Abstract
The ability to accurately track Listeria monocytogenes strains involved in outbreaks is essential for control and prevention of listeriosis. Because current typing techniques are time-consuming, cost-intensive, technically demanding, and difficult to standardize, we developed a rapid and cost-effective method for typing of L. monocytogenes. In all, 172 clinical L. monocytogenes isolates and 20 isolates from culture collections were typed by high-resolution melting (HRM) curve analysis of a specific locus of the internalin B gene (inlB). All obtained HRM curve profiles were verified by sequence analysis. The 192 tested L. monocytogenes isolates yielded 15 specific HRM curve profiles. Sequence analysis revealed that these 15 HRM curve profiles correspond to 18 distinct inlB sequence types. The HRM curve profiles obtained correlated with the five phylogenetic groups I.1, I.2, II.1, II.2, and III. Thus, HRM curve analysis constitutes an inexpensive assay and represents an improvement in typing relative to classical serotyping or multiplex PCR typing protocols. This method provides a rapid and powerful screening tool for simultaneous preliminary typing of up to 384 samples in approximately 2 hours.
Listeria monocytogenes, the causative agent of listeriosis, is a facultative intracellular pathogen of humans and animals. It is widespread in the environment and has the ability to survive and grow under extreme conditions. Patients with listeriosis show symptoms such as gastroenteritis, encephalitis, meningitis, and septicemia. The high case-fatality proportion of approximately 20% to 30% makes L. monocytogenes an important human pathogen.1–3
In listeriosis outbreaks and for epidemiological investigations, a fast and accurate protocol for subtyping L. monocytogenes is essential.4 In outbreak situations the L. monocytogenes serotyping scheme based on somatic (O) and flagellar (H) antigens5 has limited value for tracking isolates. Of the 13 serotypes of L. monocytogenes, only a small fraction (serotypes 4b, 1/2a, and 1/2b) account for more than 96% of reported human listeriosis cases in Austria.2 Insufficient reproducibility of serotyping,6 relatively low discriminatory power, and antigen sharing among serotypes all impede the value of serotyping in outbreak investigations and so demand more accurate molecular-based typing solutions.7 Research toward development of molecular typing protocols based on virulence gene analysis, ribotyping, and microarray analysis revealed that L. monocytogenes comprises five phylogenetic groups (PG).8–11 Recently developed multiplex PCR serotyping methods allow a differentiation of isolates on the PG level.12,13 Phylogenetic groups have been correlated with serotypes: PG I.1 with serotype 1/2a, 3a; I.2 with 1/2c, 3c; II.1 with 4b, 4d, 4e; II.2 with 1/2b, 3b, 7; and III with 4a, 4c.10 In cases of outbreak, the discriminatory power of multiplex PCR serotyping methods is insufficient, and it is necessary to type isolates by pulsed-field gel electrophoresis (PFGE), which is the current gold standard for L. monocytogenes typing. The PFGE method is time-consuming, however, and is difficult to standardize, which hampers interlaboratory exchange and comparison of typing results.14
Typing methods by DNA sequence analysis and single nucleotide polymorphism (SNP) detection appear more promising for fast and accurate strain typing. Recently developed multilocus sequence typing and multivirulence locus sequence typing protocols are accurate and highly discriminative procedures for subtyping of L. monocytogenes strains.14–17 The improved discriminatory power of multivirulence locus sequence typing, compared with multilocus sequence typing and PFGE, was demonstrated by subtyping of genetically diverse L. monocytogenes isolates.18,19
For rapid identification and typing of isolates in routine diagnostics, a PCR-based typing method targeting a single genetic region would be preferable in terms of cost, simplicity, turnaround time, and potential for standardization,20 because both multilocus sequence typing and multivirulence locus sequence typing still represent time-consuming and cost-intensive approaches.
High-resolution analysis of DNA melting curves represents a new, simple, rapid, and precise mutation detection method applicable to genotyping.21–23 The specificity of SNP detection using high-resolution melting (HRM) curve technology is comparable to DNA sequencing and surpasses specific probe-dependent classical PCR genotyping methods. Gene scanning by HRM allows the detection of unknown sequence alterations within the amplification product, in addition to known mutations.21 The potential of HRM curve technology to detect diverse mutations in a single, simple PCR step makes the method very powerful for SNP-based typing applications.
The aim of the present study was the development of a rapid typing method based on polymorphisms of the internalin B gene (inlB).24–26 Preliminary sequence analysis of various virulence genes (prfA, inlB, inlC, dal, clpP, lisR, inlA, actA), as well as housekeeping genes (abcZ, dat, ldh, sod, cat, dapE, pgm, bglA, lhkA) on an arbitrary selection of isolates revealed that inlB showed the highest genetic variability (unpublished data). This observation indicated that inlB could be a promising candidate for HRM curve-based typing.
Materials and Methods
Microorganisms
One hundred seventy-two clinical isolates were provided by the Austrian Reference Center for Listeria, isolated from human cases from 2004 to 2009. Twenty reference strains of serotypes 1/2a (DSMZ 20600), 1/2b (DSMZ 19094, CIP 105449), 1/2c (CIP 103573, ATCC 19112), 3b (CIP 80.10, CIP 78.35), 3c (CIP 78.36), 4a (ATCC 19114, CIP 105457, CIP 61.4), 4b (LMG 23189, LMG 23905, ATCC 13932, LMG 23356, DSMZ 15675), 4d (CIP 105458, ATCC 19117), 4e (ATCC 19118), and 7 (CIP 78.43) were purchased from culture collections, as follows: American Type Culture Collection (ATCC, Manassas, VA), the German Collection of Microorganisms and Cell Cultures [Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany], the Collection of the Laboratory for Microbiology and Microbial Genetics [Laboratorium voor Microbiologie en Microbiele Genetica (LMG), Gent, Belgium], and the Collection of the Pasteur Institute [Collection de l'Institut Pasteur (CIP), Paris, France]. Isolates of serotypes 4c and 4ab were not available and therefore were not tested.
Genomic bacterial DNA (gDNA) was extracted from bacterial cells grown overnight at 37°C on RAPID′L.mono agar (Bio-Rad, Vienna, Austria) using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Purified gDNA was quantified spectrophotometrically at 260 nm and the gDNA quality was assessed by the 260/280 ratio.27
HRM Analysis
A 500-bp fragment located in the virulence gene internalin B (inlB) was amplified for subsequent HRM analysis using the forward primer inlB-forward (5′-CATGGGAGAGTAACCCAACC-3′) and the reverse primer inlB-reverse (5′-GCGGTAACCCCTTTGTCATA-3′).17
Both PCR and HRM were performed in a single run on a LightCycler 480 instrument (Roche Diagnostics, Penzberg, Germany) in a reaction mix containing 10 ng of genomic DNA, 0.25 pmol of each primer, and 3 mmol/L MgCl2 in the LightCycler 480 high-resolution melting master mix containing ResoLight dye (Roche Diagnostics) with PCR-grade water adjusted to a final volume of 10 μL. Reaction conditions included an activation step at 95°C for 10 minutes followed by 50 cycles at 95°C for 10 seconds, 60°C for 10 seconds, and 72°C for 40 seconds. Before HRM, the amplification products were heated to 95°C for 1 minute and then cooled to 40°C for 1 minute. The HRM was performed over a range from 60°C to 95°C, rising at 1°C/s with 25 acquisitions per 1°C step. All reactions were performed in quadruplicate using an epMotion workstation (Eppendorf, Hamburg, Germany) for automatic sample preparation in 384-well microtiter plates (LC plate; Roche Diagnostics, Vienna, Austria).
For generating HRM curves, LC480 gene scanning software version 1.5 was used with manual settings for sensitivity to 0.30, for temperature shift to threshold four, and a pre-melt normalization range from 70°C to 78°C and a postmelt normalization range from 85°C to 90°C. Difference plots were generated by selecting a negative control, converting the melting profile to a horizontal line, and normalizing the melting profiles of the other samples against this sample. The HRM was performed only on amplification products that reached the plateau phase.
Sequencing
For all samples, inlB was subsequently sequenced from position 934 to 1433. The PCR products for sequencing were amplified with primers inlB-F and inlB-R containing a respective M13 sequence attached to the 5′ end. Sequence analysis was performed using a SequiTherm Excel II DNA sequencing kit (Epicentre Biotechnologies, Madison, WI) with fluorescent-labeled primers M13 universal (5′-TGTAAAACGACGGCCAGT) and M13 reverse (5′-CAGGAAACAGCTATGACC) (MWG-Biotech, Ebersberg, Germany) on a Li-Cor 4300 automated DNA sequencer v2.0 (LI-COR Bioscience, Lincoln, NE), according to the manufacturer's instructions. All sequences obtained were assembled, edited and compared using a Li-Cor AlignIR software package to determine sequence variations.
Results
Gene scanning of a 500-bp amplification product of inlB on all 192 L. monocytogenes isolates yielded 15 distinct HRM curve profiles (Figure 1). Subsequent sequence analysis of the amplification products revealed that these 15 specific HRM curve profiles originated from 18 different inlB sequence types (inlB STs) (Table 1).
Figure 1.
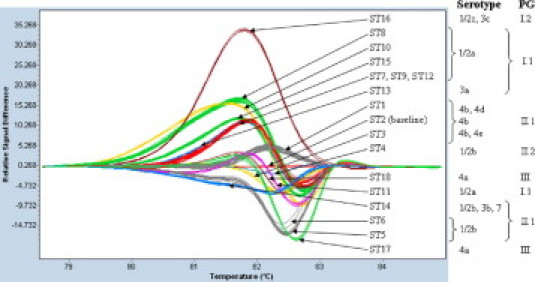
Difference plot of 18 diverse inlB sequence types (ST) obtained from 192 L. monocytogenes isolates of all five phylogenetic groups (PG) comprising serotypes 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4b, 4d, 4e, and 7. The baseline is represented by an inlB ST-2 curve profile.
Table 1.
Single-Nucleotide Polymorphisms and Mutations Detected by HRM Curve Analysis among 192 Investigated L. monocytogenes Isolates from Position 934 to 1433 of the Coding Region of the Internalin B Gene (inlB)
Phylogenetic group II.1 comprises inlB sequence types ST-1 through ST-3. Phylogenetic group II.2 comprises ST-4 through ST-6 (all serotype 1/2b) and ST-14 (serotypes 1/2b, 3b, and 7). Phylogenetic group I.1 comprises ST-7 through ST-12 and also ST-15 (all serotype 1/2a), as well as ST-13 (serotype 3a). Phylogenetic group 1.2 comprises ST-16. Phylogenetic group III comprises ST-17 and ST-18.
—, unchanged from reference sequence (ie, ST-1); HRM, high-resolution melting; PG, phylogenetic group; ST, sequence type.
The correlation of the HRM curve profiles and inlB STs with the investigated serotypes and the five phylogenetic groups PG I.1 (serotypes 1/2a, 3a), PG I.2 (serotypes 1/2c, 3c), PG II.1 (serotypes 4b, 4d, 4e), PG II.2 (serotypes 1/2b, 3b, 7), and PG III (serotypes 4a, 4c) is presented in Figure 1 and in Tables 1 and 2. Six specific HRM curve profiles were obtained for 80 PG I.1 (1/2a, 3a) isolates, one for seven PG I.2 (1/2c, 3c) isolates, three for 76 PG II.1 (4b, 4d, 4e) isolates, four for 26 PG II.2 (1/2b, 3b, 7) isolates, and two for three PG III (4a) isolates (Figure 1).
Table 2.
Consensus of Serotypes to inlB HRM Curve Profiles and inlB Sequence Types in the Tested Reference Strains and Clinical Isolates of Listeria monocytogenes
| PG | Serotype | inlB HRM curve profile | inlB ST | Reference strains | No. of clinical isolates |
|---|---|---|---|---|---|
| I.1 | 1/2a | ST8 | 8 | — | 10 |
| I.1 | 1/2a | ST10 | 10 | — | 5 |
| I.1 | 1/2a | ST11 | 11 | — | 3 |
| I.1 | 1/2a | ST15 | 15 | — | 28 |
| I.1 | 1/2a | ST7-9-12-13 | 7 | — | 11 |
| I.1 | 1/2a | ST7-9-12-13 | 9 | DSMZ 20600 | 15 |
| I.1 | 1/2a | ST7-9-12-13 | 12 | — | 4 |
| I.1 | 3a | ST7-9-12-13 | 13 | — | 3 |
| I.2 | 1/2c | ST16 | 16 | CIP 103573, ATCC 19112 | 3 |
| I.2 | 3c | ST16 | 16 | CIP 78.36 | 1 |
| II.1 | 4b | ST1 | 1 | LMG 23189, LMG 23905 | 27 |
| II.1 | 4b | ST2 | 2 | ATCC 13932, LMG 23356, DSMZ 15675 | 32 |
| II.1 | 4b | ST3 | 3 | — | 9 |
| II.1 | 4d | ST1 | 1 | CIP 105458, ATCC 19117 | — |
| II.1 | 4e | ST3 | 3 | ATCC 19118 | — |
| II.2 | 1/2b | ST4 | 4 | — | 4 |
| II.2 | 1/2b | ST5 | 5 | — | 14 |
| II.2 | 1/2b | ST6 | 6 | — | 2 |
| II.2 | 1/2b | ST14 | 14 | CIP 105449, DSMZ 19094 | 1 |
| II.2 | 3b | ST14 | 14 | CIP 80.10, CIP 78.35 | — |
| II.2 | 7 | ST14 | 14 | CIP 78.43 | — |
| III | 4a | ST17 | 17 | ATCC 19114, CIP 105457 | — |
| III | 4a | ST18 | 18 | CIP 61.4 | — |
ATCC, American Type Culture Collection, Manassas, VA; CIP, Collection of the Pasteur Institute, Paris, France; DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; HRM, high-resolution melting; inlB, internalin B gene; LMG, Collection of the Laboratory for Microbiology and Microbial Genetics, Gent, Belgium; PG, phylogenetic group; ST, sequence type.
On the phylogenetic group level, PG I.1 differed from PG I.2 by at least 1 specific SNP (ACC351ACT), PG I.1 from PG II.1 by at least 24 SNPs, PG I.1 from PG II.2 by at least 21 SNPs, and PG I.1 from PG III by at least 23 SNPs. PG II.1 differed from PG II.2 by at least 11 SNPs, and PG II.1 from PG III by at least two SNPs. PG II.2 differed from PG III by at least 11 SNPs (Table 1).
The 77 investigated serotype 1/2a isolates displayed seven distinct inlB STs and five distinct HRM profiles (Tables 1 and 2, Figure 1). The HRM curve profiles of serotype 1/2a isolates of inlB ST-7, inlB ST-9, and inlB ST-12 were indistinguishable from each other. Sequence analysis showed two SNPs (GAG339GAA and CGA343CGG) differentiating inlB ST-9 from inlB ST-12, four SNPs (GTG390GCG, GCA396ACA, AAA439AGA, and GGG445GGA) differentiating inlB ST-7 from inlB ST-12, and all six SNPs differentiating inlB ST-7 from inlB ST-9 (Table 1). The three serotype 3a isolates exhibited a specific inlB ST-13. The inlB ST-13 HRM curve profile was indistinguishable from the common curve profile of inlB ST-7, inlB ST-9, and inlB ST-12 (Figure 1). The inlB ST-13 serotype 3a isolates differed from serotype 1/2a inlB ST-9 by a single SNP (ACC400ACT), from inlB ST-12 by three SNPs (GAG339GAA, CGA343CGG, and ACC400ACT), and from inlB ST-7 by seven SNPs (GAG339GAA, CGA343CGG, GTG390GCG, GCA396ACA, ACC400ACT, AAA439AGA, and GGG445GGA) (Table 1).
Serotype 1/2b isolates belonged to inlB ST-4, inlB ST-5, inlB ST-6, inlB ST-14 and differed from each other by specific SNPs in positions 401, 427, and 435 (Table 1). Each sequence type had a characteristic HRM curve profile (Figure 1, Table 2). The inlB ST-14 comprised three serotype 1/2b isolates, two serotype 3b isolates, and the serotype 7 reference strain (Table 2).
The five serotype 1/2c and the two serotype 3c isolates shared a common inlB ST-16 with a unique HRM curve profile (Figure 1, Table 2).
The three investigated serotype 4a isolates displayed two characteristic HRM curve profiles, derived from inlB ST-17 and inlB ST-18 (Figure 1). The two serotype 4a HRM curve profiles differed from each other by five SNPs (GCA396GCG, GTG397AAC, ACA400ACC, GGG411GGA, and CTT427CTA) (Table 1).
The 73 serotype 4b isolates had three characteristic HRM curve profiles, derived from three sequence types (inlB ST-1, inlB ST-2, and inlB ST-3) (Figure 1, Tables 1 and 2). The inlB ST-1 contained, in addition to the 29 serotype 4b isolates, the two investigated 4d isolates (Table 2). The inlB ST-3 harbored nine serotype 4b isolates and the single serotype 4e reference isolate (Table 2).
To verify the reproducibility of HRM curve profiles, all 192 isolates were tested in four separate experiments. The profiles of all HRM curves that achieved the standards for HRM analysis were assignable to the corresponding STs (100% reproducibility). A subset of 32 arbitrarily chosen isolates from all inlB STs was blindly retested in quadruplicate. The curve profiles of all isolates could be correctly assigned to the corresponding STs (again 100% reproducibility).
Discussion
In an outbreak situation, the availability of a rapid and accurate typing method is crucial.20 Mutation detection through HRM curve analysis is an attractive method, because it is a closed-tube method that requires only a simple PCR, saturating double-strand DNA dye, and a HRM instrument.21 We evaluated the feasibility of gene scanning to detect specific SNPs within a particular fragment of the inlB and so to improve the rapid typing of L. monocytogenes in comparison with currently available PCR typing protocols.
A 500-bp inlB fragment was amplified for 192 L. monocytogenes isolates comprising 11 of the 13 described L. monocytogenes serotypes. Only isolates of serotype 4c and 4ab were not included in the present study, because of unavailability.
The resulting HRM curve profiles exhibit a clear correlation with both phylogenetic groups and the investigated serotypes, and allow differentiation beyond the serotype level for isolates of serotypes 1/2a, 1/2b, 4a, and 4b. In contrast to several recently described molecular serotyping approaches, which separate L. monocytogenes isolates of diverse serotypes into four groups,12,13,28 the inlB HRM curve profiling separated the 11 investigated L. monocytogenes serotypes into at least 15 specific HRM curve profiles comprising 18 different sequence types. Nonetheless, as described for other PCR-based typing methods,12,13,28 HRM curve profiling does not allow differentiation of serotype 3a isolates from serotype 1/2a inlB ST-7, inlB ST-9, and inlB ST-12 isolates, of serotype 1/2c from serotype 3c isolates, of some serotype 1/2b isolates from serotype 3b and from serotype 7 isolates, of some serotype 4b isolates from serotype 4d isolates, and of some serotype 4b isolates from serotype 4e isolates. Nevertheless, the failure of HRM curve profiling in differentiating certain serotypes does not affect the usability and accuracy of inlB gene scanning as a fast preliminary typing technique, given the rare occurrence of L. monocytogenes serotypes 3a, 3b, 7, 4d, and 4e isolates as causative pathogens of clinical listeriosis.3 De facto, all serotype 3a, 3b, 7, 4d, and 4e isolates investigated in this study were from culture collections.
An advantage of typing by HRM curve profiling is the differentiation of the three most frequent serotype isolates 1/2a, 1/2b, and 4b (responsible for more than 96% of human cases of listeriosis in Austria2) into 13 distinct subgroups. This explicit advancement, relative to classical serotyping, renders inlB gene scanning a first choice for gross typing, ahead of more sophisticated and time-consuming typing methods such as PFGE, multilocus sequence typing, or multivirulence locus sequence typing.
A challenge for the successful development of the described assay was the large amplicon size (500 bp), because a single mutation affects the melting characteristics of a small amplicon more than the melting characteristics of a large amplicon.29 According to Wittwer et al,21 the amplicon size should not exceed 250 bp; however, the HRM curve profiles of the developed gene scanning assay covering 500 bp of the inlB allowed accurate detection of the single SNP class 1 mutations ACC351ACT, AAT371GAT, GTC397GCC, and CGC401CGT, distinguishing inlB ST-15 from inlB ST-16, inlB ST-2 from inlB ST-3, inlB ST-8 from inlB ST-10, and inlB ST-5 from inlB ST-14. Using DINAMelt Server software,30 the predicted difference in melting temperature (Tm) was as low as 0.2°C between inlB ST-15 and inlB ST-16, between inlB ST-2 and inlB ST-3, between inlB ST-8 and inlB ST-10, and between inlB ST-5 and inlB ST-14. Furthermore, even a SNP class 4 ATA427TTA mutation differentiating inlB ST-6 from inlB ST-14 displaying a Tm of <0.2°C was accurately detectable.
Note, however, that the four sequence types inlB ST-7, inlB ST-9, inlB ST-12, and inlB ST-13 share identical HRM curve profiles, although distinguishable by two to seven SNPs or mutations. An explanation for these indistinguishable HRM curve profiles could be the erasing effect on the Tm caused by these diverse SNPs or mutations. When we tested the experimental results through DINAMelt Server software,30 the analysis verified identical Tm values for the four sequence types (ie, inlB ST-7, inlB ST-9, inlB ST-12, and inlB ST-13). A differentiation of these identical HRM curve profiles from diverse inlB STs may be achieved by adjacent sequence analysis of the inlB amplicons extending the total assay time by approximately 2 hours. Nevertheless, a differentiation by a single PCR step may be achieved through the addition of a reference DNA or of unlabeled probes (spiking) to the reaction, which will form heteroduplex amplicons that might exhibit differentiable HRM curve profiles.23,31,32
In conclusion, mutation scanning by HRM curve analysis of a 500-bp inlB fragment identified the clinically most relevant L. monocytogenes serotypes and enabled rapid, reliable, simple, and cost-effective typing of L. monocytogenes. Application of this technique differentiated 15 HRM subtypes and enabled the analysis of 384 samples in less than 2 hours using an LightCycler 480 instrument. This method was successfully used for screening more than 100 food isolates during a multinational outbreak of listeriosis in 2009 and 2010 in Austria, Germany, and the Czech Republic.4 The fast availability of results by HRM curve analysis made it an indispensable tool for tentative and rapid assignment of human listeriosis cases to the outbreak. The HRM curve profiling allowed unambiguous differentiation between the two outbreak clones, as well as immediate identification of new cases as either outbreak-related or unrelated sporadic clones.4 The HRM curve analysis of the inlB does not, however, have the discriminatory power of PFGE pattern analysis, which still constitutes the gold standard for L. monocytogenes typing. Nevertheless, the length of analysis time of at least 3 days for PFGE typing impedes its value in an outbreak situation, given tremendous political and media pressure and clinical need, and points to the merit of a rapid typing method like HRM curve profiling.
Acknowledgments
We thank the Austrian Reference Center for Listeria for supplying strains and serotype data.
References
- 1.Vazquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W., Gonzalez-Zorn B., Wehland J., Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasper S., Huhulescu S., Auer B., Heller I., Karner F., Würzner R., Wagner M., Allerberger F. Epidemiology of listeriosis in Austria. Wien Klin Wochenschr. 2009;121:113–119. doi: 10.1007/s00508-008-1130-2. [DOI] [PubMed] [Google Scholar]
- 3.Allerberger F., Wagner M. Listeriosis: a resurgent infection. Clin Microbiol Infect. 2010;16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 4.Fretz R., Pichler J., Sagel U., Much P., Ruppitsch W., Pietzka A.T., Stöger A., Huhulescu S., Heuberger S., Appl G., Werber D., Stark K., Prager R., Flieger A., Karpísková R., Pfaff G., Allerberger F. Update: Multinational listeriosis outbreak due to “Quargel,” a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009–2010. Euro Surveill. 2010;15 pii=19543. [PubMed] [Google Scholar]
- 5.Seeliger H.P.R., Höhne K. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 1979;13:31–48. [Google Scholar]
- 6.Schönberg A., Bannerman E., Courtieu A.L., Kiss R., McLauchlin J., Wilhelms D. Serotyping of 80 strains from the WHO multicentre international typing study of Listeria monocytogenes. Int J Food Microbiol. 1996;32:279–287. doi: 10.1016/s0168-1605(96)01142-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu D. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol. 2006;55:645–659. doi: 10.1099/jmm.0.46495-0. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen O.F., Skouboe P., Dons L., Rossen L., Olsen J.E. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- 9.Wiedmann M., Bruce JL, Keating C., Johnson A.E., McDonough P.L., Batt C.A. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Call D.R., Borucki M.K., Besser T.E. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J Clin Microbiol. 2003;41:632–639. doi: 10.1128/JCM.41.2.632-639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borucki M.K., Call D.R. Listeria monocytogenes serotype identification by PCR. J Clin Microbiol. 2003;41:5537–5540. doi: 10.1128/JCM.41.12.5537-5540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai S., Kabuki D.Y., Kuaye A.Y., Cargioli T.G., Chung M.S., Nielsen R., Wiedmann M. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J Clin Microbiol. 2002;40:3319–3325. doi: 10.1128/JCM.40.9.3319-3325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salcedo C., Arreaza L., Alcalá B., de la Fuente L., Vázquez J.A. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J Clin Microbiol. 2003;41:757–762. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Revazishvili T., Kotetishvili M., Stine O.C., Kreger A.S., Morris J.G., Jr., Sulakvelidze A. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J Clin Microbiol. 2004;42:276–285. doi: 10.1128/JCM.42.1.276-285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W., Jayarao B.M., Knabel S.J. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl Environ Microbiol. 2004;70:913–920. doi: 10.1128/AEM.70.2.913-920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Zhang W., Knabel S.J. Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J Clin Microbiol. 2007;45:835–846. doi: 10.1128/JCM.01575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomonaco S., Chen Y., Knabel S.J. Analysis of additional virulence genes and virulence gene regions in Listeria monocytogenes confirms the epidemiologic relevance of multi-virulence-locus sequence typing. J Food Prot. 2008;71:2559–2566. doi: 10.4315/0362-028x-71.12.2559. [DOI] [PubMed] [Google Scholar]
- 20.van Belkum A., Tassios P.T., Dijkshoorn L., Haeggman S., Cookson B., Fry N.K., Fussing V., Green J., Feil E., Gerner-Smidt P., Brisse S., Struelens M. M; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM): Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(Suppl 3):S1–S46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 21.Wittwer C.T., Reed G.H., Gundry C.N., Vandersteen J.G., Pryor R.J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 22.Reed G.H., Kent J.O., Wittwer C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 23.Pietzka A.T., Indra A., Stöger A., Zeinzinger J., Konrad M., Hasenberger P., Allerberger F., Ruppitsch W. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J Antimicrob Chemother. 2009;63:1121–1127. doi: 10.1093/jac/dkp124. [Erratum appears in J Antimicrob Chemother 2009, 64:436] [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen O.F., Beck T., Olsen J.E., Dons L., Rossen L. Listeria monocytogenes isolates can be classified into two major types according to the sequence of the listeriolysin gene. Infect Immun. 1991;59:3945–3951. doi: 10.1128/iai.59.11.3945-3951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ericsson H., Unnerstad H., Mattsson J.G., Danielsson-Tham M.L., Tham W. Molecular grouping of Listeria monocytogenes based on the sequence of the inlB gene. J Med Microbiol. 2000;49:73–80. doi: 10.1099/0022-1317-49-1-73. [DOI] [PubMed] [Google Scholar]
- 26.Unnerstad H., Ericsson H., Alderborn A., Tham W., Danielsson-Tham M.L., Mattsson J.G. Pyrosequencing as a method for grouping of Listeria monocytogenes strains on the basis of single-nucleotide polymorphisms in the inlB gene. Appl Environ Microbiol. 2001;67:5339–5342. doi: 10.1128/AEM.67.11.5339-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. ed 3. Cold Spring Harbor Laboratory Press; New York: 2001. pp. 19–21. A8. [Google Scholar]
- 28.Tamburro M., Ripabelli G., Fanelli I., Grasso G.M., Sammarco M.L. Typing of Listeria monocytogenes strains isolated in Italy by inlA gene characterization and evaluation of a new cost-effective approach to antisera selection for serotyping. J Appl Microbiol. 2010;108:1602–1611. doi: 10.1111/j.1365-2672.2009.04555.x. [DOI] [PubMed] [Google Scholar]
- 29.Reed G.H., Wittwer C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 30.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palais R., Liew M., Wittwer C. Quantitative heteroduplex analysis for single nucleotide polymorphism genotyping. Anal Biochem. 2005;346:167–175. doi: 10.1016/j.ab.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Hoek K.G., Gey van Pittius N.C., Moolman-Smook H., Carelse-Tofa K., Jordaan A., van der Spuy G.D., Streicher E., Victor T.C., van Helden P.D., Warren R.M. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J Clin Microbiol. 2008;46:1369–1373. doi: 10.1128/JCM.02343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


