Abstract
Acute promyelocytic leukemia (APL) is a malignancy of the bone marrow, in which there is a deficiency of myeloid cells and an excess of immature cells called promyelocytes. APL is most commonly caused by a translocation (15:17) and expression of the promyelocytic leukemia and the retinoic receptor α (PML-RARA) fusion product; however, the events that cooperate with PML-RARA in APL pathogenesis are not well understood. In this issue of the JCI, Wartman and colleagues use an innovative approach to find other relevant mutations in APL. They performed whole genome sequencing and copy number analysis of a well-characterized APL mouse model to uncover somatic mutations in Jak1 and lysine (K)-specific demethylase 6A (Kdm6a, also known as Utx) in mice with APL and validated the ability of Jak1 mutations to cooperate with PML-RARA in APL. The findings implicate the JAK/STAT pathway in the pathogenesis of APL and illustrate the power of whole genome sequencing to identify novel disease alleles in murine models of disease.
Acute promyelocytic leukemia (APL) is a clinically and molecularly distinct subtype of acute myeloid leukemia that is distinguished by a recurrent chromosomal translocation fusing chromosomes 15 and 17. The t(15:17) translocation results in the fusion of the promyelocytic leukemia (PML) gene and the retinoic receptor α (RARA) gene (PML-RARA). The PML-RARA fusion protein is thought to contribute to APL pathogenesis by dimerizing and binding DNA and repressing the transcription of RARA target genes through recruitment of corepressors. More recent work indicates that PML-RARA also is able to bind to alternate DNA sites and to interact with chromatin remodeling complexes involved in stem cell maintenance and that the PML-RARA protein undergoes posttranslational modifications (sumoylation and phosphorylation) that are required for APL initiation (1, 2). A detailed understanding of the role of the PML-RARA oncogenic fusion in APL pathogenesis has allowed investigators to elucidate the molecular basis by which retinoic acid and arsenic trioxide offer dramatic efficacy in human patients with APL. Importantly, current therapeutic approaches combining retinoic acid/arsenic with anthracyclines allow as many of 90% of patients with APL to be cured (1).
The incidence of APL is reported to be constant over a human life span, which has led to models suggesting there is a single genetic rate-limiting step involved in the development of APL (3). However, mouse models of APL suggest that PML-RARA expression in vivo leads to APL with long latency and incomplete penetrance. This suggests that the acquisition of other genetic alterations in addition to PML-RARA contributes to the development of APL (4). Indeed, data from transgenic mouse models expressing PML-RARA indicate that mice that progress to APL acquire additional genetic alterations (4–6). Studies in patients with APL have also demonstrated the occurrence of activating FLT3 mutations and NRAS, KRAS, and MYC mutations (2). Collectively, these studies indicate that other genetic alterations may contribute to APL pathogenesis, even if PML-RARA is the disease-initiating event.
The utility of mouse models of APL
Although there exist key genetic and biologic differences in the development of malignancies between mice and humans, mouse models of APL have proven to be very useful in both modeling APL pathogenesis and in investigating specific therapies. Such models have been used to elucidate the effects and mechanisms of retinoic acid and arsenic trioxide therapy, with mouse APL cells demonstrating similar in vivo responses to both compounds when compared to human APL cells (7).
Several approaches have been used to generate mouse APL models, including transgenic PML-RARA expression, xenograft models, and adoptive transfer strategies. Transgenic models placing PML-RARA under the control of cathepsin G or migration inhibitory factor-related protein 8 (MRP8) promoters resulted in APL; however, in each case, APL developed with relatively long latency (6 months or longer) with incomplete penetrance (8). These data suggest additional mutations are required for the development of APL. The role of cooperating disease alleles in APL pathogenesis is underscored by mutational studies of primary human APL samples. Candidate gene studies have shown that mutations in NRAS, KRAS, FLT3–internal tandem duplication (FLT3-ITD), and FLT3–tyrosine kinase domain (FLT3-TKD) as well as trisomy 8 are observed in a subset of patients who present with APL (2). The functional significance of the Kras mutation in particular was elucidated by Chan et al., who found that coexpressing oncogenic Kras from the endogenous Ras locus with PML-RARA resulted in APL with shorter latency and near complete penetrance (9). Taken together, the human and murine genetic data suggest that additional disease alleles cooperate with PML-RARA in APL pathogenesis.
To elucidate potential cooperating events in murine APL models, previous studies performed karyotypic analysis. Zimonjic et al. used spectral karyotype analysis to identify recurrent abnormalities in murine APL cells, including interstitial or terminal deletion of one copy of chromosome 2, gains of chromosome 15, and loss of chromosome 11, X, and Y (5). Le Beau et al. performed spectral karyotyping analysis in hMRP8-PML-RARA mice and identified trisomies 8, 15, and 16 and monosomies X or Y as recurrent somatic alterations in murine APL cells (4). These results collectively indicate that PML-RARA fusion is necessary but not sufficient to produce APL in murine transgenic models. However, in the majority of humans with APL and in most murine APL models, the identity of cooperating disease alleles has not been revealed.
Whole genomic sequencing
In this issue of the JCI, Wartman and colleagues used massive parallel DNA sequencing in an effort to perform systematic mutational analysis of the murine APL genome (10). Until recently, whole genome sequencing of primary murine and human tumors was not feasible due to cost and the requirement for large amounts of tumor material. However, these limitations have largely been overcome due to improved sequencing technology and analytic tools (11). Indeed, previous efforts by the current investigators using whole genome sequencing in human patients with acute myelogenous leukemia (AML) has allowed them to identify novel clinically and biologically relevant AML mutations, demonstrating the power of massive parallel sequencing (11, 12).
Wartman et al. used an innovative strategy to find additional mutations in this APL model (10). An inbred mouse strain was used in an attempt to reduce the number of variants, as many of the variants found in sequencing a murine genome may not be relevant to disease pathogenesis. Mice expressing the PML-RARA transgene under the control of the murine cathepsin G promoter were backcrossed to the Black 6/Taconic background for 10 generations. These mice developed an APL-like disease with a relatively long latency (9–12 months), suggesting the acquisition of additional genetic events is required for APL development in this model. In previous human studies, tumor whole genome sequencing data were compared with sequencing data from matched germline DNA to assess whether candidate mutations were present in the germline or were bona fide somatic mutations acquired during tumorigenesis. In contrast, here the authors compared the spectrum of single nucleotide variants present in murine APL cells with a sequenced genome of the initial mouse strain. An alternative, and perhaps more discriminating, strategy might have compared the APL mutational data with DNA from littermate controls or compared the murine APL genome with hematopoietic DNA from the same mouse from an earlier time point, before APL development.
Six nonsynonymous mutations were identified and validated as being present in the APL genome; the authors then performed secondary mutational analysis of 89 additional mouse APL samples for these 6 mutations. Importantly, this approach allowed them to identify that one mutation, Jak1 V658F, was present as a recurrent alteration in murine APL. Of note, the Jak1 V658F mutation occurs at the homologous position to JAK2 V617, which is commonly mutated in patients with myeloproliferative neoplasms (MPNs) (13), and has been observed previously in patients with high-risk acute lymphoblastic leukemia (ALL) (14). They then ectopically expressed Jak1 V658F in mCG-PML-RARA bone marrow, followed by transplantation into lethally irradiated recipients, which resulted in a short latency, completely penetrant APL phenotype. In addition, they also performed high-resolution copy number analysis of the murine tumors and identified a somatic deletion of a histone demethylase, lysine (K)-specific demethylase 6A (Kdm6a, also known as Utx), in the same murine APL genome, a deletion also observed in human APL. However, its functional contribution to APL pathogenesis and to APL development in this mouse model has not been elucidated.
Mutations in the JAK/STAT pathway and targeted therapies
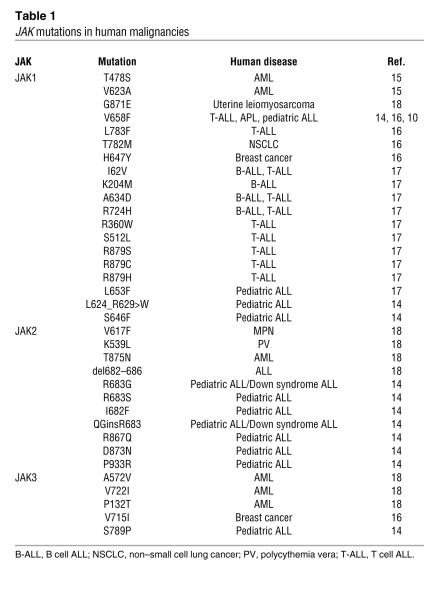
The JAKs are involved in the transduction of cytokine receptor signaling. These kinases (JAK1, JAK2, JAK3, and Tyk2) bind to the cytosolic domains of cytokine receptors. Conformational changes occur in the cytokine receptor as a result of binding to cytokines, allowing them to recruit JAKs. Somatic mutations in JAKs have been described in human malignancy (Table 1). Most notable has been the elucidation of the role of JAK2 V617F mutation in MPN pathogenesis. Somatic JAK1 mutations have been recently described in AML (15, 16) and in ALL (17). These genetic observations have led to the development of JAK inhibitors, including inhibitors with JAK1 inhibitory activity; these agents have now entered late-stage clinical trials in patients with MPN. Here, the investigators used a pan-JAK inhibitor in the context of a methylcellulose assay to show that JAK1 inhibition exhibited similar efficacy to all-trans retinoic acid (a standard treatment for APL) in reducing APL colony formation. Furthermore, the use of the pan-JAK inhibitor decreased STAT5 phosphorylation, indicating on-target effects with regards to the JAK/STAT pathway. Collectively, these data indicate a role for the JAK/STAT pathway in the pathogenesis of APL in this model that extends beyond the presence of a JAK mutation. The role of the JAK/STAT pathway in APL and in other AML subtypes thus warrants further investigation. In addition, whether JAK1-mutant APL constitutes a specific clinicopathologic subtype of APL with prognostic or therapeutic relevance remains to be delineated.
Table 1.
JAK mutations in human malignancies
Conclusions
Wartman and colleagues successfully employed an elegant use of whole genome sequencing as a dragnet to ensure broad coverage of the genome. It is possible that this strategy may not identify all relevant disease alleles, due to the sequencing approach, the analytic platform, or the presence of large mutations that may be missed by short-read parallel sequencing. However, the strength of this approach is demonstrated in this report, as the authors show how whole genome sequencing can be used to provide pathogenetic insight in murine cancer models. We predict that subsequent utilization of this approach will allow investigators to add further to the list of recurrent driver mutations that contribute to malignant transformation.
Acknowledgments
R.K. Rampal is supported by the Memorial Sloan-Kettering Cancer Center Clinical Scholars Program. R.L. Levine is a Geoffrey Beene Junior Faculty Chair at the Memorial Sloan-Kettering Cancer Center.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(4):1255–1258. doi:10.1172/JCI57200.
See the related article beginning on page 1445.
References
- 1.de The H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10(11):775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- 2.Akagi T, et al. Hidden abnormalities and novel classification of t(15;17) acute promyelocytic leukemia (APL) based on genomic alterations. Blood. 2009;113(8):1741–1748. doi: 10.1182/blood-2007-12-130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vickers M, Jackson G, Taylor P. The incidence of acute promyelocytic leukemia appears constant over most of a human lifespan, implying only one rate limiting mutation. Leukemia. 2000;14(4):722–726. doi: 10.1038/sj.leu.2401722. [DOI] [PubMed] [Google Scholar]
- 4.Le Beau MM, Bitts S, Davis EM, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice parallel human acute promyelocytic leukemia. Blood. 2002;99(8):2985–2991. doi: 10.1182/blood.V99.8.2985. [DOI] [PubMed] [Google Scholar]
- 5.Zimonjic DB, Pollock JL, Westervelt P, Popescu NC, Ley TJ. Acquired, nonrandom chromosomal abnormalities associated with the development of acute promyelocytic leukemia in transgenic mice. Proc Natl Acad Sci U S A. 2000;97(24):13306–13311. doi: 10.1073/pnas.97.24.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Beau MM, Davis EM, Patel B, Phan VT, Sohal J, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice identify cooperating events and genetic pathways to acute promyelocytic leukemia. Blood. 2003;102(3):1072–1074. doi: 10.1182/blood-2003-01-0155. [DOI] [PubMed] [Google Scholar]
- 7.Lallemand-Breitenbach V, Zhu J, Kogan S, Chen Z, de The H. Opinion: how patients have benefited from mouse models of acute promyelocytic leukaemia. Nat Rev Cancer. 2005;5(10):821–827. doi: 10.1038/nrc1719. [DOI] [PubMed] [Google Scholar]
- 8.Kogan SC. Mouse models of acute promyelocytic leukemia. Curr Top Microbiol Immunol. 2007;313:3–29. doi: 10.1007/978-3-540-34594-7_2. [DOI] [PubMed] [Google Scholar]
- 9.Chan IT, et al. Oncogenic K-ras cooperates with PML-RAR alpha to induce an acute promyelocytic leukemia-like disease. Blood. 2006;108(5):1708–1715. doi: 10.1182/blood-2006-04-015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wartman LD, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121(4):1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. . N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355(23):2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 14.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106(23):9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Z, et al. Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood. 2008;111(9):4809–4812. doi: 10.1182/blood-2007-05-090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong EG, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14(12):3716–3721. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 17.Flex E, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205(4):751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci. 2008;33(3):122–131. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]