Abstract
Purpose of review
Host genetic factors influencing HCV transmission outcomes are incompletely defined. However, vast differences observed in rates of spontaneous clearance between individuals infected with the same parental HCV strain strongly indicate a role for genetic determinants in the host immune response to HCV. This review discusses genetic association studies, particularly those published in the past year, that show gene linkages with spontaneous and treatment-induced HCV clearance. The valuable role that blood collection centers can play in increasing the sample size of HCV confirmed seropositive donors with resolved versus persistent infections for large-scale genetic association studies is highlighted.
Recent findings
Recent groundbreaking genome-wide association study (GWAS) and targeted single-nucleotide polymorphism (SNP) analysis from independent groups have demonstrated immune response gene polymorphisms, and particularly in the interleukin (IL)-28B gene, that are strongly linked to HCV clearance. The IL-28B gene encodes interferon lambda 3, an innate immune response cytokine. SNPs in the promoter region of IL-28B were first shown to be associated with HCV treatment-induced viral clearance and subsequently to be a key determinant of spontaneous HCV resolution in infected individuals. Samples from blood donors with resolved and chronic HCV infections have contributed to these findings.
Summary
These genetic studies have provided the strongest evidence so far of a host genetic determinant linked to HCV clearance. Such large-scale genetic association studies will promote better understanding of HCV disease pathogenesis and assist in effective prognosis of HCV in the future. Continued and preferably expanded participation of blood centers in this research is encouraged.
Keywords: Hepatitis C virus, HCV, HCV host genetics, HCV clearance, role for blood centers
Introduction
The factors associated with spontaneous and treatment-induced hepatitis C virus (HCV) clearance are not well defined, but likely involve a combination of host and viral factors. Studies have reported correlations between rate of resolution and host HLA polymorphisms, host demographics (including age at infection, gender, race-ethnicity), coexisting viral infections, and other cofactors. Neither the strength of these effects nor the extent to which they are generalizable to different infected populations are well defined. In most instances genetic analyses that study the host immune response genes in HCV spontaneous clearance are separate from studies that look at genetic factors associated with treatment-induced response to HCV. The factors influencing both these conditions could be distinct and controlled by different genes, or could be similar, which is suggested by similar demographic correlates (gender; race/ethnicity) of spontaneous and treatment-induced clearance. However, recent groundbreaking genetic association studies summarized in this review link polymorphisms in the IL-28B gene to virus elimination in both spontaneous and treatment-induced HCV clearance. Therefore, in this review we will discuss the host genetic correlates pertaining to both instances, spontaneous and treatment-induced HCV clearance. There has been extensive research that has led to compelling evidence that HCV clearance is also influenced by viral factors, including genotype, inoculum, route, quasispecies diversity and evolution, and by timing and strength of adaptive immune response in newly infected individuals. However, viral factors and adaptive immune responses associated with HCV clearance have not proven to be as important as originally hypothesized, and are not discussed in this review; interested readers are directed to other recent reviews [1-5].
Epidemiology and Clinical Course of HCV Infection
In the 20 years since its identification, HCV has emerged as a major etiological agent of liver disease throughout the world. Globally, there are an estimated 170 million people infected with HCV, including nearly 4 million cases in the USA [6, 7]. As a result of donor screening and behavioral changes related to the AIDS epidemic, HCV incidence has declined in the USA over the past decade [8, 9]. However, there are still an estimated 17, 000 new infections occurring annually [10]. In Europe and North America HCV genotypes 1, 2 and 3 are prevalent with a predominance of genotype 1 infected individuals.
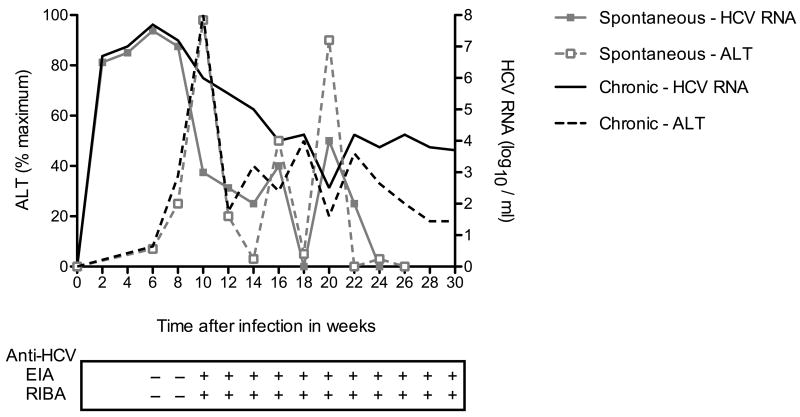
Most of what is known about the very early events in HCV infection has been learned from studies of blood and plasma donors identified by RNA screening, archived samples from recipients in transfusion-associated infections, and from experimentally infected chimpanzees [2, 11-15]. Figure 1 is a schematic representation of the clinical and virological course of HCV in spontaneous resolvers versus chronically infected individuals. Of persons acutely infected with HCV, 20-30% recover spontaneously and clear viremia shortly following seroconversion, while 70-80% develop persistent infection. Of the chronically infected individuals, 30% will have progressively worsening liver disease potentially culminating in cirrhosis or hepatocellular carcinoma, 40% will have slowly evolving infections with only modest liver damage and 30% will have viremia in the absence of clinically detectable liver damage. However, for a given infected individual there is currently no way of predicting prognosis.
Figure 1.
Schematic representation of clinical and virological course of HCV in spontaneous resolvers versus chronically infected individuals. HCV RNA typically becomes detectable within 7-14 days after exposure and specific antibodies appear within 20 to 150 days, with a mean of approximately 60 days. Elevation of alanine aminotransferases (ALT) correlates with early immune responses and occurs after week 6, but may occur as early as the second week following exposure. Enzyme immunoassay (EIA) detects antibodies to HCV. The recombinant immunoblot assay (RIBA) identifies the specific antigens to which antibodies are reacting in EIA.
Although new HCV anti-viral drugs, including protease inhibitors, are in development and clinical trials appear promising, the current standard of care treatment consists of pegylated-interferon-α in combination with ribavirin that must be given for 24-48 weeks depending on genotype and virological response [16-18]. Added to the lengthy treatment course is the complication of extensive side-effects associated with these drugs. Using this regime, 54-63% of individuals with persistent infection, exhibited a sustained virological response (SVR) which is believed to represent true eradication of HCV infection [19, 20]. However, the SVR rates for genotype 1 infected individuals are lower - 42-52% as compared to individuals infected with genotype 2 or 3 at 76-84%.
Host immunogenetic correlates of HCV clearance and “candidate gene” analysis
A study in 2000 by Thomas et al. showed a clear difference in rates of HCV clearance based on race, with Caucasian subjects more likely to clear virus spontaneously than African-Americans subjects [21]. This was also found to be true for treatment-induced viral elimination, with African-Americans less likely to achieve a SVR to therapy than Caucasians [22, 23]. In particular, HCV displays vastly different prognosis to interferon-α therapy in chronically infected individuals from different racial/ethnic groups who are infected with genotype 1 virus. The differences observed in either spontaneous or treatment-induced HCV elimination suggested that there may be a strong genetic contribution, perhaps manifested phenotypically in more or less robust host immune responses against HCV. The examination of HCV infection and immune response genes has been hampered in part by the lack of in-bred and out-bred animal models for HCV where host genetics could be studied in the context of acute infection and longitudinal outcome studies of genetically heterogenous groups of animals. Consequently investigators have mostly studied HCV infected human cohorts in an effort to identify specific gene variants (“candidate genes”) with demonstrable anti-viral effects for immune correlates of HCV recovery.
As with many infectious agents, investigators first looked for associations between HCV resolution and polymorphisms at both class I HLA and class II HLA loci. Class I HLA presents antigen to CD8+ cytolytic T-cells while class II HLA mediates the CD4+ T-cell response to infection. Several class I HLA alleles have been found to be associated with viral clearance including HLA-B*57 and HLA-Cw*0102. Conversely, others are associated with persistence, such as HLA-A*2301 and HLA-Cw*04 [24-26]. Class II HLA genes DQB1*0301 and DRB1*11 have been associated with viral clearance and the studies that support these observations have been repeated using several independent cohorts [25, 26]. However, while an association to spontaneous clearance has been observed, the effect is modest and DQB1*0301 or DRB1*1101 expression in the host has not been associated with treatment-induced HCV elimination.
A significant study by Khakoo et al., (2004) showed that individuals homozygous for inhibitory NK cell receptor (KIR2DL3) and the ligand HLA-C1 genes are more likely to clear HCV spontaneously compared to those with other NK receptor-ligand combinations [27]. Improved NK cell activity is postulated to play an important role in reducing HCV replication. However, spontaneous HCV clearance associated with NK cells was observed only in individuals infected via intravenous drug use (IDU) and not those infected by blood transfusion. It is currently unknown whether it is the amount of virus inocula, route of infection or other factors that are responsible for this difference.
Several studies have examined cytokine genes or expression as immune mediators of spontaneous HCV resolution [3, 5, 28, 29]. The most studied of these are IFN-γ, TNF-α, IL-10, IL-12 and TGF-β [30-35]. IFN-γ and IL-10 show low-level associations with HCV spontaneous clearance, and to some extent SVR to treatment [3, 32, 35, 36]. There is also increasing interest in understanding the role played at the molecular level of genes and regulatory elements downstream of important cytokines, such as interferon-stimulated genes (ISGs). Increased expression in hepatocytes of ISGs prior to treatment has been correlated with poorer response rate [37]. However, ISG as a predictive genetic association factor for HCV clearance has not been fully evaluated. A recent large-scale genetic analysis approach screened 1426 SNP in 112 candidate genes for spontaneous clearance of HCV. They documented four immune response genes, TNFSF18 (tumor necrosis factor superfamily, member 18), TANK (TRAF family member associated NF-κB activator), HAVCR1 (Hepatitis A virus cellular receptor 1), and IL18BP (IL-18 binding protein), that appeared to play a role in HCV spontaneous clearance [38]. Many of the candidate gene studies described above give leads as to the host immune response genes that might be involved in predicting spontaneous clearance or treatment outcomes. However, other than the HLA polymorphism results, most studies are based on small samples and will require further validation involving the use of larger cohorts of HCV infected individuals.
Genome-wide association studies that link IL-28B to HCV clearance
The genetic tools and the knowledge derived from the human genome project, as well as availability of powerful and relatively inexpensive whole human genome analysis systems from companies such as Illumina and Affymetrix, have allowed researchers to embark on studies that were not possible a decade ago. In a powerful genetic approach called genome-wide association study (GWAS) one can examine common genetic variations across large spans of the human genome by analyzing hundreds of thousands of SNP spanning all chromosomes. The SNP may be directly involved with or physically linked to genes responsible for a distinct phenotype or trait. Performing GWAS involves four major steps. First, the selection of the interest group with a well characterized phenotype (e.g., HCV clearance) and a corresponding control group. Second, DNA isolation, GWAS genotyping and data analysis. Third, the data is used in complex statistical analysis to determine if there are associations of SNP with the selected trait versus the control group. Fourth, the data is applied to design studies of other independent groups of clinical samples to ascertain whether or not the association is corroborated, as well as studies to elucidate if the genetic determinant identified by the SNP analysis is responsible for functional characteristics of the trait that is biologically plausible.
In 2009 work from three independent groups reported a SNP in the IL-28B (interferon lambda 3) promoter region associated with treatment response to HCV (Table 1). All three studies reported SVR of HCV genotype 1 infected participants to treatment with pegylated interferon-α in combination with ribavirin. Ge et al. [39**] reported a genetic polymorphism in the IL-28B gene on chromosome 19; the SNP rs12979860 associated with HCV treatment-induced clearance. The CC genotype at the SNP was associated with higher response rates to treatment compared to CT or TT genotypes. This association to treatment response held true for all races within the group, including Europeans, African-Americans and Hispanics. However, there was a clear difference in prevalence of the CC genotype in people of European ancestry versus African-Americans that was postulated to account for approximately half the difference seen from previous studies that described lower response rates to treatment in African-Americans. In the GWAS by Suppiah et al. [40**] a well-characterized homogeneous cohort of Australians was analyzed in their initial study and results were confirmed using a second heterogeneous cohort of Australians. The advantage with the primary Australian cohort is that it is a homogeneous population in which differences in age, body mass index (BMI) and viral load were the same between treatment responders and non-responders (control group). This study design excluded these factors from having played a role in determining the outcome. They found a SNP rs8099917 in the intergenic region between IL-28A and IL-28B was associated with positive response to treatment-induced recovery. The minor G allele at SNP rs8099917 as opposed to the major T allele was strongly associated with poor virological response to treatment. In a third GWAS paper, Tanaka et al. [41**] reported results of a study in a Japanese cohort and found that the same SNP as Suppiah et al. [40**] (rs8099917) was associated with response to treatment. Once again the minor G allele was shown to be associated with poor response. In addition they describe another minor allele at SNP rs12980275 corresponding with poor response. Both the Suppiah et al. [40**] and Tanaka et al. [41**] studies documented reduced expression by real-time PCR of IL-28B mRNA in the blood of individuals who were either predicted to, or did, fail to respond to anti-viral treatment. In particular, the homozygous and heterozygous carriers of the minor G allele had significantly reduced IL-28B mRNA expression compared to the homozygous carriers of the major T allele. Thus three independent studies that sampled different population groups have unequivocally shown that the IL-28B genotype and expression is linked to treatment-induced clearance of HCV, albeit with identified SNP residues that are different (probably reflecting genetic linkage to the etiological polymorphism).
Table 1. Genetic association studies on IL-28B gene polymorphism and HCV clearance.
| References | Country | Number of patients | Ethnic origin | HCV Disease outcome | SNP identified |
|---|---|---|---|---|---|
| Ge et al. [39] | North America | 1137 | African-Americans 191 European-Americans 871 Hispanics 75 |
Treatment-induced (IFN-α + ribavirin) clearance and chronics | rs12979860 (∼ 3kb upstream of IL-28B) linkage disequilibrium with rs 8099917 |
| Suppiah et al. [40] | Australia Europe |
848 | Australians 293 European 555 |
Treatment-induced (IFN-α + ribavirin) clearance and chronics | rs8099917 (∼8kb upstream of IL-28B) 16 other SNPs identified |
| Tanaka et al. [41] | Japan | 314 | Japanese 314 | Treatment-induced (IFN-α + ribavirin) clearance and chronics | rs8099917 (∼8kb upstream of IL-28B) rs12980275rs1188122rs8105790 |
| Thomas et al. [42] | North America | 1008 | African-Americans 290 European-Americans 642 Other 76 |
Spontaneous clearance and chronics | rs12979860 (∼ 3kb upstream of IL-28B) |
| Rauch et al. [43] | Switzerland | 1362 | Swiss whites 1362 | Spontaneous clearance, Treatment-induced clearance and chronics | rs8099917 (∼8kb upstream of IL-28B) |
Following the identification of the association of the IL-28B SNP rs12979860 with treatment-induced HCV clearance, Thomas et al. [42**] examined the association of SNP rs12979860 with spontaneous HCV clearance (Table 1). They genotyped individuals for the SNP rs12979860 who spontaneously cleared infection compared to a control group of persistently infected individuals (this study included a large number of blood donors with resolved and persistent HCV infections). Interestingly the same IL-28B SNP rs12979860 shown to be associated with treatment-induced viral clearance was also associated with spontaneous clearance of HCV. Individuals with CC as opposed to CT or TT genotypes had a three fold higher chance of clearing the virus spontaneously irrespective of race (Figure 2). The investigators also examined the global pattern of the rs12979860 C allele frequency in order to gain insight into the geographic frequency distribution. Interestingly the comparison of the C allele frequency across six different regions of the world showed that the C allele is a minor allele with reduced frequency in Africa compared to most other regions. This might explain why East Asians and Europeans with higher C allele frequency have higher rates of treatment-induced and spontaneous HCV clearance compared to individuals with African ancestry.
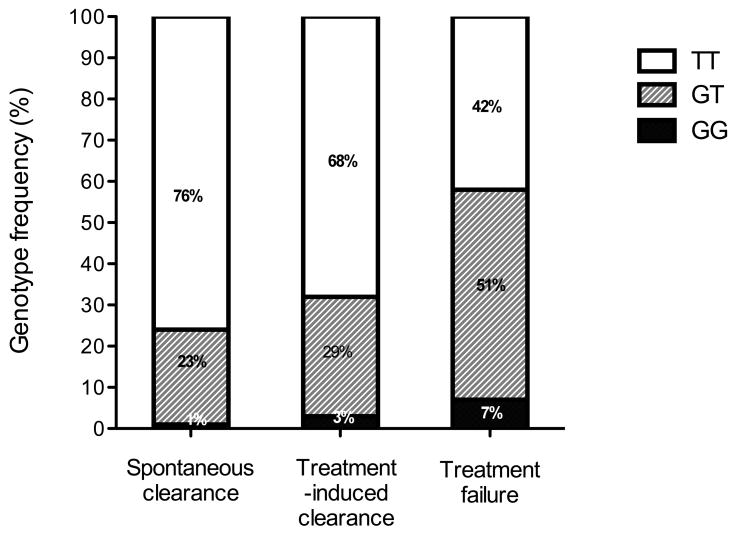
Figure 2.
Percentage of HCV clearance by rs12979860 genotype. Individuals with CC as opposed to CT or TT genotypes have a higher chance of clearing the virus spontaneously irrespective of race. Figure redrawn with permission from Thomas et al.[42].
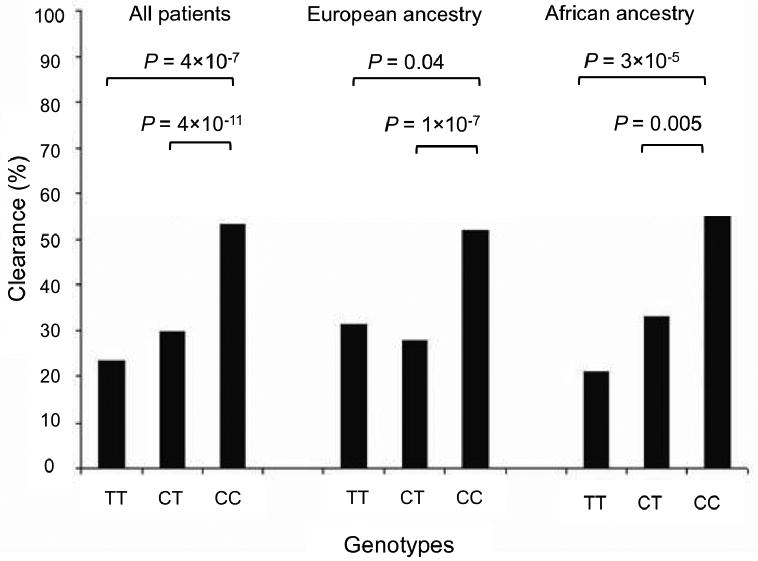
Rauch et al. [43*] performed a GWAS for individuals with different HCV disease outcomes that included those who spontaneously cleared the virus, responded to treatment (pegylated interferon-α and ribavirin) or were chronically infected (Table 1). This study helped answer some of the IL-28B gene polymorphism related questions raised by the four papers described above. In this study the population analyzed was exclusively Swiss Caucasians infected with HCV. The SNP rs8099917 (in the IL-28B promoter region) gave the strongest association with spontaneous HCV clearance and no other SNP outside the IL-28B/A locus reached genome-wide significance. The rs8099917 SNP is a T/G allele with the minor G allele identified as the risk allele. The genotypes TT, GT and GG were analyzed and the results showed that the risk allele frequencies increased from spontaneous clearance followed by treatment-induced clearance to chronic infection indicating treatment failure (Figure 3).
Figure 3.
Distribution of rs8099917- TT, GT and GG genotypes in an HCV infected population. Among the HCV infected patients, those with treatment-failure have higher rates of carriage of the G-risk allele than patients with treatment-induced clearance and patients with spontaneous clearance. Figure redrawn with permission from Rauch et al.[43].
All five papers described here demonstrate the IL-28B gene polymorphism to be one of the strongest genetic effects to be associated with spontaneous and treatment-induced HCV recovery. However, the underlying mechanism that IL-28B uses to clear HCV and its functional effect needs further work. Interferon-λ is a newly discovered family of interferons that includes IL-29 (lambda type 1), IL-28A (lambda type 2) and IL-28B (lambda type 3) [44, 45]. These interferons use the JAK-Stat signal cascade in a similar fashion to interferon-α and induce interferon-stimulated genes [46]. ISGs have been shown to play an important role in HCV treatment outcomes and spontaneous clearance [37]. Interferon lambda 1 (IL-29) has a lower anti-viral activity than interferon-α in vitro but was shown to function in conjunction with interferon-α for inhibition of HCV [47, 48]. IL-29 is currently in phase II clinical trials as a treatment for HCV. However, the function of IL-28B and its anti-viral effect on HCV is less well understood but will undoubtedly be the focus of extensive further work to understand its role in immunopathogenesis and potential therapeutics.
Importance of HCV infected sample collection by blood centers for large-scale genetic studies
The Thomas et al. [42**] paper describing the association of the IL-28B SNP with spontaneous HCV clearance used samples from various HCV patient cohorts as well as HCV positive samples collected from blood centers for large-scale genetic analysis. Individuals with a history of HCV infection identified by donor screening represent an immense and potentially unique subject pool from which to obtain the sample sizes necessary to identify and subsequently confirm or reject the genetic correlates while minimizing selection bias. Universal implementation of serological screening of blood donors for HCV antibody in 1990 and nucleic acid amplification testing (NAT) for HCV RNA in 1999 has led to the identification of large numbers of antibody confirmed donors with presumptive resolved (RNA-neg/anti-HCV-pos) and chronic (RNA-pos/anti-HCV-pos) infections (Figure 4) [49, 50]. Blood centers in the USA collect approximately 2.6 million units of whole blood and apheresis components from unique first-time (FT) donors every year. Given the current seroprevalence of HCV among FT donors, there are approximately 5000 newly diagnosed HCV infections made in the US blood donor setting annually [50, 51]. The blood donation system therefore represents the largest systematic screening program in the US. Since implementation of NAT screening, US donor screening programs have detected over 50 donors per year in the viremic pre-seroconversion phase of HCV infection who can be followed to observe and study the outcome of infection (clearance versus persistence) [52, 53]. NAT has also facilitated routine discrimination of RIBA-confirmed seropositive donors into those with probable resolved or persistent viremia. At the time of identification and deferral, these persons are notified of their HCV status, including confirmatory test results.
Figure 4.
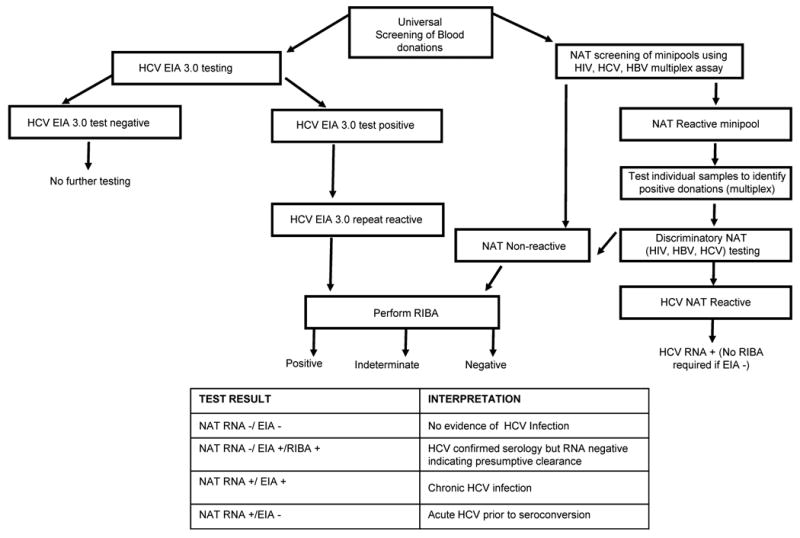
Flow chart represents routine testing for HCV at blood centers in the US and classification of disease outcomes. NAT- Nucleic acid amplification testing; EIA-Enzyme immunoassay; RIBA-Recombinant immunoblot assay.
The HCV infected donor samples (with data to sub-classify the infections as viremic pre-serconversion window phase, chronic carrier and resolved infections) obtained from blood centers can include information as to the donor age, gender and self-identified ethnicity. The blood centers also screen for HIV-1 and HBV by NAT and antibody assays that allows the HCV genetic studies to either include or exclude these samples from co-infected individuals if desired. On a global level blood banks collaborate extensively, and most of the world's blood supply is now routinely screened for HCV antibodies and RNA, such that sourcing of infected donor samples could be expanded dramatically now that this approach as been demonstrated to be useful in the U.S. Table 2, lists the advantages and limitations of blood center derived samples for inclusion in future HCV genetic association studies.
Table 2. Advantages and limitations of using blood center derived HCV positive samples for large-scale genetic studies.
Advantages
|
Limitations
|
Conclusion
In this review we summarized groundbreaking genetic association studies conducted in the past year that have revealed host genes associated with HCV treatment-induced and spontaneous clearance. These studies are indispensable to understanding the immune correlates of HCV disease pathogenesis and developing new treatment options for HCV infected persons. The power of large-scale genetic association studies related to HCV can be improved significantly by increasing the number of samples analyzed. Blood collection centers can play a valuable role in this regard. The blood centers are a rich source of uniquely pedigreed infected donor samples that could substantially increase the number of samples represented in genetic studies examining spontaneous clearance versus chronic HCV infection. Transfusion medicine scientists should strive to actively participate in these important studies, which will likely lead to novel diagnostic, prognostic and therapeutic strategies for HCV.
References
- 1.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–54. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. Lancet. 2008;372:321–32. doi: 10.1016/S0140-6736(08)61116-2. [DOI] [PubMed] [Google Scholar]
- 3.Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12:713–26. xi. doi: 10.1016/j.cld.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sklan EH, Charuworn P, Pang PS, Glenn JS. Mechanisms of HCV survival in the host. Nat Rev Gastroenterol Hepatol. 2009;6:217–27. doi: 10.1038/nrgastro.2009.32. [DOI] [PubMed] [Google Scholar]
- 5.Post J, Ratnarajah S, Lloyd AR. Immunological determinants of the outcomes from primary hepatitis C infection. Cell Mol Life Sci. 2009;66:733–56. doi: 10.1007/s00018-008-8270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong GL, Wasley A, Simard EP, et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–14. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- 7.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 1:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 8.Alter MJ, Kruszon-Moran D, Nainan OV, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–62. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 9.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–41. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control. Surveillance of Acute Viral Hepatitis-US, 2007. MMWR. 2009;50(No. SS-3) [Google Scholar]
- 11.Busch MP. Insights into the epidemiology, natural history and pathogenesis of hepatitis C virus infection from studies of infected donors and blood product recipients. Transfus Clin Biol. 2001;8:200–6. doi: 10.1016/s1246-7820(01)00125-2. [DOI] [PubMed] [Google Scholar]
- 12.Marcellin P. Hepatitis C: the clinical spectrum of the disease. J Hepatol. 1999;31 1:9–16. doi: 10.1016/s0168-8278(99)80368-7. [DOI] [PubMed] [Google Scholar]
- 13.Alter HJ, Conry-Cantilena C, Melpolder J, et al. Hepatitis C in asymptomatic blood donors. Hepatology. 1997;26:29S–33S. doi: 10.1002/hep.510260705. [DOI] [PubMed] [Google Scholar]
- 14.Mosley JW, Operskalski EA, Tobler LH, et al. The course of hepatitis C viraemia in transfusion recipients prior to availability of antiviral therapy. J Viral Hepat. 2008;15:120–8. doi: 10.1111/j.1365-2893.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 15.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–29. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 16.Pereira AA, Jacobson IM. New and experimental therapies for HCV. Nat Rev Gastroenterol Hepatol. 2009;6:403–11. doi: 10.1038/nrgastro.2009.92. [DOI] [PubMed] [Google Scholar]
- 17.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292–303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 18.Flisiak R, Horban A, Gallay P, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–26. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 19.Maheshwari A, Thuluvath PJ. Management of acute hepatitis C. Clin Liver Dis. 2010;14:169–76. x. doi: 10.1016/j.cld.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Farnik H, Mihm U, Zeuzem S. Optimal therapy in genotype 1 patients. Liver Int. 2009;29 1:23–30. doi: 10.1111/j.1478-3231.2008.01969.x. [DOI] [PubMed] [Google Scholar]
- 21.Thomas DL, Astemborski J, Rai RM, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–6. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 22.Jeffers LJ, Cassidy W, Howell CD, et al. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39:1702–8. doi: 10.1002/hep.20212. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle JH, Wahed AS, Brown RS, Jr, et al. Early changes in hepatitis C virus (HCV) levels in response to peginterferon and ribavirin treatment in patients with chronic HCV genotype 1 infection. J Infect Dis. 2009;199:1112–20. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 24.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–45. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 26.Thio CL, Thomas DL, Goedert JJ, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 27.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 28.Lemon SM. Induction and Evasion of Innate Antiviral Responses by Hepatitis C Virus. J Biol Chem. 2010 doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauch A, Gaudieri S, Thio C, Bochud PY. Host genetic determinants of spontaneous hepatitis C clearance. Pharmacogenomics. 2009;10:1819–37. doi: 10.2217/pgs.09.121. [DOI] [PubMed] [Google Scholar]
- 30.Thio CL, Goedert JJ, Mosbruger T, et al. An analysis of tumor necrosis factor alpha gene polymorphisms and haplotypes with natural clearance of hepatitis C virus infection. Genes Immun. 2004;5:294–300. doi: 10.1038/sj.gene.6364072. [DOI] [PubMed] [Google Scholar]
- 31.Kimura T, Saito T, Yoshimura M, et al. Association of transforming growth factor-beta 1 functional polymorphisms with natural clearance of hepatitis C virus. J Infect Dis. 2006;193:1371–4. doi: 10.1086/503436. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Yang H, Borg BB, et al. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci U S A. 2007;104:985–90. doi: 10.1073/pnas.0609954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller T, Mas-Marques A, Sarrazin C, et al. Influence of interleukin 12B (IL12B) polymorphisms on spontaneous and treatment-induced recovery from hepatitis C virus infection. J Hepatol. 2004;41:652–8. doi: 10.1016/j.jhep.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Lio D, Caruso C, Di Stefano R, et al. IL-10 and TNF-alpha polymorphisms and the recovery from HCV infection. Hum Immunol. 2003;64:674–80. doi: 10.1016/s0198-8859(03)00080-6. [DOI] [PubMed] [Google Scholar]
- 35.Oleksyk TK, Thio CL, Truelove AL, et al. Single nucleotide polymorphisms and haplotypes in the IL10 region associated with HCV clearance. Genes Immun. 2005;6:347–57. doi: 10.1038/sj.gene.6364188. [DOI] [PubMed] [Google Scholar]
- 36.Selzner N, McGilvray I. Can genetic variations predict HCV treatment outcomes? J Hepatol. 2008;49:494–7. doi: 10.1016/j.jhep.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Shackel NA, McCaughan GW. Intrahepatic interferon-stimulated gene responses: can they predict treatment responses in chronic hepatitis C infection? Hepatology. 2007;46:1326–8. doi: 10.1002/hep.22006. [DOI] [PubMed] [Google Scholar]
- 38.Mosbruger TL, Duggal P, Goedert JJ, et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis. 2010;201:1371–80. doi: 10.1086/651606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]; The authors use GWAS to identify genetic polymorphism in the IL-28B gene, that represents an important outcome predictor for pegylated interferon and ribavirin treatment of HCV-1 patients. A clear difference in prevalence of the major allele in people of European ancestry versus African-Americans was postulated to account for approximately half the difference seen from previous studies that described lower response rates to treatment in African-Americans.
- 40**.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–4. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]; The authors independently did GWAS as in reference 39 but in this case the analysis was done using a cohort of Australians. The study design excluded factors such as differences in age, body mass index (BMI) and viral load from having played a role in determining the outcome of HCV infection in treatment responders and non-responders.
- 41**.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]; The authors use GWAS to identify SNP associated with treatment-induced HCV clearance in a Japanese cohort. This study was published at the same time as the reports referenced in 39 and 40. The SNP rs8099917, in the IL-28B promoter region (the same one described by authors in reference 40) is associated with treatment-induced HCV clearance.
- 42**.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]; The investigators examined the association of IL-28B SNP rs12979860 with spontaneous HCV clearance using SNP analysis. The IL-28B SNP was found to be the strongest predictor of spontaneous HCV clearance reported to date.
- 43*.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL-28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45. 1345 e1–7. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]; The authors performed GWAS for individuals with different HCV disease outcomes that included those who spontaneously cleared the virus, responded to treatment or were chronically infected. They used a cohort of Swiss whites for their analysis.
- 44.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 45.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 46.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–72. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Pagliaccetti NE, Eduardo R, Kleinstein SH, et al. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079–89. doi: 10.1074/jbc.M804296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–98. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 49.Stramer SL. Nucleic acid testing for transfusion-transmissible agents. Curr Opin Hematol. 2000;7:387–91. doi: 10.1097/00062752-200011000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Zou S, Dorsey KA, Notari EP, et al. Prevalence, incidence, and residual risk of human immunodeficiency virus and hepatitis C virus infections among United States blood donors since the introduction of nucleic acid testing. Transfusion. 2010 doi: 10.1111/j.1537-2995.2010.02622.x. [DOI] [PubMed] [Google Scholar]
- 51.Murphy E, Fang J, Tu Y, Cable R. Hepatitis c virus prevalence and clearance among US blood donors, 2006-2007: associations with birth cohort, multiple pregnancies and body mass index. J Infect Dis. 2010 doi: 10.1086/654882. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stramer SL, Glynn SA, Kleinman SH, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–8. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 53.Bernardin F, Tobler L, Walsh I, et al. Clearance of hepatitis C virus RNA from the peripheral blood mononuclear cells of blood donors who spontaneously or therapeutically control their plasma viremia. Hepatology. 2008;47:1446–52. doi: 10.1002/hep.22184. [DOI] [PubMed] [Google Scholar]