Abstract
Colorectal cancer (CRC) is the second most common tumor in developed countries. The present study was undertaken to determine the expression of the sperm-associated antigen 9 gene (SPAG9) as a possible biomarker in CRC, to investigate its correlation with humoral immune response and different stages and grades in CRC patients, and to explore its possible role in colon tumorigenesis in vitro and in an in vivo mouse model. SPAG9 expression was determined by RT-PCR, in situ RNA hybridization, and immunohistochemistry. Humoral response against SPAG9 was detected by enzyme-linked immunosorbent assay and Western blotting. SPAG9 gene silencing was performed using plasmid-based small interfering RNA to study various malignant properties of colon cancer cells in vitro and in vivo. The majority of CRC patients showed SPAG9 expression and generated humoral response. There was a close relationship between SPAG9 protein expression and humoral immune response in the majority of early-stage CRC patients, indicating that anti-SPAG9 antibodies could be a novel serum biomarker for early diagnosis. The down-regulation of SPAG9 (mediated by small interfering RNA) inhibited malignant properties in in vitro and significantly suppressed tumor growth in vivo. These findings collectively suggest that SPAG9 may have a role in tumor development and early spread and thus could serve as a novel target for early detection and for cancer immunotherapy.
Colorectal carcinoma (CRC) is the third most common cause of cancer and is the second leading cause of cancer-related death.1 Surgical resection is the primary treatment modality for CRC. Chemotherapy and/or radiotherapy may be recommended, depending on the individual patient's staging and other medical factors. Current chemotherapy for advanced CRC yields only a modest improvement in survival.2 It has been reported, however, that CRC patients have a higher survival rate if the cancer is detected at early stages, before lymphovascular invasion.3 Thus, there is an urgent need to identify tumor biomarkers that may be used for early detection and diagnosis of CRC.
The underlying principle for pursuing colon cancer screening is that detection of early-stage cancer may help reduce the risk of the disease. Because CRC develops slowly from precancerous lesions that can be removed, and because it is curable at early stages, screening for CRC has the potential to reduce both incidence and mortality of the disease.4 However, testing for fecal occult blood and colonoscopy (the primary tools in current use) have the defects of poor compliance and lack of widespread access. A noninvasive biomarker would therefore be of great clinical benefit and could be useful in the early diagnosis of CRC patients.
A unique class of testis proteins known as cancer/testis (CT) antigens has been found to be expressed in various malignancies.5 Based on their restricted expression in cancerous tissues and in view of their immunogenicity in cancer patients, CT antigens are the ideal targets for cancer immunotherapy and serum-based biomarkers.5
Recently, we characterized the sperm-associated antigen 9 gene (SPAG9; HGNC: 14524), newly identified as a member of the CT antigen family, as associated with various malignancies.6 The SPAG9 gene generates eight protein isoforms by alternative splicing, and all differ from the canonical sequence of the full-length protein c-Jun-amino-terminal kinase-interacting protein 4 (JIP4 isoform 1; UniProt O60271-1). It has been reported that approximately three-quarters of all human genes undergo alternative splicing,7–9 which may affect functional role, cellular localization, binding and interacting properties, and stability of the encoded proteins.10 In earlier studies, we found that SPAG9 protein (JIP4 isoform 5; UniProt O60271-5) is involved in the c-Jun N-terminal kinase (JNK) signaling module6,11 and functions as a scaffolding protein for binding to JNKs that play an important regulatory role in several physiological processes, including cell survival, proliferation, apoptosis, and tumor development.6,12
Cancer-specific alternatively spliced mRNAs and protein isoforms may be used as cancer biomarkers. Our recent study demonstrated SPAG9 expression and its association with clinicopathological characteristics of tumors in renal cell carcinoma,13 epithelial ovarian cancer,14 breast cancer,15 cervical carcinoma,16 and thyroid cancer.17 These findings suggest that SPAG9 may have a role in early spread of disease. Furthermore, SPAG9 expression was also found to be associated with circulating anti-SPAG9 antibodies in early stages and in low grade of breast cancer15 and cervical cancer patients,16 suggesting its potential use in early detection of disease.
In the present study, we systematically investigated SPAG9 mRNA and protein expression, as well as in vivo immunogenicity, in human CRC patients and their role in the tumorigenicity of colon cancer. The findings demonstrated a close relationship between SPAG9 expression and humoral immune response in early stages, pointing to a functional role in tumorigenesis of colon cancer. SPAG9 could be considered as a potential noninvasive biomarker and target molecule for developing antigen-based vaccine and immunotherapeutic approaches for treatment of CRC.
Materials and Methods
Patients and Samples
The present investigation was conducted with clinical specimens obtained from CRC patients who underwent surgical resection of the tumor, in accordance with the Institutional Ethics Committee and after obtaining informed consent. SPAG9 gene and protein expression was analyzed in 78 CRC tissues, and humoral response against SPAG9 was investigated in the 54 blood samples available from these patients. The study included 52 men and 26 women, with a median age of 54 ± 15.90 years (range, 25 to 86 years). In 26 cases, paired adjacent noncancerous tissue specimens were collected. We also investigated SPAG9 protein expression in 40 control colon tissue samples, obtained from the archives of the Department of Pathology. Clinicopathological characteristics are given in Table 1. Blood samples were also obtained from 50 normal healthy donors.
Table 1.
Clinicopathological Characteristics of Colorectal Carcinoma Patients and SPAG9 Expression
| Clinicopathological characteristics | SPAG9 expression⁎ |
|
|---|---|---|
| ELISA, n/N (%) | RT-PCR/IHC, n/N (%) | |
| Tumor, total | 38/54 (70) | 58/78 (74) |
| ANCT, total | 0/26 (0) | |
| Normal colon tissue, total | 0/40 (0) | |
| Tumor stages | ||
| Stage I | 1/1 (100) | 7/10 (70) |
| Stage II | 11/11 (100) | 16/16 (100) |
| Early stages (I + II) | 12/12 (100) | 23/26 (88) |
| Stage III | 20/33 (61) | 29/43 (67) |
| Stage IV | 6/9 (67) | 6/9 (67) |
| Late stages (III + IV) | 26/42 (62) | 35/52 (67) |
| Histologic grades | ||
| Well differentiated | 17/23 (74) | 28/37 (76) |
| Moderately differentiated | 17/23 (74) | 26/33 (79) |
| Mucinous | 4/8 (50) | 4/8 (50) |
| Lymph node involvement | ||
| Positive | 26/41 (63) | 30/45 (67) |
| Negative | 12/13 (92) | 28/33 (85) |
| Metastasis | ||
| Positive | 7/10 (70) | 11/14 (79) |
| Negative | 31/44 (70) | 47/64 (73) |
ANCT, adjacent noncancerous tissue; ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry.
The ELISA analysis detected circulating autoantibodies against SPAG9. The RT-PCR and IHC analyses detected SPAG9 mRNA expression.
Cell Lines and Transient Transfection
Human colon cancer cell lines COLO 205 and HCT 116 were procured from the American Type Culture Collection (ATCC, Manassas, VA) and were maintained according to standard procedures. COLO 205 or HCT 116 cells (1 × 105) were plated in 6-well plates, allowed to attach, and were transiently transfected using Lipofectamine reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions and as described previously.13
Detection of SPAG9 Transcript in Cell Lines and CRC Specimens
RT-PCR was performed to investigate the expression of SPAG9 mRNA in colon cancer cells and in CRC tissues. RNA was isolated from colon cancer cells and from 78 colon cancer tissues and 26 paired adjacent noncancerous tissue specimens. RT-PCR was performed using SPAG9 gene-specific primers, as described previously.14 The PCR product thus obtained was cloned into TOPO vector (Invitrogen, Life Technologies) for confirming the SPAG9 DNA sequence.
Immunofluorescence Microscopy and Flow Cytometric Analysis
SPAG9 protein expression in colon cancer cells was examined using indirect immunofluorescence microscopy and flow cytometric analysis using anti-SPAG9 antibody, as described previously.13 Antibodies to pan cadherin (CH-19, ab6528; Abcam, Cambridge, MA), calnexin (6D195, sc-70481; Santa Cruz Biotechnology, Santa Cruz, CA), GM130 (B-10, sc-55591; Santa Cruz Biotechnology), mitochondria (MTCO2 mitochondrial marker, ab3298; Abcam), and lamin A/C (636, sc-7292; Santa Cruz Biotechnology) were used for colocalization studies. Fluorescence microscopy was performed with a Zeiss confocal microscope.
In Vitro Synthesis of Riboprobes and in Situ RNA Hybridization
Sectioning of paraffin-embedded CRC tissue was performed in RNase-free water (water treated with diethyl pyrocarbonate) for probing SPAG9 mRNA expression, as described previously.14
Immunohistochemical Detection of SPAG9 Protein Expression
Paraffin-embedded serial sections of CRC specimens were analyzed for the localization of SPAG9 protein using anti-SPAG9 antibody, as described previously.15 We assessed the immunostaining of SPAG9 by counting >500 cells from each CRC specimen in five random fields under ×400 magnification in each tissue section. The SPAG9 immunoreactivity score was defined as the percentage of CRC cells expressing SPAG9 protein. We considered a distinct positive immunoreactivity in a specimen as showing >10% of cancer cells stained for SPAG9 protein. To confirm SPAG9 immunoreactivity, a neutralization experiment was performed by including recombinant SPAG9 protein (15 μg/ml) in the incubation with primary antibody, as described previously.13
Down-Regulation of SPAG9 Using RNA Interference
In gene silencing experiments, two independent siRNA target sequences directed against SPAG9 were used (designated SPAG9 siRNA-I and SPAG9 siRNA), along with control siRNA (scrambled SPAG9), as described previously.13 COLO 205 or HCT 116 cells were transfected with SPAG9 siRNA-I, SPAG9 siRNA, or control siRNA using Invitrogen Lipofectamine transfection reagent according to the manufacturer's instructions. After 48 hours, cells were harvested and processed for immunoblotting to determine SPAG9 protein expression.
Cell Growth and Colony Formation Assay
The effects of down-regulation of SPAG9 expression using SPAG9 siRNA on cellular growth and colony forming ability in COLO 205 or HCT 116 cells were investigated as described previously.18
Cell Invasion, Migration, and Wound Healing Assay
The effects of SPAG9 siRNA on cell migration, invasion, and wound healing were assessed as described previously.18
In Vivo Tumor Xenograft Studies
Male Nii/NIH (S) (nu/nu) nude mice (5 to 6 weeks old) were used and maintained in pathogen-free conditions. Exponentially growing COLO 205 cells (1 × 107) were injected subcutaneously. Once the mice attained a tumor volume of 50 to 100 mm3, SPAG9 siRNA or control siRNA (50 μg) was administered intratumorally, followed by a booster injection with 25 μg of plasmid twice a week for 6 weeks. Kinetics of tumor formation was estimated by measuring tumor size and volume at 3-day intervals. Tumor size was measured with calipers, and tumor volume was determined using the following formula: volume = 0.5 × width2 × length. At the end of the experiment, animals were sacrificed and tumors were dissected, weighed, and processed for immunohistochemical analysis.
Inhibition of Cellular Proliferation in Vivo in a Xenograft Model of Human Colon Cancer
To detect in vivo inhibition of cellular proliferation by an siRNA approach, murine tumor tissues were processed for histological H&E staining and for immunohistochemistry using anti-Ki-67 and anti-SPAG9 antibody, as described previously.18
ELISA and Western Blot Analysis for Detection of Circulating Autoantibodies against SPAG9
The anti-SPAG9 antibody in CRC patients and in normal healthy donors was detected by enzyme-linked immunosorbent assay (ELISA) and Western blotting using recombinant SPAG9 protein, as described previously.13 Western blot analysis for detecting circulating anti-SPAG9 antibodies was also performed by resolving recombinant SPAG9 protein (0.5 μg/lane) in SDS-polyacrylamide gel electrophoresis and transferring onto polyvinylidene fluoride membrane, as described previously.13
Statistical Analysis
Statistical analyses were performed using Student's t-test, Mann-Whitney U-test and Pearson's χ2 test (Table 2). Results are expressed as means ± SE and represent the results of at least three experiments. Statistical calculations were performed using SPSS version 16.0 (SPSS, Chicago, IL). A two-sided significance level of 0.05 was used for all statistical tests.
Table 2.
Statistical Analysis
| Clinicopathologic features | Mann-Whitney U-test (P value) |
Pearson's χ2 test (P value) |
||
|---|---|---|---|---|
| RT-PCR/IHC | ELISA | RT-PCR/IHC | ELISA | |
| Tumor stage | ||||
| I + II | 0.349 | 0.008⁎ | 0.020⁎ | 0.020⁎ |
| II + III | 0.981 | 0.281 | 0.009⁎ | 0.013⁎ |
| III + IV | 0.358 | 0.051 | 0.740 | 0.740 |
| I + II & III + IV | 0.332 | 0.572 | 0.044⁎ | 0.011⁎ |
| Histological grade | ||||
| WD, MD + mucinous | — | — | 0.239 | 0.393 |
| WD + MD | 0.617 | 0.011⁎ | 0.757 | 0.900 |
| WD + mucinous | 0.912 | 0.091 | 0.146 | 0.213 |
| MD + mucinous | 0.340 | 0.135 | 0.099 | 0.213 |
| Lymph node positivity | 0.493 | 0.338 | 0.069 | 0.047⁎ |
| Metastasis positivity | 0.691 | 0.018⁎ | 0.690 | 0.977 |
ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry; MD, moderately differentiated; WD, well differentiated.
P < 0.05 (statistically significant).
Results
SPAG9 mRNA Expression in CRC Specimens and Cell Lines
Our RT-PCR analysis showed that 58 of 78 (74%) CRC patients expressed SPAG9 mRNA. Both COLO 205 and HCT 116 colon cancer cells also expressed SPAG9 mRNA. No SPAG9 expression was detected in adjacent noncancerous tissue specimens. The size of the PCR product in tumor tissues, colon cancer cells, and in testis was the same (Figure 1A) and showed no mutations, which was confirmed by DNA sequence analysis. The relationship between SPAG9 mRNA expression and clinicopathological features is summarized in Table 1. SPAG9 expression was detected in 23 of 26 (88%) early stage (I and II) and in 35 of 52 (67%) late stage (III and IV) categories of CRC specimens. Based on histopathological grading categories, SPAG9 mRNA expression was detected in 28 of 37 (76%) well differentiated, 26 of 33 (79%) moderately differentiated, and 4 of 8 (50%) mucinous type of CRC specimens. Further, SPAG9 expression was detected in 30 of 45 (67%) CRC specimens with positive lymph node involvement as compared to 28 of 33 (85%) CRC specimens with negative lymph node involvement. In addition, our data indicated that 11 of 14 (79%) metastatic CRC patients revealed SPAG9 expression as compared to 47 of 64 (73%) negative metastatic CRC patients. Statistical analysis using Pearson's χ2 test revealed a significant association between SPAG9 expression and various tumor stages of CRC patients, (early stages I + II, P = 0.020; late stages II + III, P = 0.009; and both early and late stages (P = 0.044) (Table 2), suggesting a close relationship between SPAG9 expression and tumor stage. However, SPAG9 mRNA expression was independent of tumor histological grade, lymph node, and metastasis positivity, indicating no correlation between SPAG9 expression and tumor grade (P = 0.239), lymph node positivity (P = 0.069) and metastasis (P = 0.690). These results suggest that SPAG9 may participate in early cancer growth.
Figure 1.
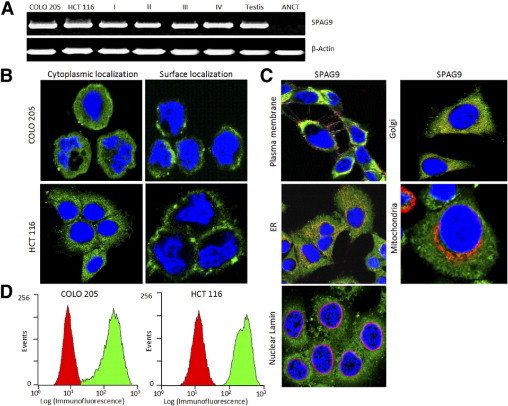
Expression of SPAG9 in CRC specimens and colon cancer cells. A: RT-PCR analysis showed SPAG9 mRNA expression in COLO 205 and HCT 116 human colon cancer cells and in CRC tissues of stage I, II, III, and IV and in adjacent noncancerous tissue (ANCT). β-actin served as the internal control and testis as the positive control. B: Detection of endogenous SPAG9 protein expression in COLO 205 and HCT 116 cells (cytoplasmic localization in fixed and permeabilized cells and surface localization in live cells). Nuclei are stained with DAPI. C: SPAG9 colocalization with various cell organelles. HCT 116 cancer cells were probed for SPAG9 and various markers. Localization of SPAG9 was examined by indirect immunofluorescence assay, and immunofluorescence staining was detected by a laser-scanning confocal microscope. The images are merged for colocalization of SPAG9 (green) and marker costaining (red), using antibodies to plasma membrane (pan cadherin), Golgi (GM130), endoplasmic reticulum (calnexin), mitochondria (MTCO2), and nuclear marker (lamin A/C). Original magnification, ×630; objective, 63×). D: Flow cytometric analysis of SPAG9 surface expression in COLO 205 and HCT 116 colon cancer cells. The control secondary antibody profile is red; the profile for surface localization of SPAG9 protein is green.
SPAG9 Protein Is Expressed in Colon Cancer Cell Lines
Indirect immunofluorescence assays showed strong SPAG9 immunoreactivity in virtually all of the cancer cells, with predominant expression in cytoplasm in fixed and permeabilized cells (Figure 1B) and surface localization in live cells (Figure 1B). To further examine the SPAG9 colocalization with various cell organelles, multiple markers were used, including a plasma membrane marker (pan cadherin), Golgi marker (GM130), an endoplasmic reticulum marker (calnexin), a mitochondrial marker (MTCO2), and nuclear marker (lamin A/C). SPAG9 colocalized with the plasma membrane, Golgi marker, and endoplasmic reticulum markers (Figure 1C), but did not appear to colocalize directly with the mitochondria marker or with nuclear lamin (Figure 1C). Thus, SPAG9 has distinct cytoplasmic and surface localization and colocalization with Golgi, endoplasmic reticulum, and plasma membrane. The surface localization was further confirmed by flow cytometric analysis in COLO 205 and HCT 116 cells (Figure 1D), indicating that SPAG9 may be a potential target molecule for antigen-based vaccine and immunotherapy.
SPAG9 Expression in CRC Specimens
Our in situ RNA hybridization studies using antisense riboprobe further confirmed the expression of SPAG9 mRNA in all stages (Figure 2), whereas sense riboprobe failed to show SPAG9 expression. Subsequently, SPAG9 protein expression was validated by immunohistochemistry in serial tissue sections of CRC and adjacent noncancerous tissue specimens (Figure 3). Notably, cytoplasmic localization of SPAG9 protein expression was observed in 58 of 78 (74%) CRC specimens expressing SPAG9 mRNA, but no expression was detected in 26 paired adjacent noncancerous tissue specimens or in 40 control colon tissue specimens.
Figure 2.

In situ RNA hybridization in different stages of CRC specimens for localization of SPAG9 gene expression. Representative H&E-stained sections of stage I, II, III, and IV are shown. Tissue sections from different CRC stages showed SPAG9 gene expression when probed with anti-sense riboprobe (chocolate brown color), but probing with sense riboprobe failed to show any reactivity. Original magnification, ×200; objective, 20×.
Figure 3.

Immunohistochemical staining of CRC specimens for SPAG9 protein expression. Representative H&E- stained sections of stage I, II, III, and IV are shown. Serial tissue sections at different stages revealed SPAG9 cytoplasmic localization; however, no SPAG9 immunoreactivity was observed in ANCT and control colon specimens or with control IgG. The H&E-stained sections for stages I-IV are identical to those shown in Figure 2. Original magnification, ×200; objective, 20×.
SPAG9 Immunoreactivity Score and Its Association with Clinicopathologic Variables
A significant association was found between SPAG9 expression and various stages [stage I + II (P = 0.020), stages II + III (P = 0.009), and stages I + II & III + IV (P = 0.044)], using Pearson's χ2 test, indicating that SPAG9 expression is strongly associated with tumor stage (Table 2). However, no significant association was found between SPAG9 expression and histological grades (P = 0.239) using the Pearson's χ2 test. Furthermore, based on SPAG9 immunoreactivity score in colorectal tumors, two groupings were analyzed: i) a moderate-score group, with ≤50% tumor cells expressing SPAG9 protein; and ii) a high-score group, with >50% tumor cells expressing SPAG9 protein. Our results indicated that 46 of 58 (79%) CRC patients had high SPAG9 immunoreactivity scores (70.48 ± 1.8), the remainder (12 of 58) having moderate SPAG9 immunoreactivity scores (38.58 + 2.28). When we compared the moderate and high SPAG9 immunoreactivity score groups in terms of stage of disease, using the Mann-Whitney U-test, a significant difference (P < 0.0001) was found, with a greater number of CRC patients demonstrating high SPAG9 expression at early than at late stages. This finding suggests that a majority of CRC patients exhibited elevated SPAG9 protein expression during early stages of the disease. Thus, our analysis on SPAG9 immunoreactivity scores in tumors of early stages indicates that this biomarker may have good predictive value for the early detection of cancer.
CRC Patients Generate Humoral Response against SPAG9
The circulating anti-SPAG9 antibodies were determined in sera of 54 CRC patients, using an enzyme-linked immunosorbent assay (Figure 4A). The enzyme-linked immunosorbent assay titers greater than the mean + 2 SD (0.240 + 0.06) of the 50 normal healthy sera were considered positive. Antibodies against SPAG9 were detected in 70% (38 of 54) in various stages and histological grades of CRC patients (Table 1). The presence of autoantibodies against SPAG9 in the sera of CRC patients found positive in enzyme-linked immunosorbent assay was further examined by Western blotting, which exhibited immunoreactivity against SPAG9 protein regardless of tumor stages. A representative Western blot is shown in Figure 4B. Sera from 50 normal healthy donors revealed no reactivity by Western blotting. It is interesting to mention that all the early-stage CRC patients [Stage I + II: 12 of 12 (100%)] revealed anti-SPAG9 antibody although the sample size was small as compared to late-stage CRC patients [Stage III + IV: 26 of 42 (62%)]. The statistical analysis revealed that there was significant association between circulating anti-SPAG9 antibodies and lymph node involvement (P = 0.047) indicating its potential use as a biomarker in early spread of cancer.
Figure 4.

Circulating anti-SPAG9 antibodies in CRC patients. A: Enzyme-linked immunosorbent assay analysis of sera from CRC patients of different tumor stages (I, II, III, and IV) and grades [well differentiated (WD), moderately differentiated (MD), and mucinous type], as well as 50 normal healthy donors. The horizontal line and X indicate the cutoff (mean + 2 SD of 50 normal healthy donors), with positivity above and negativity below the line. Significant difference in the seroreactivity was observed between normal healthy donors and CRC patients (P < 0.0001). B: Western blotting analysis. Lane 1: Affinity-purified recombinant SPAG9 stained with Coomassie Brilliant Blue. Serum from tumor stage I (lane 2), stage II (lane 3), stage III (lane 4), and stage IV (lane 5) showed specific immunoreactivity with recombinant SPAG9. Neutralization experiments were done by including recombinant SPAG9 (15 μg/ml) in the incubation with sera from stage I (lane 6), stage II (lane 7), stage III (lane 8), and stage IV (lane 9) patients, which resulted in loss of immunoreactivity. Western blot analysis with sera from 50 normal healthy donors revealed no reactivity (lanes 10–13).
Inhibition of Cellular Growth and Colony Forming Ability Using Gene Silencing Approach
Two independent siRNA target sequences directed against SPAG9 (SPAG9 siRNA I; SPAG9 siRNA) were used for gene silencing studies. Our data indicated that SPAG9 siRNA target treatment resulted in ablation of SPAG9 protein expression, but the other target, SPAG9 siRNA-I, did not ablate SPAG9 expression (Figure 5A). We therefore used the SPAG9 siRNA target for all our subsequent experiments (although a weak protein band, representing residual SPAG9 protein expression, was always observed). At 72 hours after transfection with SPAG9 siRNA, cell growth was significantly (P < 0.0001) reduced, to 31% and 33% for COLO 205 and HCT 116, respectively (Figure 5B). In addition, significant reduction in colony-forming ability was observed in SPAG9 siRNA-transfected cells (Figure 5C). Statistical analysis revealed that colony-forming ability was significantly decreased for cell numbers seeded for COLO 205 (P < 0.0001) (range, 29% to 31% for 400 to 1000 cells) and HCT 116 (P < 0.0001) (range, 29% to 30% for 400 to 1000 cells) cells. These data indicate that SPAG9 expression is apparently involved in cell growth and colony-forming ability of colon cancer cells.
Figure 5.

Down-regulation of SPAG9 expression using plasmid-based siRNA in colon cancer cells in vitro. Two independent siRNA target sequences directed against SPAG9 (designated SPAG9 siRNA-I and SPAG9 siRNA) and control siRNA (scrambled SPAG9) were used. A: COLO 205 and HCT 116 cells treated with SPAG9 siRNA revealed ablation of SPAG9 protein, compared with SPAG9 siRNA-I, by Western blotting analysis. No reduction in SPAG9 protein was detected with control siRNA. B and C: Reduced SPAG9 expression suppressed the cellular proliferation (B) and colony forming ability (C) of COLO 205 and HCT 116 cells. Cells were counted, and the results are representative of three independent experiments performed in triplicate. Data are reported as means ± SE of triplicate plates. *P < 0.0001.
SPAG9 Knockdown Suppresses Migration and Invasion Abilities of HCT 116 and COLO 205 Cells
To determine whether SPAG9 is involved in early spread of colorectal carcinoma, the migration and invasion abilities of COLO 205 and HCT 116 cells were evaluated. SPAG9 knockdown suppressed migration and invasion abilities of COLO 205 and HCT 116 cells (Figure 6), with 69% inhibition of migration abilities for COLO 205 (P < 0.0001) and 70% inhibition for HCT 116 (P < 0.0001) (Figure 6A, histograms) (P < 0.0001). Decreased SPAG9 expression also led to the inhibition of invasion by 71% in COLO 205 and 68% in HCT 116 (Figure 6B, histograms) (P < 0.0001). Wound healing assay demonstrated a similar pattern of reduced motility. The reduction in cellular motility of SPAG9 siRNA-transfected colon cancer cells, compared with cells transfected with control siRNA or untreated cells, can be seen distinctly in Figure 6C.
Figure 6.
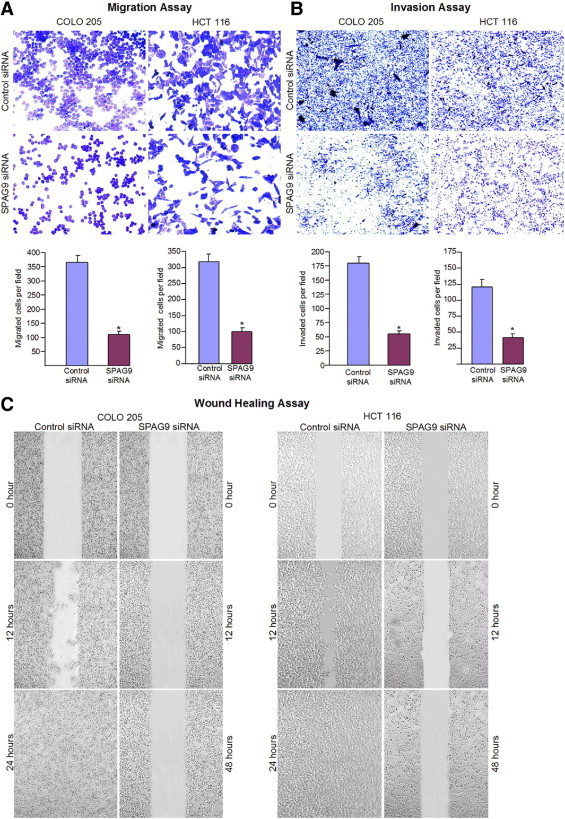
SPAG9 gene silencing significantly inhibited migration and invasion of human COLO 205 and HCT 116 cells. A: At 24 hours after transfection, the cells transfected with SPAG9 siRNA showed reduced migration through Transwell chambers, compared with control siRNA-transfected cells (observations based on three experimental triplicates). B: SPAG9 siRNA significantly impaired the ability of COLO 205 and HCT 116 cells to invade through Matrigel, compared with control siRNA. Data are reported as means ± SE of triple determinations. *P < 0.0001. C: Wound healing assay. Confluent COLO 205 and HCT 116 cells transfected with either SPAG9 siRNA or control siRNA were wounded, and images were acquired at 0, 12, 24, and 48 hours. These results are representative of three independent experiments performed in triplicate.
SPAG9 siRNA Inhibited the Growth of COLO 205 Cell Xenograft
Our in vitro data showed that SPAG9 siRNA reduced SPAG9 protein expression and resulted in suppression of cellular proliferation of COLO 205 and HCT 116 cells. We further validated the effect of SPAG9 siRNA on inhibition of cellular growth of COLO 205 cell xenograft in athymic mice. By day 44, there was significant reduction in the average size (Figure 7A) and volume (Figure 7B) of tumors in SPAG9 siRNA-treated animals, compared with control siRNA-treated animals. On average, SPAG9 siRNA treatment resulted in 70% decreased tumor growth and 71% decreased mass at day 44 (Figure 7C) (P < 0.0001).
Figure 7.

SPAG9 silencing inhibits the growth of colon tumors in vivo. A: Inhibition of subcutaneous solid tumor development in nude mice after treatment with SPAG9 siRNA, compared with control siRNA. At the end of the experiment, the tumors were surgically removed, weighed, and processed for immunohistochemical analysis. B: Tumor volume of nude mice treated with control siRNA (blue diamonds) or with SPAG9 siRNA (pink squares). C: Tumor weight in nude mice was reduced after treatment with SPAG9 siRNA, compared with control siRNA. Data are reported as means ± SE of 6 animals in each group. *P < 0.0001. D: Immunohistochemical analysis for SPAG9 protein expression in tumor specimens revealed strong SPAG9 immunoreactivity in tumors treated with control siRNA, compared with SPAG9 siRNA. Tumor sections probed for cellular proliferation marker Ki-67 demonstrated inhibition of cellular proliferation in tumors treated with SPAG9 siRNA, compared with control siRNA. In histograms, data are reported as positive reactivity of cells for SPAG9 protein or Ki-67, means ± SE. *P < 0.0001.
We examined the association of SPAG9 protein expression with reduced tumor growth. The xenograft tumors were resected and tumor tissue sections were probed for SPAG9 protein expression by immunohistochemistry. It was noticeable that the reduced growth of COLO 205 cell xenograft in vivo was associated with decreased SPAG9 protein expression (Figure 7D). In addition, tumor tissue sections were also probed with cellular proliferating Ki-67 marker, which revealed distinctly reduced tumor cell proliferation in SPAG9 siRNA-treated animals, compared with control siRNA-treated animals (Figure 7D). Our in vivo data further confirm the significant effect of SPAG9 suppression on colon cancer growth and suggest that SPAG9 may be an important target molecule for cancer treatments.
Discussion
CRC is one of the most common malignancies in developed countries, and represents a significant public health issue; worldwide, CRC is reported to be the fourth most common cancer in men and the third most common cancer in women.1 Moreover, significant variations in the geographic distribution of CRC have also been observed.19 The gold standard for colorectal cancer detection is colonoscopy. Because of its invasive nature, however, and for lack of medical resources in developing countries, the majority of CRC patients are diagnosed only at advanced stages. Early detection of CRC and high-risk precursor lesions will certainly help to improve cure rates.
An important issue in carcinogenesis is the migration and invasion of cancer cells, resulting in distant metastasis. Cell migration is a critical feature of numerous physiological and pathological phenomena, including development, wound repair, angiogenesis, and metastasis. Distant metastasis has been reported as the main cause of death in CRC patients.20 Identification of specific target molecules that are associated with tumor aggressiveness may lead to useful new biomarkers for assessing the early spread of CRC. In the present study, we examined the clinical relevance and the potential role of SPAG9 in early spread of CRC. SPAG9 expression was found in both of the colon cancer cell lines examined. Our data revealed that SPAG9 expression was found in CRC specimens, and its expression level was associated with the characteristics of the tumors. Because the invasiveness of cancer cells is dependent on increased migratory and invasive properties, we examined in colon cancer cell lines the potential role of SPAG9 expression in promoting migration and invasion of colon cancer cells. Ablation of SPAG9 protein by using SPAG9 siRNA resulted in significant reduction and alteration of cell migratory and invasive properties of colon cancer cells. The close relationship of SPAG9 expression with early clinical tumor stages (stages I and II) (Table 1) indicates that SPAG9 may play an important role in cellular growth, colony formation (Figure 3), cell migration, and invasion (Figure 4).
Ablation of SPAG9 protein in vivo significantly retarded xenograft tumor growth, which confirms SPAG9 as having a potential role in colorectal carcinogenesis (Figure 5). Our findings further demonstrated that SPAG9 is not only expressed at the mRNA level but is also translated to its protein, as evident from immunohistochemistry analysis of various stages and different grades of CRC. Our recent studies demonstrated that SPAG9 is associated with epithelial ovarian cancer,14 renal cell carcinoma,13 breast carcinoma,15 cervical cancer,16 and thyroid cancer.17 CRC remains a major cause of cancer mortality in developed countries, because most patients are diagnosed at advanced stages. We note that the present results seem applicable to early stages of CRC, supporting the potential role of SPAG9 as a novel biomarker for early detection and diagnosis in cancer management.
Numerous approaches have been used to characterize tumor-associated antigens in cancer, including natural protein arrays prepared with fractions obtained from two-dimensional liquid chromatography separations of cancer samples21 and using high-density protein microarrays containing 8000 human proteins, especially in CRC.22 The majority of these proteins have not been investigated as a potential cancer biomarkers. Only a few proteins have been described as biomarkers in CRC (carcinoembryonic antigen, CA19.9 and CA125,23, and none of these are recommended for clinical screening and cancer management.24
A unique class of tumor-associated antigens encoded by the CT gene family is expressed in various malignancies5,25 and has been proposed to contribute to a variety of malignant properties.26 In CRC, limited data are available regarding the expression pattern of CT genes and their relationship with pathological characteristics for clinical use. A novel finding from the present investigation is expression of the CT antigen SPAG9 in a majority of CRC patients, compared with other known CT antigens. Recently, Li et al27 reported the expression of various CT antigen genes in CRC specimens: SCP1, 1.7%; SSX2, 2.5%; SSX4, 2.5%; SSX1, 5.0%; MAGEC2 (alias CT10), 6.6%; CTAG1B (alias NY-ESO-1), 9.9%; MAGE1, 11.6%; CTAG2 (alias LAGE-1), 15.7%; MAGE4, 22.3%; and MAGE3, 27.3%. In another study, the presence of CTAG1B mRNA was determined by reverse transcription PCR, which showed only 11.9% CTAG1B mRNA positive expression in gastric cancer specimens.28 Although the total numbers of gastric cancer specimens showing positive CTAG1B mRNA expression were not great, among these positive specimens the CTAG1B mRNA expression was detected in more gastric cancer patients in advanced stages (III and IV; 11 of 69, or 15.9%) than in patients in early stages (I and II; 1 of 30, or 3.3%). In contrast, SPAG9 mRNA was detected in a greater number of CRC patients in early stages (I and II; 23 of 26, or 88%), compared with patients in late or advanced stages (III and IV; 35 of 52, or 67%). This finding represents an important step toward identifying tumor-specific gene expression for early detection, monitoring, or prognostic prediction of CRC and warrants large-scale validation studies.
At present, no noninvasive methods for early detection are available for monitoring CRC in clinical practice. Our demonstration of SPAG9 protein expression, especially in early stages of CRC, is an important step toward developing a tumor biomarker. Our finding of SPAG9 protein expression in all of the SPAG9 mRNA-positive CRC specimens is consistent with our earlier findings in various other cancers.13–17 Moreover, the present study revealed a significantly close relationship of higher SPAG9 protein expression (higher immunoreactivity scores) with early and late stages in CRC development (P < 0.0001).
Because surgical resection remains the only potentially curative option for patients with metastatic CRC, our study reports an important finding, in that we demonstrate a specific SPAG9 protein expression in CRC patients with early stages of disease, suggesting an important role for this biomarker in early diagnosis of CRC malignancies for surgical resection. The present study provides a foundation for large-scale validation. SPAG9 is thus a potential protein target for studies that may contribute to the discovery process for new cancer biomarkers for diagnostic, prognostic, and immunotherapeutic purposes.
Survival of a cancer patient depends mainly on early detection, which calls for a practical tumor biomarker—ideally, a simple blood-based test. Serum and plasma are easily obtained and thus are the optimal source for diagnostic screening based on noninvasive markers, posing no adverse effects per se. In this context, noninvasive methods for the early detection of CRC should be based on detecting tumor-specific proteins or autoantibodies against tumor-specific antigens in serum or plasma.29 Several CRC-specific antigens have been identified, but none are in clinical use.23 In contrast, the use of autoantibodies against specific CRC antigens for early cancer detection could be more useful and beneficial in cancer management, because the immune system can respond and amplify even a low-abundance antigen by generating a very specific and sensitive antibody response. That fact makes quantification easier.
We compared the in vivo immunogenicity against SPAG9 in CRC patients with other CT antigens. One of the most immunogenic CT antigens (NY-ESO-1, encoded by the CTAG1B gene) generated humoral immune response in only 6 of 12 (50%) of gastric cancer patients.28 Subsequently, another study revealed only 1 of 82 (1.2%) of the CRC patients showed humoral immune response against NY-ESO-1.27 In the present study, however, a majority of CRC patients (70%) generated humoral response against SPAG9 protein: CRC patients with higher SPAG9 protein expression (>50% cells expressing SPAG9 protein) exhibited significantly higher immune response (P < 0.0001) against SPAG9, compared with moderate SPAG9 protein expression (≤50% cells expressing SPAG9 protein) in CRC patients. Moreover, our demonstration of humoral response in a significant proportion of CRC patients is consistent with the known immunogenicity of this antigen. Our earlier studies demonstrated that SPAG9 is highly immunogenic in vivo (eg, 67% of epithelial ovarian cancer,14 80% of breast cancer,15 77% of renal cell carcinoma,13 80% of cervical carcinoma,16 and 78% of thyroid cancer).17 Thus, the generation of immune response against SPAG9 in a majority of CRC patients supports our hypothesis for the clinical relevance of this biomarker, with great promise as an ideal serum-based biomarker for early detection and diagnosis and warranting further large-scale investigation.
The mitogen-activated protein kinases (MAPKs) play a crucial functional role in signaling pathways involved in various physiological processes. JNK interacting proteins (JIPs), which function as scaffolding proteins, bind components of MAPK signaling modules and regulate many cellular responses.30 A recent report on IB1/JIP-1 protein showed that IB1/JIP-1 interacts with JNK through the JNK binding domain (JBD) and prevents apoptosis of pancreatic β-cell lines induced by IL-1β. The JBD thus acts as a crucial regulator of survival in insulin-secreting cells.31 SPAG9 also harbors a JBD that exhibits a significant sequence identity to JBD of IB1/JIP-1. Our earlier studies have shown that a SPAG9 mutant that lacks the JBD (SPAG9ΔLZΔT) fails to interact with the JNK pathway, suggesting that the JBD of SPAG9 is involved in JNK interaction6 and may regulate the signaling modules in different aspects of cell growth and oncogenesis. This needs further investigation.
Two members of the JIP group of scaffold proteins [splice variants of the SPAG9 gene: JIP432 and SPAG96 (JIP4_Human; isoform 5, UniprotKB/Swiss-Prot O60271-5)] have different expression in various tissues, with possibly different functional roles in the MAPK signaling module. Studies on JIP4 indicate that it does not appear to function as a scaffold for the JNK signaling pathway;however, JIP4 was found to be involved in activation of p38 MAPK module.32 In contrast, our earlier MAPK interaction studies demonstrated that SPAG9 functions as a scaffolding protein and exhibits higher binding affinity to JNK2 and JNK3 than to JNK1, and does not interact with p38 MAPK.6 It is not fully understood how the interaction between JNKs and scaffolding proteins regulates the JNK signaling pathways toward specific physiological processes. SPAG9 protein expression has characteristics that are different from those of JIP4 protein. Thus, JIP4 is a broadly distributed cytoplasmic protein with no colocalization with endoplasmic reticulum, mitochondria, and Golgi, which suggests no association with these organelles.32 We note that JIP4 colocalization studies were performed not in cancer cells but in transfected culture COS cells that expressed recombinant JIP4 protein. In contrast, SPAG9 protein was expressed on the cancer cell surface and in the cytoplasm and also colocalized with Golgi and endoplasmic reticulum, indicating an association with these organelles. It is quite possible that SPAG9 protein serves the needs of cancer cells, in that overexpression of this gene product may alter the specificity and efficiency of MAPK signaling pathways and so may facilitate the processes of cellular growth, migration, and invasion. This warrants further study.
In conclusion, SPAG9 represents a novel member of the CT antigen family with high expression in CRC. To our knowledge, this is the first report demonstrating a CT antigen playing pivotal role in CRC cellular growth, migration, and invasion. We have put forth a proof of concept, wherein SPAG9 as a novel gene product participating in various malignant features could represent a candidate molecule target for CRC and could eventually provide new research leads for immunotherapeutic approaches to CRC. Most importantly, our study on CRC early detection and diagnosis based on a serum biomarker should be of great benefit for new modalities of cancer treatment. Thus, the present study lays a foundation for further investigation into validation and manipulation of SPAG9 in CRC treatment.
Acknowledgments
We thank Avadhesha Surolia (National Institute of Immunology, New Delhi, India) for his constant encouragement of this work. We thank Confocal Laser Scanning Microscope Core Facility at the National Institute of Immunology for immunofluorescence imaging.
Footnotes
Supported by grants from the Cancer Research Program, Associated Cancer Centre for Immunotherapy (ACCI), Centre for Molecular Medicine, Department of Biotechnology, Government of India.
References
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M.J. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Gill S., Thomas R.R., Goldberg R.M. New targeted therapies in gastrointestinal cancers. Curr Treat Options Oncol. 2003;4:393–403. doi: 10.1007/s11864-003-0040-9. [DOI] [PubMed] [Google Scholar]
- 3.Compton C.C. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16:376–388. doi: 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- 4.Walsh J.M.E., Terdiman J.P. Colorectal cancer screening. JAMA. 2003;289:1288–1296. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 5.Suri A. Cancer testis antigens-their importance in immunotherapy and in the early detection of cancer. Expert Opin Biol Ther. 2006;6:379–389. doi: 10.1517/14712598.6.4.379. [DOI] [PubMed] [Google Scholar]
- 6.Jagadish N., Rana R., Selvi R., Mishra D., Garg M., Yadav S., Herr J.C., Okumura K., Hasegawa A., Koyama K., Suri A. Characterization of a novel human sperm-associated antigen 9 (SPAG9) having structural homology with c-Jun N-terminal kinase-interacting protein. Biochem J. 2005;389:73–82. doi: 10.1042/BJ20041577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 8.Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Tammana H., Gingeras T.R. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Shankar S., Mohapatra B., Suri A. Cloning of a novel human testis mRNA specifically expressed in testicular haploid germ cells, having unique palindromic sequences and encoding a leucine zipper dimerization motif. Biochem Biophys Res Commun. 1998;243:561–565. doi: 10.1006/bbrc.1997.7943. [DOI] [PubMed] [Google Scholar]
- 12.Qi M., Elion E.A. MAP kinase pathways. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 13.Garg M., Kanojia D., Khosla A., Dudha N., Sati S., Chaurasiya D., Jagadish N., Seth A., Kumar R., Gupta S., Gupta A., Lohiya N.K., Suri A. Sperm-associated antigen 9 is associated with tumor growth, migration, and invasion in renal cell carcinoma. Cancer Res. 2008;68:8240–8248. doi: 10.1158/0008-5472.CAN-08-1708. [DOI] [PubMed] [Google Scholar]
- 14.Garg M., Chaurasiya D., Rana R., Jagadish N., Kanojia D., Dudha N., Kamran N., Salhan S., Bhatnagar A., Suri S., Gupta A., Suri A. Sperm-associated antigen 9, a novel cancer testis antigen, is a potential target for immunotherapy in epithelial ovarian cancer. Clin Cancer Res. 2007;13:1421–1428. doi: 10.1158/1078-0432.CCR-06-2340. [DOI] [PubMed] [Google Scholar]
- 15.Kanojia D., Garg M., Gupta S., Gupta A., Suri A. Sperm-associated antigen 9, a novel biomarker for early detection of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:630–639. doi: 10.1158/1055-9965.EPI-08-0629. [DOI] [PubMed] [Google Scholar]
- 16.Garg M., Kanojia D., Salhan S., Suri S., Gupta A., Lohiya N.K., Suri A. Sperm-associated antigen 9 is a biomarker for early cervical carcinoma. Cancer. 2009;115:2671–2683. doi: 10.1002/cncr.24293. [DOI] [PubMed] [Google Scholar]
- 17.Garg M., Kanojia D., Suri S., Gupta S., Gupta A., Suri A. Sperm-associated antigen 9: a novel diagnostic marker for thyroid cancer. J Clin Endocrinol Metab. 2009;94:4613–4618. doi: 10.1210/jc.2009-0703. [DOI] [PubMed] [Google Scholar]
- 18.Garg M., Kanojia D., Suri S., Suri A. Small interfering RNA-mediated down-regulation of SPAG9 inhibits cervical tumor growth. Cancer. 2009;115:5688–5699. doi: 10.1002/cncr.24658. [DOI] [PubMed] [Google Scholar]
- 19.Center M.M., Jemal A., Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18:1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 20.Penna C., Nordlinger B. Colorectal metastasis (liver and lung) Surg Clin North Am. 2002;82:1075–1090. doi: 10.1016/s0039-6109(02)00051-8. x–xi. [DOI] [PubMed] [Google Scholar]
- 21.Bouwman K., Qiu J., Zhou H., Schotanus M., Mangold L.A., Vogt R., Erlandson E., Trenkle J., Partin A.W., Misek D., Omenn G.S., Haab B.B., Hanash S. Microarrays of tumor cell derived proteins uncover a distinct pattern of prostate cancer serum immunoreactivity. Proteomics. 2003;3:2200–2207. doi: 10.1002/pmic.200300611. [DOI] [PubMed] [Google Scholar]
- 22.Babel I., Barderas R., Díaz-Uriarte R., Martínez-Torrecuadrada J.L., Sánchez-Carbayo M., Casal J.I. Identification of tumour-associated autoantigens for the diagnosis of colorectal cancer in serum using high-density protein microarrays. Mol Cell Proteomics. 2009;8:2382–2395. doi: 10.1074/mcp.M800596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carpelan-Holmström M., Louhimo J., Stenman U.H., Alfthan H., Haglund C. CEA, CA 19–9 and CA 72–4 improve the diagnostic accuracy in gastrointestinal cancers. Anticancer Res. 2002;22:2311–2316. [PubMed] [Google Scholar]
- 24.Locker G.Y., Hamilton S., Harris J., Jessup J.M., Kemeny N., Macdonald J.S., Somerfield M.R., Hayes D.F., Bast R.C., Jr, ASCO ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 25.Scanlan M.J., Simpson A.J., Old L.J. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 26.Simpson A.J., Caballero O.L., Jungbluth A., Chen Y.T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 27.Li M., Yuan Y., Han Y., Liu Y.X., Yan L., Wang Y., Gu J. Expression profile of cancer-testis genes in 121 human colorectal cancer tissue and adjacent normal tissue. Clin Cancer Res. 2005;11:1809–1814. doi: 10.1158/1078-0432.CCR-04-1365. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Wu X.J., Zhao A.L., Yuan Y.H., Chen Y.T., Jungbluth A.A., Gnjatic S., Santiago D., Ritter G., Chen W.F., Old L.J., Ji J.F. Cancer/testis antigen expression and autologous humoral immunity to NY-ESO-1 in gastric cancer. Cancer Immun. 2004;4:11. [PubMed] [Google Scholar]
- 29.Hanash S.M., Pitteri S.J., Faca V.M. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 30.Willoughby E.A., Collins M.K. Dynamic interaction between the dual specificity phosphatase MKP7 and the JNK3 scaffold protein beta-arrestin 2. J Biol Chem. 2005;280:25651–25658. doi: 10.1074/jbc.M501926200. [DOI] [PubMed] [Google Scholar]
- 31.Haefliger J.A., Tawadros T., Meylan L., Gurun S.L., Roehrich M.E., Martin D., Thorens B., Waeber G. The scaffold protein IB1/JIP-1 is a critical mediator of cytokine-induced apoptosis in pancreatic beta cells. J Cell Sci. 2003;116:1463–1469. doi: 10.1242/jcs.00356. [DOI] [PubMed] [Google Scholar]
- 32.Kelkar N., Standen C.L., Davis R.J. Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signalling pathways. Mol Cell Biol. 2005;25:2733–2743. doi: 10.1128/MCB.25.7.2733-2743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


