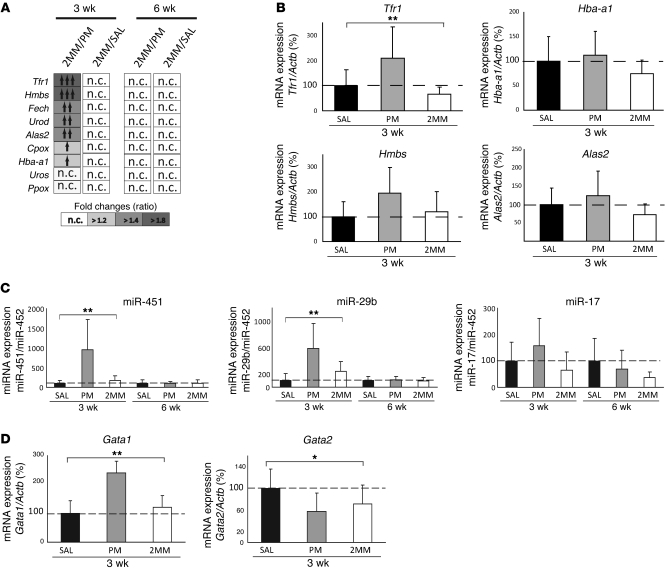
To investigate how hepatic miR-122 inhibition leads to elevated spleen iron levels (Table 2), we analyzed mRNA expression of iron-related genes on a specialized microarray platform (IronChip; mouse version 9). Interestingly, we observed increased mRNA expression of Tfr1 and several enzymes involved in heme biosynthesis — hydroxymethylbilane synthase (Hmbs), ferrochelatase (Fech), uroporphyrinogen decarboxylase (Urod), coproporphyrinogen oxidase (Cpox), and delta-aminolevulinate synthase 2 (Alas2) — as well as the hemoglobin α adult chain 1 (Hba-a1) in the spleen of PM_anti–miR-122–injected mice at 3, but not 6, weeks after treatment (Figure 6A). Increased mRNA expression of Tfr1, Hmbs, Alas2, and Hba-a1 as confirmed by qPCR analysis (Figure 6B). Additionally, miChip analysis of splenic total RNA indicated that 3 miRNAs (miR-451, miR-17, and miR-29b) were selectively increased in the spleen of PM_anti–miR-122–injected mice (Supplemental Figure 8), a result confirmed by miRNA qPCR (Figure 6C). miR-451 was previously linked to murine and human erythroid differentiation (53–55), in that its transcription is activated by the erythroid transcription factor Gata-binding factor–1 (Gata1), which drives erythroid differentiation to a late stage (53–55). miR-451 in turn suppresses Gata2, which is responsible for maintaining the erythroid precursor stage. Consistent with enhanced erythropoiesis in the spleen of PM_anti–miR-122–injected mice, we observed increased miR-451 and Gata1. Gata2 mRNA levels only showed lower expression in PM_anti–miR-122–injected mice compared with the saline control (Figure 6D). Collectively, these data suggest that inhibition of miR-122 expression leads to iron deficiency, which in turn increases extramedullary hematopoiesis.
Figure 6. Extramedullary hematopoiesis in PM_anti–miR-122–injected mice.
(A) Heat map representing genes with increased mRNA expression in the spleen of PM_anti–miR-122 treated mice, as assessed by IronChip. n.c., not changed. (B) Splenic Tfr1 (P = 0.007), Hmbs (P = 0.08), Hba-a1 (P = 0.24), and Alas2 (P = 0.14) mRNA expression, as assessed by qPCR 3 weeks after injection of anti-miRs. Data were normalized to mRNA expression of reference gene Actb. (C) Splenic expression of miR-451 (3 weeks, P = 0.0016; 6 weeks, P = 0.979), miR-29b (3 weeks, P = 0.0019; 6 weeks, P = 0.844), and miR-17 (3 weeks, P = 0.093; 6 weeks, P = 0.19) by miRNA qPCR 3 and 6 weeks after injection of anti-miRs. Data were normalized to expression of miR-452, which remained unchanged in miRNA expression profiles of splenic total RNAs of the same mice. (D) Splenic Gata1 (P = 0.003) and Gata2 (P = 0.041) mRNA expression 3 weeks after injection of anti-miRs. Data are mean ± SD (n = 8), and values from the saline-treated group were set to 100%. *P < 0.05, **P < 0.01; 1-way ANOVA.
Discussion
Hepatocytes sense systemic iron availability and regulate systemic iron fluxes. Plasma iron levels are controlled by 2 major hepatic signaling pathways: the HH-associated proteins (Hfe, Hjv, and Tfr2) and the Bmp/Smad signaling pathway, which control the hepcidin response to systemic iron availability, and proinflammatory cytokines and the Jak/Stat pathway, which mount the inflammatory response of hepcidin (56, 57). Activation of either of these signaling pathways increases hepcidin levels, which leads to decreased duodenal iron absorption and iron release from macrophages. In contrast, TMPRSS6 (58) and the inhibitory Smad protein Smad7 (59) suppress hepcidin levels.
In this study, we showed for the first time to our knowledge that systemic iron homeostasis is also controlled by a miRNA. Hepatic miR-122 expression was critical to prevent iron deficiency in plasma and liver, impairment of hematopoiesis caused by low iron availability, and extramedullary erythropoiesis in the spleen. Thus, decreased miR-122 expression, which was observed in Hfe–/– mice and HH patients (Figure 1, A and D), may be a compensatory response of the liver to limit iron uptake and counteract liver iron overload. However, the biological significance of the decreased miR-122 levels in Hfe–/– mice needs to be established, because hepatic miR-122 reduction in Hfe–/– mice failed to increase mRNA expression of the known miR-122 target gene Aldoa or to affect molecular pathways that reduce cholesterol levels in miR-122–depleted mice (data not shown). Thus, it may simply be a marker rather than a functionally consequential response.
Our findings demonstrated that hepatic miR-122 inhibition by LNA-modified PM_anti–miR-122 was highly specific: miR-122, and no other miRNA, was exclusively inhibited in the liver upon PM_anti–miR-122 injection (Supplemental Figure 2), and 2 mismatches within the LNA-modified anti–miR (2MM_anti–miR-122) were sufficient to prevent hepatic miR-122 inhibition and alterations in systemic iron homeostasis (Figure 2 and Table 1). Interestingly, hepatic miR-122 inhibition in the liver of WT mice and in primary murine hepatocytes caused increased mRNA expression of Hamp and 3 of its transcriptional activators (Hfe, Hjv, and Bmpr1a; Figures 3 and 4). Notably, increased Hfe, Hjv, and Bmpr1a mRNA expression was also reported in 3 previous studies on miR-122 antagonism in mice (31, 32, 60). This suggests that the observed stimulation of Hfe and Hjv mRNA expression is specific for miR-122 depletion and independent of the antisense chemistry applied. Elevated expression of Hfe, Hjv, and Bmpr1a would be expected to stimulate the Bmp/Smad signaling pathway in mice (44). However, increased Smad1/5/8 phosphorylation in protein extracts from PM_anti–miR-122–injected mice was not detected (Supplemental Figure 6). Consistently, mRNA expression of Id1, a target gene of the Bmp/Smad signaling pathway (48), remained unchanged in the liver of PM_anti–miR-122–injected mice (Supplemental Figure 3). High hepcidin levels would be expected to cause iron retention (e.g., in splenic macrophages) and decreased iron absorption by stimulating the internalization and degradation of the iron exporter ferroportin (4). However, ferroportin protein levels in the spleen were not decreased in PM_anti–miR-122–injected mice (Supplemental Figure 7). We speculate that the iron demand for extramedullary erythropoiesis that occurs in the spleen of PM_anti–miR-122–injected mice needs to be satisfied, and thus hepcidin-independent regulatory mechanisms, such as BACH1- or NRF2-mediated transcriptional control of ferroportin (61), may override the hepcidin response of ferroportin.
Extramedullary hematopoiesis in PM_anti–miR-122–injected mice is characterized by increased splenic mRNA expression of Tfr1, enzymes involved in heme biosynthesis (Hmbs, Fech, Urod, Cpox, and Alas2), and Hba-a1 as well as the hematopoietic miR-451. Interestingly, overexpression of Hba-a1 mRNA in anemic mice correlates with rapid recovery from anemia (62), which suggests that similar mechanisms may apply to PM_anti–miR-122–injected mice. Also of note is the elevated expression of miR-451, which was previously reported to be essential for erythropoiesis in mammals and zebrafish and shown to depend on Gata1 (53–55). Elevated miR-451 expression is responsible for decreased expression of Gata2, a transcription factor required to maintain the erythroid precursor stage. Here we show that Gata1 and miR-451 expression increased and Gata2 mRNA levels tended to decrease in the spleen of PM_anti–miR-122–injected mice, which suggests that this regulatory network is maintained during murine extramedullary erythropoiesis in the spleen. It is intriguing to speculate that increased expression of miR-29b and miR-17 (Figure 6C) — which was also observed in miR-122–depleted mice and, to our knowledge, previously not linked to erythropoiesis — may also be relevant for this process. Taken together, our findings suggest that iron deficiency induced by miR-122 limitation triggers erythropoiesis in the spleen to avoid anemia.
We further uncovered a mechanistic link among miR-122 depletion, elevated hepcidin expression, and systemic iron deficiency. We showed that Hfe and Hjv, 2 genes mutated in the frequent iron overload disorder HH, contained functional miR-122 binding sites (Figure 5, Supplemental Figure 5, and Supplemental Table 3). Importantly, mutation of the predicted miR-122 binding sites prevented the suppressive effect of miR-122, which suggests that miR-122–dependent inhibition of Hfe and Hjv is specific.
The fatty acid and iron metabolic networks are interconnected; however, the identification of common regulatory elements has been elusive. Recently, several studies showed that miR-122 is required to maintain serum cholesterol levels in mice and nonhuman primates (30–32, 63, 64). However, the miR-122 target gene responsible for this phenotype has not been discovered. In addition to its role in maintaining iron and cholesterol metabolism, miR-122 is critical for efficient HCV replication (33–35, 65). Inhibition of hepatic miR-122 in chimpanzees chronically infected with HCV led to long-lasting suppression of both HCV replication and viremia (64). miR-122 expression is also reduced in hepatocellular carcinoma (28, 29) and is regulated by the circadian rhythm (60), which suggests that miR-122 activity may be required to synchronize genes within overlapping metabolic pathways. In recognition of the circadian rhythmicity of miR-122 expression, samples from the mice analyzed in this study were always collected at the same time of the day and sacrificed in a random order. Furthermore, the link between miR-122 expression and important human diseases suggests that miR-122 could be a target for therapeutic intervention.
In summary, we propose the following model for the role of miR-122 in iron homeostasis: miR-122 regulates the expression of the HH-associated proteins Hfe, Hjv, and possibly others. Overexpression of Hfe and Hjv activates Hamp mRNA expression (66). Elevated hepcidin levels will limit the iron export capacity from duodenal enterocytes and macrophages and cause plasma and liver iron deficiency. As a consequence, iron-deficient hematopoiesis develops, and extramedullary hematopoiesis is observed in the spleen. Future experiments are needed to clarify the signaling pathways that control the Hamp response to elevated Hfe and Hjv expression in anti–miR-122–treated mice.
Methods
Animal care and animal models.
All mice were housed in the European Molecular Biology Laboratory animal facility under a constant light-dark cycle, maintained on a standard mouse diet, and allowed ad libitum access to food and water. Mice were euthanized by CO2 inhalation. All mouse breeding and animal experiments were approved by and conducted in compliance with the guidelines of the European Molecular Biology Laboratory Institutional Animal Care and Use Committee. A total of 48 female WT mice on the C57BL/6J genetic background were injected at 10–12 weeks of age. 16 mice were injected i.p. with 25 mg/kg PM_anti–miR-122 (31) in isotonic solution (0.9% NaCl), with perfect complementarity to miR-122. As controls, 16 mice were injected i.p. with 400 μl NaCl or with 25 mg/kg 2MM_anti–miR-122 (designed with 2 mismatches toward the miR-122 seed sequence; ref. 31) in isotonic solution. Hfe–/– mice were described previously (36). Induction of iron overload by a dietary regimen (67) or iron-dextran injection (8) has been described previously.
Hematological parameters, serum and tissue iron content, and total cholesterol measurement.
Heparinized blood was collected by cardiac puncture. Hematological parameters were determined using a blood counter instrument. Plasma iron content was assayed using the Iron Kit (Thermo Electron GmbH) in a 96-well format using a serial dilution of iron atomic absorption standard solution (1,000 mg/ml iron in HCl; Sigma-Aldrich). The unsaturated iron binding capacity was measured using the U.I.B.C. kit (Biolabo) according to the manufacturer’s instructions. Tissues isolated from anti-miR–treated mice were analyzed for non-heme iron as previously described (68, 69). Total plasma cholesterol was analyzed using ABX Pentra Cholesterol CP (Horiba Group, Horiba ABX Diagnostics) according to the manufacturer’s instructions. The measurements were carried out in duplicate and were correlated to a 2-fold diluted standard curve generated from an ABX Pentra MultiCal solution (Horiba ABX Diagnostics).
RNA extraction, reverse transcription, and mRNA qPCR.
Tissue was disrupted using a Tissue Lyzer (Qiagen), and total RNA was isolated using TRIzol (Invitrogen). RNA quality and concentration was assessed using a 2100 Bioanalyzer (Agilent Technologies) and Nanodrop ND-1000 (Nanodrop Technologies), respectively. For mRNA analysis by qPCR, 2 μg total RNA was reverse transcribed with random hexamers using Superscript II Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions. qPCR was carried out using 10 ng cDNA in a 20-μl reaction volumes using SYBR Green I (Applied Biosystems) on a Prism 7500 (Applied Biosystems) and gene-specific primers. Relative qPCR expression of individual genes was normalized to the expression of reference genes Gapdh or actin B (Actb) using qBASE (70) and the ΔΔCq method (71). See Supplemental Table 4 for primer sequences.
Extraction of total RNA from formalin-fixed, paraffin-embedded biopsies.
Pseudonymized male human tissue samples were provided by the Tissue Bank of the National Center for Tumor Diseases Heidelberg (project no. 564) after approval by the ethics committee (no. 206/2005, Medical Faculty, Heidelberg, Germany). Because only long-term archived (>5 years), pseudonymized formalin-fixed, paraffin-embedded (FFPE) tissues were used, informed consent was not required for this study. 3 7-μm-thick FFPE sections were dewaxed in 500 μl xylene by incubation at 65°C (3 g) for 3 5-minute periods. The dewaxed sections were washed 3 times in 500 μl 100% ethanol (2 minutes; 15,700 g). After discarding the supernatant, samples were dried for 1 hour at 37°C. Tissue was digested using 10 μl proteinase K in 150 μl PKD buffer (Rneasy Kit FFPE, Qiagen) for 15 minutes at 55°C, followed by proteinase inactivation for 15 minutes at 80°C. Total RNA was extracted using the miRneasy kit (Qiagen).
IronChip, miChip, and miRNA qPCR in anti-miR mice.
For IronChip analyses, 5 μg total RNA was reverse transcribed, amplified, and labeled as described previously (72). Expression of miR-122 in the anti-miR–injected mice was analyzed by miRNA-specific TaqMan assays according to the manufacturer’s instructions (Applied Biosystems).
For miRNA expression profiling by miChip, we pooled equal amounts of total RNA extracted from each mouse within the individual groups and analyzed RNA as previously described (37–39). Array data were deposited in the Gene Expression Omnibus repository (GEO; accession no. GSE24786; ref. 73). Hepatic miR-122 levels in Hfe–/– mice were analyzed by miRCURY-LNA specific miRNA assays according to the manufacturer’s instructions (Exiqon). miRNA measurements in the spleen (miR-451, miR-29b, and miR-17) and liver of anti–miR-122–injected mice, in patient biopsies (miR-122 and RNU6), and in primary hepatocytes (miR-122 and RNU6) were analyzed by miRNA qPCR (see Supplemental Methods). Independent of the method of qPCR analysis, relative miRNA expression levels were calculated using qBase (70) and determined relative to the appropriate reference gene(s).
3′-RACE, 3′-UTR cloning, and site-directed mutagenesis.
cDNA for the 3′–rapid amplification of cDNA ends (3′-RACE) reaction was synthesized as described above, except that reverse transcription was primed using an oligo(dT)-adapter primer (Qiagen). 3′-RACE was carried out in a final volume of 50 μl containing 1 pmol of gene-specific primers (Supplemental Table 3), 20 ng cDNA, 25 mM dNTP mix, and 2.5 U Thermo-Start Taq polymerase (Thermo-Fisher). Amplification was performed with an initial activation step at 95°C for 15 minutes, followed by 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 3 minutes. Primers were designed to amplify the full-length 3′-UTRs of mouse Hamp (bp 1–bp 106), Hjv (bp 1–bp 519), Aldoa (bp 1–bp 183), Gapdh (bp 1–bp 178), and Hfe (bp 1–bp 545). Predicted miR-122 binding sites are shown in Supplemental Table 3.
pMIR is a derivative of the pGL3 vector (Promega) that was engineered to accommodate a multiple cloning site 3′ of the firefly luciferase gene. PCR products were cloned into the SacI/NheI sites and sequenced. To generate a positive (pMIR-122+) and a negative (pMIR-122–) control vector, a double-stranded DNA oligo nucleotide with the identical sequence of miR-122 (see Supplemental Table 3) was inserted in the pMIR-SacI/NheI cloning sites either in the sense (+) or antisense (–) orientation.
Site-directed mutagenesis of the predicted miR-122 seed sequence (see Supplemental Table 3) within the 3′-UTR of the genes of interest was performed using GeneTailor System (Invitrogen) following the manufacturer’s protocol.
Preparation of murine primary hepatocytes, transfections, and luciferase assays.
Murine primary hepatocytes were isolated according to an optimized protocol described previously (ref. 74 and Supplemental Methods) and transiently transfected in 6-well plates using RNAiMAX (Invitrogen) with PM_anti–miR-122 (500 ng; Santaris Pharma) or a scrambled oligo control sequence (500 ng; Santaris Pharma). Total RNA was extracted 24 and 48 hours after transfection using TRIzol (Invitrogen).
For miR-122 overexpression, Hepa1-6 cells were transiently transfected in 24-well plates using Lipofectamine 2000 (Invitrogen) and 5 μM pre–miR-122 (Ambion) as well as 0.05 ng renilla and 10 ng firefly luciferase vectors. Cells were lysed at the indicated time points using 100 μl of 1× passive lysis buffer (Promega). To assay luciferase counts, we used a microplate luminometer (Microlumat plus LB96V; Berthold), 25 μl cell lysate, and 10 μl of both LAR II (Promega) and Stop and Glo (Promega).
Statistics.
Data are expressed as mean ± SD. Results compared between groups were analyzed by 2-tailed Student’s t test when 2 samples were considered or by 1-way ANOVA for 3 or more samples. When groups were compared using 1-way ANOVA, we assumed that data were normally distributed. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Bruno Galy for his excellent advice, Katarzyna Mleczko for sharing her data, Sandra Manthey for helping with the primary hepatocyte preparation, and Steven Dooley and Rolf Gebhardt for sharing their knowledge of RNAi knockdown in primary hepatocytes. We thank the Tissue bank of the National Center for Tumor Diseases Heidelberg for providing the biopsies of hemochromatosis patients. The authors gratefully acknowledge John D. Ryan for critical reading of the manuscript. M. Castoldi is supported by an Excellence Fellowship of The Medical Faculty of the University of Heidelberg. This work was supported by a NIH grant to R.E. Fleming (R01 DK 063016); a Network HepatoSys/Virtual Liver grant (BMBF) to M.U. Muckenthaler, L.A. D’Alessandro, and U. Klingmüller; and a DFG grant to M.U. Muckenthaler and M. Castoldi.
Footnotes
Conflict of interest: J. Elmén, M. Lindow, and S. Kauppinen are employed by Santaris Pharma, a biopharmaceutical company engaged in the development of RNA-based medicines.
Citation for this article: J Clin Invest. 2011;121(4):1386–1396. doi:10.1172/JCI44883.
References
- 1.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142(1):24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Krause A, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480(2–3):147–150. doi: 10.1016/S0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 3.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276(11):7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 4.Nemeth E, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 5.Finberg KE, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40(5):569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietrangelo A. Hereditary hemochromatosis. Biochim Biophys Acta. 2006;1763(7):700–710. doi: 10.1016/j.bbamcr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Origa R, et al. Liver iron concentrations and urinary hepcidin in beta-thalassemia. Haematologica. 2007;92(5):583–588. doi: 10.3324/haematol.10842. [DOI] [PubMed] [Google Scholar]
- 8.Muckenthaler M, et al. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34(1):102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- 9.Bridle KR, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361(9358):669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 10.Papanikolaou G, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 11.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115(8):2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115(8):2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camaschella C, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25(1):14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 14.Fleming RE, et al. Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis. Proc Natl Acad Sci U S A. 2002;99(16):10653–10658. doi: 10.1073/pnas.162360699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andriopoulos B, Jr, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 17.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med. 2009;87(5):471–480. doi: 10.1007/s00109-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang RH, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2(6):399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Nemeth E, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113(9):1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Meister G. miRNAs Get an Early Start on Translational Silencing. Cell. 2007;131(1):25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 26.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padgett KA, et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. . J Autoimmun. 2009;32(3–4):246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutay H, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. . J Cell Biochem. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48(4):648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 34.Jopling CL. Regulation of hepatitis C virus by microRNA-122. Biochem Soc Trans. 2008;36(pt 6):1220–1223. doi: 10.1042/BST0361220. [DOI] [PubMed] [Google Scholar]
- 35.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15(1):31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 36.Vujic Spasic M, et al. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7(2):173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Castoldi M, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). Rna. 2006;12(5):913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castoldi M, Benes V, Hentze MW, Muckenthaler MU. miChip: A microarray platform for expression profiling of microRNAs based on locked nucleic acid (LNA) oligonucleotide capture probes. Methods. 2007;43(2):146–152. doi: 10.1016/j.ymeth.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Castoldi M, Schmidt S, Benes V, Hentze MW, Muckenthaler MU. miChip: an array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat Protoc. 2008;3(2):321–329. doi: 10.1038/nprot.2008.4. [DOI] [PubMed] [Google Scholar]
- 40.Bala M, Sosna J, Appelbaum L, Israeli E, Rivkind AI. Enigma of primary aortoduodenal fistula. World J Gastroenterol. 2009;15(25):3191–3193. doi: 10.3748/wjg.15.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trenor CC 3rd, Campagna DR, Sellers VM, Andrews NC, Fleming MD. The molecular defect in hypotransferrinemic mice. Blood. 2000;96(3):1113–1118. [PubMed] [Google Scholar]
- 42.Brugnara C, Schiller B, Moran J. Reticulocyte hemoglobin equivalent (Ret He) and assessment of iron-deficient states. Clin Lab Haematol. 2006;28(5):303–308. doi: 10.1111/j.1365-2257.2006.00812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campi MG, et al. Molecular Probe Data Base (MPDB). Nucleic Acids Res. 1997;25(1):92–95. doi: 10.1093/nar/25.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babitt JL, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 45.Corradini E, et al. Bone morphogenetic protein signaling is impaired in an Hfe knockout mouse model of hemochromatosis. Gastroenterology. 2009;137(4):1489–1497. doi: 10.1053/j.gastro.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kautz L, et al. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114(12):2515–2520. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- 47.Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatology. 2009;50(6):1992–2000. doi: 10.1002/hep.23198. [DOI] [PubMed] [Google Scholar]
- 48.Kautz L, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112(4):1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 49.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 51.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69(6):2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 52.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 53.Dore LC, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A. 2008;105(9):3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113(8):1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen KD, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207(7):1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109(1):353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 57.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108(9):3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silvestri L, Pagani A, Nai A, De Domenico, I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8(6):502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mleczko-Sanecka K, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115(13):2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 60.Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marro S, et al. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position -7007 of the FPN1 promoter. Haematologica. 2010;95(8):1261–1268. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noyes HA, et al. Mechanisms controlling anaemia in Trypanosoma congolense infected mice. PLoS One. 2009;4(4):e5170. doi: 10.1371/journal.pone.0005170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 64.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133(4):1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7(3):205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez A, Luukkaala T, Fleming RE, Britton RS, Bacon BR, Parkkila S. Global transcriptional response to Hfe deficiency and dietary iron overload in mouse liver and duodenum. PLoS One. 2009;4(9):e7212. doi: 10.1371/journal.pone.0007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torrance JD, Bothwell TH. A simple technique for measuring storage iron concentrations in formalinised liver samples. S Afr J Med Sci. 1968;33(1):9–11. [PubMed] [Google Scholar]
- 69.Vujic Spasic M, et al. Physiologic systemic iron metabolism in mice deficient for duodenal Hfe. Blood. 2007;109(10):4511–4517. doi: 10.1182/blood-2006-07-036186. [DOI] [PubMed] [Google Scholar]
- 70.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Muckenthaler M, et al. Relationships and distinctions in iron-regulatory networks responding to interrelated signals. Blood. 2003;101(9):3690–3698. doi: 10.1182/blood-2002-07-2140. [DOI] [PubMed] [Google Scholar]
- 73.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klingmuller U, et al. Primary mouse hepatocytes for systems biology approaches: a standardized in vitro system for modelling of signal transduction pathways. Syst Biol (Stevenage). 2006;153(6):433–447. doi: 10.1049/ip-syb:20050067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.