Abstract
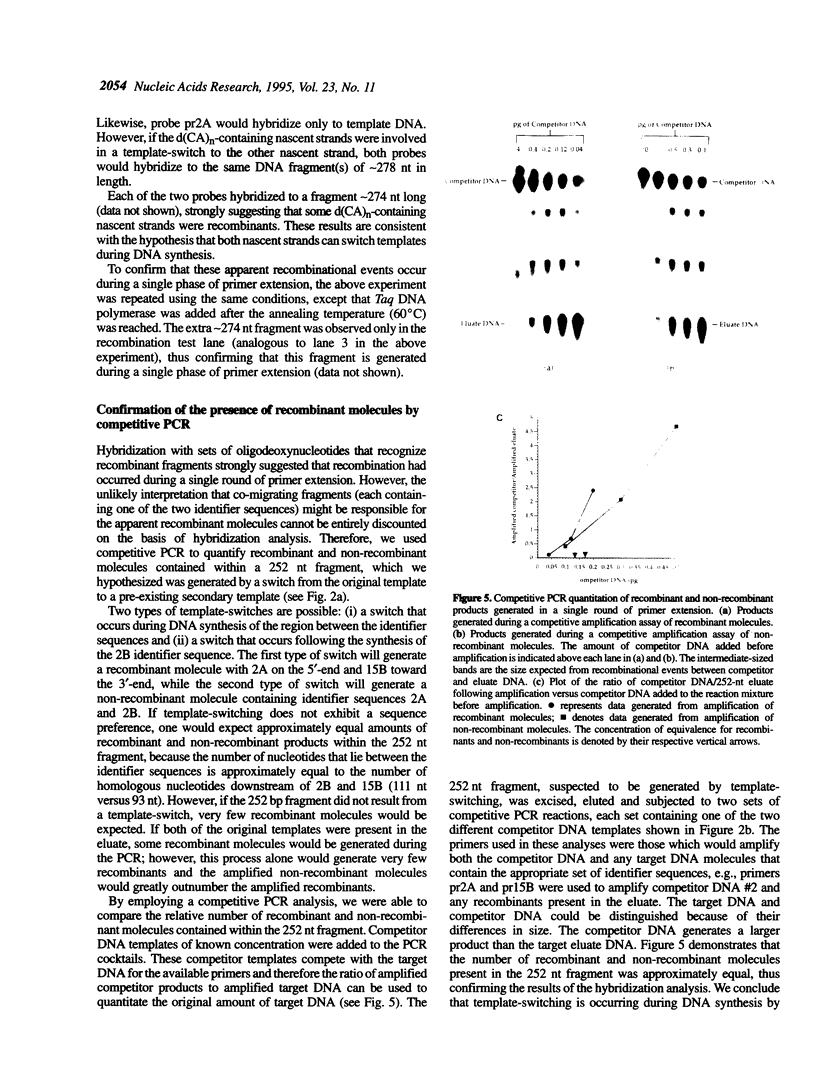
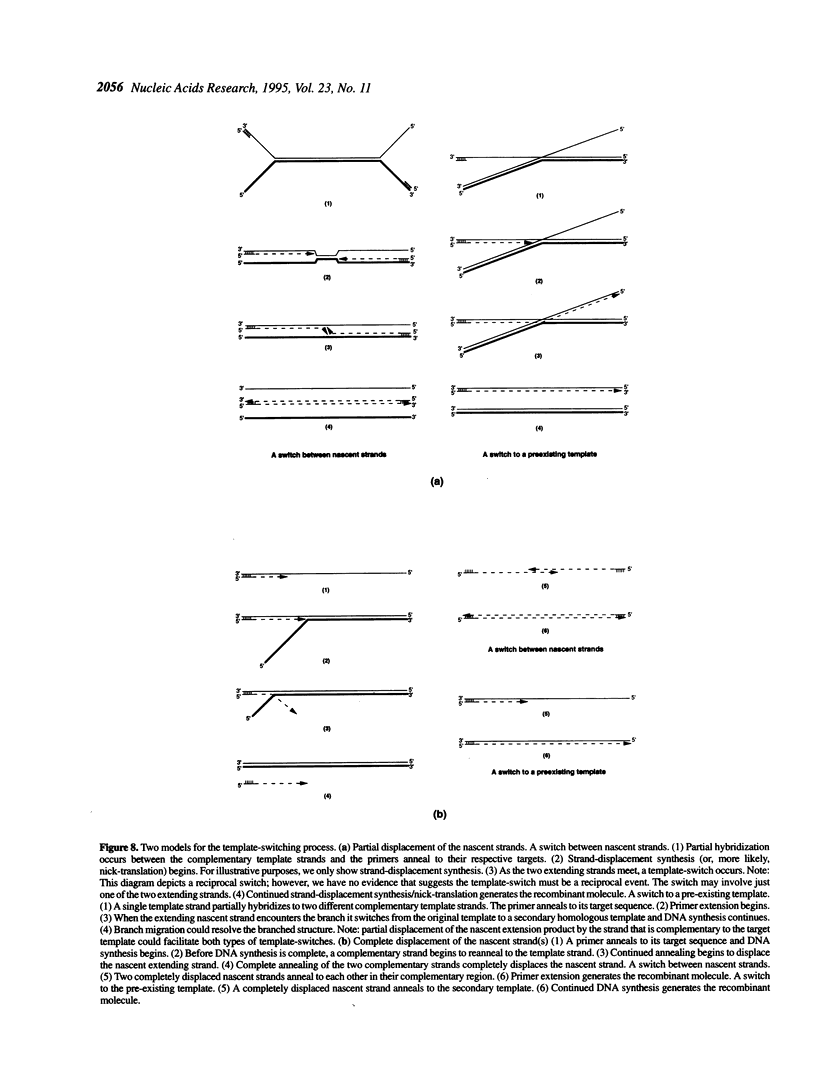
Recombinant DNA molecules are often generated during the polymerase chain reaction (PCR) when partially homologous templates are available [e.g., see Pääbo et al. (1990) J. Biol. Chem. 265, 4718-4721]. It has been suggested that these recombinant molecules are a consequence of truncated extension products annealing to partially homologous templates on subsequent PCR cycles. However, we demonstrate here that recombinants can be generated during a single round of primer extension in the absence of subsequent heat denaturation, indicating that template-switching produces some of these recombinant molecules. Two types of template-switches were observed: (i) switches to pre-existing templates and (ii) switches to the complementary nascent strand. Recombination is reduced several fold when the complementary template strands are physically separated by attachment to streptavidin magnetic beads. This result supports the hypothesis that either the polymerase or at least one of the two extending strands switches templates during DNA synthesis and that interaction between the complementary template strands is necessary for efficient template-switching.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bouzoubaa S., Niesbach-Klösgen U., Jupin I., Guilley H., Richards K., Jonard G. Shortened forms of beet necrotic yellow vein virus RNA-3 and -4: internal deletions and a subgenomic RNA. J Gen Virol. 1991 Feb;72(Pt 2):259–266. doi: 10.1099/0022-1317-72-2-259. [DOI] [PubMed] [Google Scholar]
- Bron S., Holsappel S., Venema G., Peeters B. P. Plasmid deletion formation between short direct repeats in Bacillus subtilis is stimulated by single-stranded rolling-circle replication intermediates. Mol Gen Genet. 1991 Apr;226(1-2):88–96. doi: 10.1007/BF00273591. [DOI] [PubMed] [Google Scholar]
- Buiser R. G., DeStefano J. J., Mallaber L. M., Fay P. J., Bambara R. A. Requirements for the catalysis of strand transfer synthesis by retroviral DNA polymerases. J Biol Chem. 1991 Jul 15;266(20):13103–13109. [PubMed] [Google Scholar]
- Bujarski J. J., Kaesberg P. Genetic recombination between RNA components of a multipartite plant virus. 1986 May 29-Jun 4Nature. 321(6069):528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M., Kohama T., Sugiura A. A measles virus subgenomic RNA: structure and generation mechanism. Virology. 1989 Aug;171(2):427–433. doi: 10.1016/0042-6822(89)90611-9. [DOI] [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986 Dec 5;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Fujimura R. K., Roop B. C. Characterization of DNA polymerase induced by bacteriophage T5 with DNA containing single strand breaks. J Biol Chem. 1976 Apr 10;251(7):2168–2174. [PubMed] [Google Scholar]
- Goodrich D. W., Duesberg P. H. Retroviral recombination during reverse transcription. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2052–2056. doi: 10.1073/pnas.87.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. M., Abramson R. D., Watson R., Gelfand D. H. Detection of specific polymerase chain reaction product by utilizing the 5'----3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Jansen R., Ledley F. D. Disruption of phase during PCR amplification and cloning of heterozygous target sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5153–5156. doi: 10.1093/nar/18.17.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J. G., Stohlman S. A., Soe L. H., Makino S., Lai M. M. Multiple recombination sites at the 5'-end of murine coronavirus RNA. Virology. 1987 Feb;156(2):331–341. doi: 10.1016/0042-6822(87)90413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 1988 Dec 23;16(24):11705–11723. doi: 10.1093/nar/16.24.11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986 Nov 7;47(3):433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Kawamura N., Nomoto A. Genetic variation occurring on the genome of an in vitro insertion mutant of poliovirus type 1. J Virol. 1989 Mar;63(3):1069–1075. doi: 10.1128/jvi.63.3.1069-1075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Saito I., Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986 Dec 5;192(3):473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- Lawyer F. C., Stoffel S., Saiki R. K., Myambo K., Drummond R., Gelfand D. H. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J Biol Chem. 1989 Apr 15;264(11):6427–6437. [PubMed] [Google Scholar]
- Lechner R. L., Engler M. J., Richardson C. C. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983 Sep 25;258(18):11174–11184. [PubMed] [Google Scholar]
- Leppert M., Kort L., Kolakofsky D. Further characterization of Sendai virus DI-RNAs: a model for their generation. Cell. 1977 Oct;12(2):539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton A., Delbecchi L., Bourgaux P. DNA nicking favors PCR recombination. Nucleic Acids Res. 1991 May 11;19(9):2423–2426. doi: 10.1093/nar/19.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamune Y., Richardson C. C. Strand displacement during deoxyribonucleic acid synthesis at single strand breaks. J Biol Chem. 1971 Apr 25;246(8):2692–2701. [PubMed] [Google Scholar]
- Meier E., Harmison G. G., Keene J. D., Schubert M. Sites of copy choice replication involved in generation of vesicular stomatitis virus defective-interfering particle RNAs. J Virol. 1984 Aug;51(2):515–521. doi: 10.1128/jvi.51.2.515-521.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J. P., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990 Apr 11;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima A., Inouye S., Inouye M. In vivo duplication of genetic elements by the formation of stem-loop DNA without an RNA intermediate. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1016–1020. doi: 10.1073/pnas.89.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palasingam K., Shaklee P. N. Reversion of Q beta RNA phage mutants by homologous RNA recombination. J Virol. 1992 Apr;66(4):2435–2442. doi: 10.1128/jvi.66.4.2435-2442.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E. V., Blinov V. M., Agol V. I. Gross rearrangements within the 5'-untranslated region of the picornaviral genomes. Nucleic Acids Res. 1990 Jun 11;18(11):3371–3375. doi: 10.1093/nar/18.11.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päbo S., Irwin D. M., Wilson A. C. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990 Mar 15;265(8):4718–4721. [PubMed] [Google Scholar]
- Re G. G., Morgan E. M., Kingsbury D. W. Nucleotide sequences responsible for generation of internally deleted Sendai virus defective interfering genomes. Virology. 1985 Oct 15;146(1):27–37. doi: 10.1016/0042-6822(85)90050-9. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Cabrera-Juárez E., Griffin K. Mechanism of acquisition of chromosomal markers by plasmids in Haemophilus influenzae. J Bacteriol. 1984 Nov;160(2):662–667. doi: 10.1128/jb.160.2.662-667.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]