Abstract
Questions about how and why tissue regeneration occurs capture the attention of countless biologists, biomedical engineers, and clinicians. Regenerative capacity differs greatly across organs and organisms, and a spectrum of model systems with different technical advantages and regenerative strategies are studied. Several key issues common to natural regenerative events are receiving new attention from improving models and approaches, including: the determination of regenerative capacity; the importance of stem cells, dedifferentation and transdifferentiation; how regenerative signals are initiated and targeted; and the mechanisms that control regenerative proliferation and patterning.
Regeneration is commonly defined as the replacement of body parts lost by injury. If we lose blood from a wound, it regenerates through the activity of multipotent hematopoietic stem cells. If a lobe is removed from a mouse liver, hepatocytes within remaining lobes will proliferate and regenerate the lost mass. If a newt’s forelimb is amputated, a new limb with patterned, vascularised, and innervated muscle and bone will regenerate, via formation of a mound of proliferative tissue called a blastema. Yet, regeneration is most certainly not a cloaked figure emerging only after trauma – it permeates and defines everyday adult biology. The renewal of intestinal lining, the generation of new neurons in the brain, and the maintenance of our skin, hair and bone all depend on ongoing or cyclical regeneration.
A key goal of tissue regeneration studies is to gain knowledge that will foster the broad new field of regenerative medicine. This acquired information may include clues for stimulating stem cell activity, bioengineering better scaffolds, or directly initiating regenerative programs with biological factors. We already understand some forms of regeneration sufficiently to manipulate and modify key events for therapeutic causes. For instance, the common practice of bone marrow transplantation relies on the convenient homing of hematopoietic cells to their regenerative niches. However, for most examples of regeneration, we are just beginning to acquire the knowledge and techniques to attempt to selectively block or enhance precise steps during regeneration.
Regeneration research has re-emerged on the shoulders of a wide range of model systems with different experimental advantages and regenerative prowess (Table 1). For instance, spectacular animal regeneration in planarians and hydra can now be studied using standard RNA interference approaches, and transgenic axolotls and zebrafish facilitate mechanistic studies of vertebrate limb and fin regeneration (Box 1). Although natural regenerative capacities in mouse are modest by comparison, the superb range of genetic tools available for this species are primed to address our relative deficiency in understanding mammalian regeneration.
Table 1.
Model systems and tools for regeneration studies.
| Hydra | Planarians | Zebrafish | Xenopus | Axolotl | Newt | Mouse | Drosophila | |
|---|---|---|---|---|---|---|---|---|
| Key tissues assessed in regeneration studies |
Whole animal |
Whole animal |
Fins | Tadpole tail |
Tail | Limbs | Blood | Midgut |
| Germ cells | Heart | Tadpole limbs |
Limbs | Lens | Skeletal muscle |
Germ cells | ||
| Nervous system |
Retina | Retina | Spinal cord | Heart | Liver | |||
| Spinal cord | Tail | Pancreas | ||||||
| Hair cells | Spinal cord |
Peripheral nerve |
||||||
| Retina | Skin | |||||||
| Gut epithelium | ||||||||
| Germ cells | ||||||||
| Tools: | ||||||||
| Genome sequence finished or ongoing |
√ | √ | √ | √ | - | - | √ | √ |
| Knockout by homologous recombination |
- | - | - | - | - | - | √ | √ |
| RNAi | √ | √ | - | - | - | - | √ | √ |
| Transgenesis | √ | - | √ | √ | √ | - | √ | √ |
| Recombinase driven lineage tracing |
- | - | √ | - | - | - | √ | √ |
| Cell transplantation or tissue grafting |
√ | √ | - | √ | √ | √ | √ | - |
| Forward genetics | - | - | √ | - | - | - | √ | √ |
| Potential for real-time imaging of regeneration |
√ | √ | √ (larvae) | - | - | - | - | √ |
| Stage- or age-dependent regenerative capacity |
- | - | - | √ | - | - | √ | - |
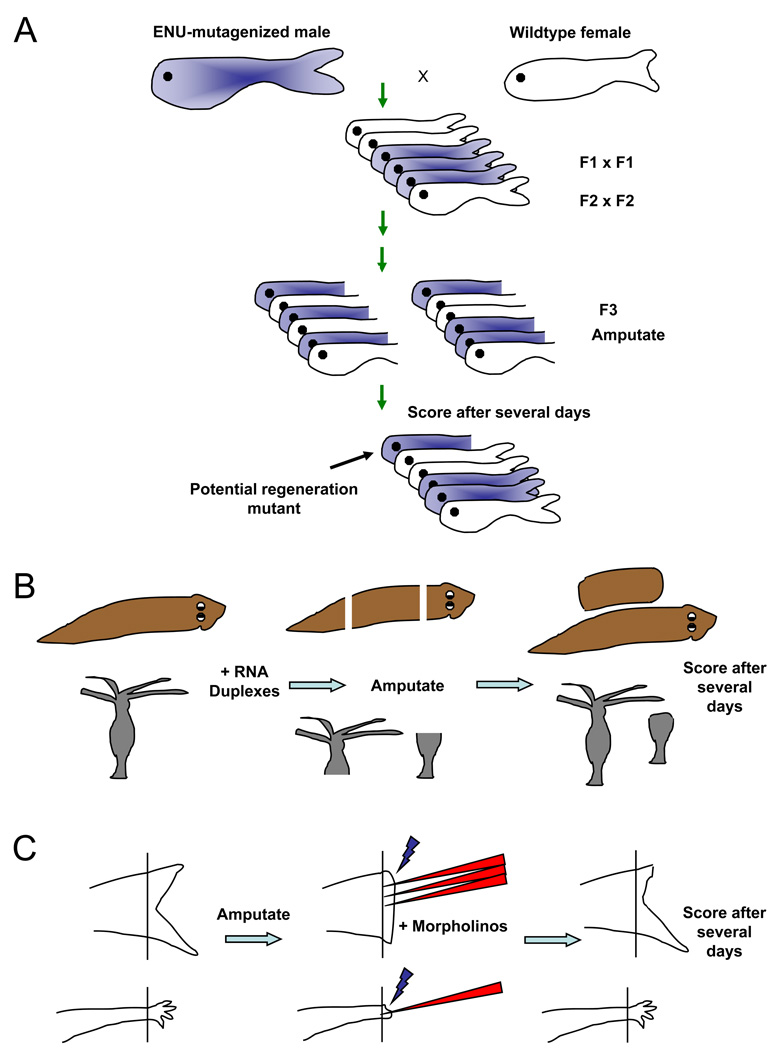
Box 1. Loss-of-function approaches in highly regenerative systems.
A. A forward genetic screen in zebrafish. Point mutations are randomly induced by chemical treatment with ENU, followed by breeding mutations to homozygosity through the F3 generation. F3 animals have their fins clipped at 2–3 months of age, and are scored for regenerative defects after 1–2 weeks. This can also be performed as a temperature-sensitive screen for conditional mutations, with animals shifted from 25–26 degrees to 33 degrees during the regeneration period7, 8. Robust mutants are chosen for genetic mapping and mutation identification.
B. RNA interference in planarians and Hydra. RNA duplexes targeting a candidate gene are introduced by injection or by feeding of bacteria containing RNAi-generating expression cassettes, in various forms, to adult animals. Animals are then amputated and scored for regeneration, sometimes followed by an additional round of RNAi and regeneration tests113.
C. Morpholino electroporation. Fluorescently labeled antisense morpholinos are injected into the blastema of the regenerating zebrafish fin114 (top) or axolotl limb115, followed by electroporation to allow nucleic acid entry into cells. The regenerate is assessed after several days; in this example, fin regeneration is delayed in morpholino-treated region.
The central questions in regeneration research remain much as they were a century ago: first, what defines and controls regenerative potential? Second, what are the cellular sources of regeneration, and is there lineage-switching to create diverse cell types in a complex structure? Third, what factors initiate regeneration and how is their activation targeted to an injured area? And finally, what signals control proliferation and patterning during regeneration and how is the process completed appropriately? Here, I synthesize studies from many model systems to broadly highlight recent insights that are energizing the field. While previous reviews have largely focused on specific animals, structures, or molecular signaling pathways, the aim here instead is to bring together common principles and key future directions in the general field of tissue regeneration.
Regenerative capacity
One mystery of regeneration is nature’s capricious distribution of this property. There is a striking hierarchy of regenerative potential among animals and organ systems. The invertebrates planaria and Hydra are at the top of this hierarchy, with the capacity to renew whole animals from tiny body pieces, or even small numbers of dissociated and re-aggregated cells1–4. They are, in essence, immortal. No animal can survive without some regenerative or self-renewal capacity, for instance in germ cells. However, many mammalian tissues, like cardiac muscle, spinal cord, and major appendages, have strikingly little regenerative capacity.
Clearly, tissues must be competent at the cellular and molecular levels to regrow patterned structures after injury. Thus, the earliest “stage” in a sequence of regenerative events is achieving or maintaining competence as an intact adult structure to respond to injury with proper regeneration. Perhaps surprisingly, the competency of a certain tissue can show differences not only between animal phyla – where significant differences at the genetic level are likely to be responsible - but also in association with what appear to be minor regulatory and epigenetic changes as described below. The latter findings are attractive as they suggest that a structure can toggle between regenerative and non-regenerative states with relatively few steps. Experimentally, and with respect to regenerative medicine, this might mean fewer manipulations to bring about regeneration.
Regeneration genes
That regenerative capacity has been occasionally lost during evolution in various species, rather than gained, is a relatively well-accepted concept. However, the basis of regenerative differences between organisms is poorly understood.
One possibility is that certain genes are present and functional in a highly regenerative species and not in a poorly regenerative species. Evidence for this model has been reported for a single gene to date: Prod1, implicated in limb regeneration as described later in this review, appears to be a salamander-specific protein not represented in fish or mammalian genomes5. It is possible that this situation will change as the genomes of increasing numbers of species are sequenced. An alternative suggestion is that, in organisms with high regenerative capacity, certain phylogenetically conserved genes function only during regeneration. In zebrafish, several screens have been carried out to identify conditional mutations in genes that have essential roles during both embryonic development and in the regeneration of amputated adult fins6–8. The results of one such screen and suggested that the Fgf ligand gene fgf20a, which is induced early after amputation and whose mammalian orthologue has not been implicated in regeneration, might function solely or primarily in tissue regeneration9. Indeed, the effects of an apparent null fgf20a mutation on embryonic zebrafish development were negligible compared to its effects on regeneration. One interpretation of these findings is that fgf20a has been specifically preserved for its role in regeneration during zebrafish evolution. However, it is equally possible that fgf20a is redundant or has minor roles during embryogenesis; there may have been more selection for backup to the function of this gene in embryogenesis compared to adult regeneration, which is less likely to be maintained by selection. Consistent with this notion, a recent study identified defects in hindbrain neurogenesis in fgf20a mutant embryonic zebrafish, indicating some additional functions for this ligand10. Future genetic screens in zebrafish may reveal additional examples with which to further examine the idea of regeneration-specific genes. Cross-referencing such genes for regenerative function in other species will also be pertinent to this discussion.
A more conservative notion is that certain gene programs are activated selectively after injury in regenerative systems, but not in non-regenerative systems. Evidence for this theory comes from studies of zebrafish fin regeneration, in which JunB proteins are phosphorylated by Jun N-terminal kinase (JNK) during fin regeneration and are required for normal regeneration11. By contrast with zebrafish JunB proteins, mammalian and Xenopus laevis JunB proteins lack a JNK phosphorylation site12, suggesting that this site may be a molecular signature of regeneration. Xenopus tadpoles in fact provide a convenient model for identifying such signatures, given that the regenerative potential of their limbs is gradually lost as development proceeds. For example, an expression program including Fgf10 is induced specifically following amputation of early stage limbs, which show regeneration, but not in later stage limbs, which are non-regenerative13. Similarly, Xenopus tails can regenerate robustly before they are resorbed during metamorphosis, with the exception of stage 45–47 animals14. This refractory period can be suppressed by experimental increases in BMP or Notch signaling, or by manipulating H+ ion flow14, 15. These data indicate again that relatively small steps in developmental progression or small changes in gene expression can toggle regenerative capacity.
There are multiple possible mechanisms by which these regeneration programs can be kept active or inactive, some of which are beginning to be explored. For instance, chromatin regulation at key genes in ES cells is critical for decisions of pluripotency or differentiation16. Early experiments suggest that removal of repressive histone methylation marks at key genes contribute to reactivating expression of regeneration genes after amputation of adult zebrafish fins17. Similarly, microRNAs provide a potential means to quickly and concurrently regulate hundreds of genes during regeneration. A recent study indicates that zebrafish fin regeneration is fine-tuned by reducing levels of miR-133, a microRNA that inhibits pro-regeneration targets, following fin amputation. Supporting this notion, artificially high levels of miR-133 blocked fin regeneration, while miR-133 antagonism accelerated the process18, 19.
Growth, aging, and regenerative capacity
As mentioned above, there is a correlation between developmental stage and regenerative capacity in Xenopus laevis. Similar results have been observed in mammals. Children show an enhanced ability to regrow lost fingertips20, 21, and fetal mice renew digit tips much more rapidly than mature animals22, 23. In these examples, regeneration can occur if the amputation occurs within the nailbed. In mice, this area has an expression domain of Msx122, a transcription factor shown to regulate regeneration in concert with BMPs24, 25. There is even recent evidence that fetal mice can regenerate cardiac muscle cells26, while by contrast, little or no regeneration occurs after cardiac injury in adult mammals. It is suspected that, as these embyonic or juvenile tissues are still in their growth phase, they might have easier access to embryonic programs necessary for regeneration than in adults that have been largely quiescent for months or years. This possibility is particularly interesting when one considers that many highly regenerative vertebrates like salamanders and fish species continue to grow for an extended period of their adult life. Multiple studies indicate that adult salamanders and zebrafish express substantial levels of developmental triggers like transcription factors and signaling ligands in uninjured tissues, the same factors active in rapidly regenerating structures, for presumed use in either growth or homeostatic maintenance27–29. While more experiments are necessary, including functional studies and more precise comparisons with corresponding mammalian structures, it is fascinating to postulate that regenerative capacity is at least in part a function of the accessibility to molecular programs that are normally employed for adult growth and maintenance.
Similarly, the extent to which the aging process impacts the regenerative capacity of adult tissues is striking. It is now clear that many mammalian tissues show a decline with age in homeostatic renewal or injury-induced regeneration. These tissues include: hematopoietic stem cells, displaying reduced capacity for long-term reconstitution of irradiated mice; the pancreas, displaying reduced islet cell proliferation and regenerative capacity with age; the brain, in which less subventricular zone neurogenesis takes place in aged mice; and skeletal muscle, displaying reduced satellite cell-mediated myogenesis with age in response to injury30–33. Such age-related changes in regenerative capacity are believed to contribute to age-related disease and decline. Thus, new insights into regenerative mechanisms and capacity stand to have enormous impact on interpreting and changing the biology of aging.
Little is known about the molecular underpinnings of the inverse relationship between age and regenerative capacity. However, recent studies made a compelling argument that age-related increases in p16INK4a levels impede regeneration in several tissues30–32. Furthermore, aging skeletal muscle appears to respond positively to a blood-born factor(s) produced in young animals33, with recent publications pointing to Wnt signaling effectors34, 35. There is currently much excitement about such age-dependent regulators of regenerative capacity, which promises to be an interesting line of research.
Cellular sources of regeneration
To understand any regenerating system, it is crucial to delineate the cellular origins of renewed tissues. This not only aids in identifying signals acting upon those cells, but is also germane to potential regenerative therapies. An ongoing rush of studies employ techniques like genetic lineage-tracing and single-cell transplantation to delineate regenerative sources (Box 2). These tools have matured first in model systems like mice and flies, but have also begun to emerge in highly regenerative animals like zebrafish. The mechanisms that provide the cellular source for regeneration can be generally classed as either involving stem cells or progenitor cells, or as requiring the dedifferentiation or transdifferentiation of cells within the tissue.
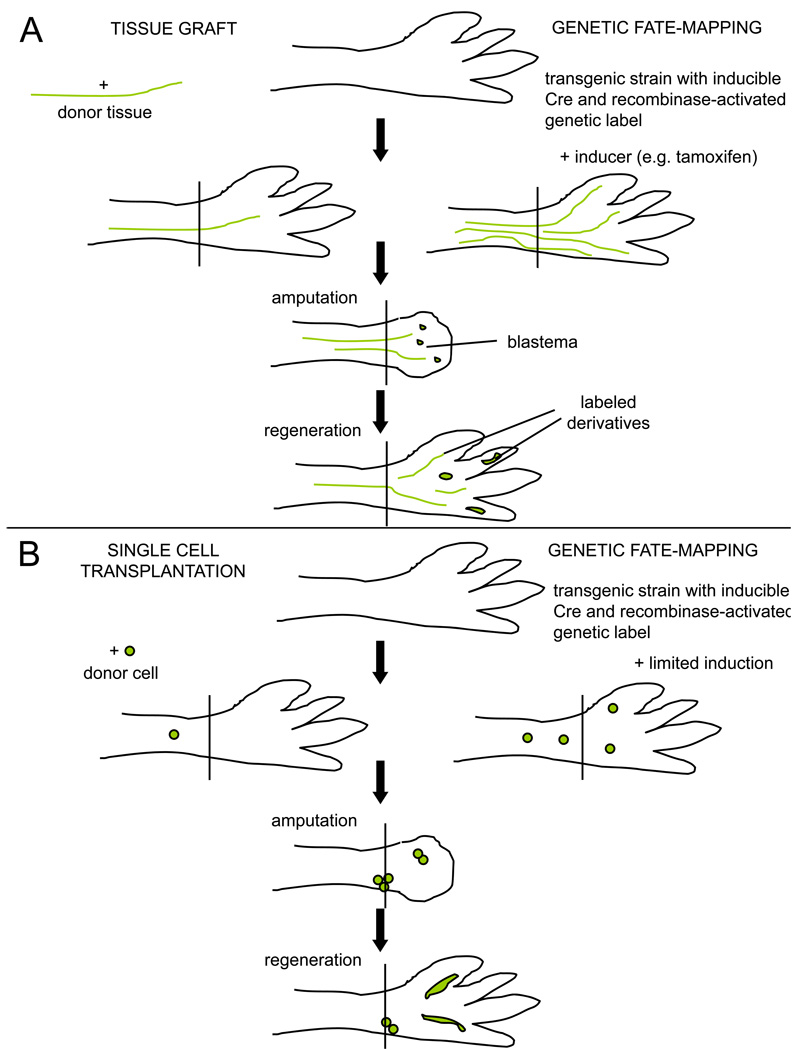
Box 2. Lineage tracing techniques used in regeneration studies.
A. Tests of tissue contributions. (Left) Tissue grafts are accepted well in salamanders, when performed either at the embryo or adult stage. A tissue type from a genetically distinct animal, e.g. a triploid animal or a transgenic animal expressing a fluorescent reporter gene in all cell types, can be dissected surgically and implanted into the intact limb of an unlabeled host. After amputation through the host limb, labeled tissues in the blastema and in the regenerate represent the derivatives of the graft. If multiple tissue contributions are detected after a transplant, issues of initial graft purity arise. (Right) Transgenic fate-mapping approaches typically use a cell type-specific promoter driving an inducible recombinase, e.g. a tamoxifen-inducible Cre fused to a mutated estrogen receptor, to trace the progeny of cells activating that promoter. After irreversibly labeling a limb cell type by tamoxifen injection, the limb is amputated and the blastema and regenerate assessed for labeled derivatives. A downside to this approach is that it relies on the availability and fidelity of a presumed tissue-specific promoter, relying on one marker for conclusions.
B. Clonal analyses can aid conclusions in lineage tracing experiments. (Left) A single labeled cell purified by flow cytometry is implanted into the intact host limb, or into the blastema (not shown). Labeled tissues in the regenerate can be traced back to that single cell, permitting rigorous tests of multipotentiality and self-renewal. (Right) A limited dose of tamoxifen will induce recombination in a small number of isolated cells, and presumably in those cells with relatively strong activation of the cell type-specific promoter driving the recombinase. Clones of cells in the regenerate can be assessed for cell type-specific markers to determine the tissue diversity of clonally related cells.
Stem-cell based regeneration
Stem or progenitor cell-based regeneration involves maintenance of a self-renewing source of differentiated cell types to be regenerated after injury. Key research directives include: 1) to identify stem cells and stem cell subpopulations, often by restricted marker expression; 2) to define the stem cell niche and mechanisms by which stem cells are activated by injury; and 3) to determine the differentiation potential of stem cells, and the regulation and dynamics of the processes by which they reconstitute tissue.
Planarians display arguably the best-understood example of stem cell-based regeneration. These mechanisms can homeostatically renew an entire animal in a week, or regenerate entire animals after repeated amputations. Their stem cells are referred to as neoblasts, and are generally classified by their low cytosolic content, their expression of markers piwi36 and bruno-like37, and their property as the only proliferating cells in the organism. Additional lines of evidence support their designation as stem cells. For instance, their transplantation after purification can rescue lethally irradiated planarians, and they can be labelled with a BrdU pulse that can be “chased” into other cell types38, 39.
The extent to which neoblasts might be a homogeneous population is unclear, as the developmental potential of a single neoblast has not yet been elucidated. Technological developments in this model system will be needed for this question to be addressed. These include: in vitro culturing and differentiation of purified clonally isolated neoblasts; implantation of single neoblasts into irradiated animals and analysis of tissue contributions; and transgenic lineage-tracing approaches as used in other model systems, often based on cell type-specific promoters and a site-specific recombinases. The study of planarian neoblasts, if indeed they are confirmed pluripotent, will be particularly powerful when it becomes possible to generate transgenic animals. One can envisage transgenic planaria with fluorescent neoblasts, imaged in real-time as these stem cells divide, differentiate, and interact with their niches.
Many vertebrate organ systems maintain adult stem cells, including blood, the frequently reviewed champion of stem cell-based regeneration40, 41, as well as skin, brain, lung, gut epithelium, and skeletal muscle. This last tissue contains undifferentiated progenitors called satellite cells that carry out one of the best-studied and most robust mammalian examples of solid tissue regeneration. These committed myogenic cells occupy a quiescent location submerged within the basal lamina of muscle fibers. They become activated upon injury and mature into myoblasts that fuse to existing myoblasts and myofibers to replace mass42. Vertebrate skeletal muscle regeneration is vigorous, but requires that a connective tissue scaffolding remains from the injured tissue43; a large mass of skeletal muscle will typically regenerate only in the setting of appendage regeneration in amphibians and certain fishes. Recent studies point to subclasses of satellite cell types that express classic satellite cell markers like Pax744 but also other indicators, such as certain cell-surface markers, that vary between subclasses45, 46. Elegant transplantation assays have demonstrated robust self-renewal and differentiation activity from single prospectively isolated satellite cells45.
Dedifferentiation and transdifferentiation
Multiple tissues regenerate through mechanisms that do not appear to require a multipotent stem cell or an undifferentiated progenitor cell. The scenarios can involve processes known as dedifferentiation and transdifferentiation. Early use of the term “dedifferentiation” at the beginning of the 20th century described ascidian stolon regeneration47, and currently it refers to a reduction in the molecular and/or functional properties of a differentiated cell type. This reduction can be minor and transient, affecting a few key genes, or can in more dramatic fashion achieve multipotency. Indeed, recent studies have clearly demonstrated that differentiated cells can be forced back into pluripotent stem cells from which complete animals can be derived48. There are also clear examples of natural dedifferentiation during regeneration - at least that which is not caused by forced stem cell conversion through gene manipulation. These include the reversion of Drosophila spermatogonia, which can dedifferentiate back to a germline stem cell and occupy the niche near the central “hub” cells, after the stem cells are transiently depleted of Jak-STAT signaling49. A recent study indicated that similar reversibility occurs in the murine spermatogenic compartment; cyst cells past a certain level of differentiation are able to re-acquire the ability for long-term repopulation of the compartment50.
Accumulating evidence suggests that heart regeneration, which is limited to non-mammalian vertebrates, involves dedifferentiation. Adult zebrafish replace most or all of the muscle when 20% of the ventricle is removed at the apex51. Recent studies in zebrafish used inducible Cre recombinase-based lineage tracing to indicate that the majority, if not all, of this regenerated cardiac muscle is derived from cells expressing the gene cmlc2, which is required for the contractile function of cardiac cells, whether cmlc2-expressing cells are irreversibly labeled in embryos52 or in adults53. Regenerating myocytes were found to induce regulatory sequence for the transcription factor gata4, which is required for embryonic heart development, and show reduced cmlc2 expression and loss of sarcomeric structure53. The most likely model to explain these results is that existing differentiated cardiomyocytes reduce their contractile state to revert to a more embryonic form in which cell division is facilitated.
A regenerative phenomenon related to dedifferentiation is transdifferentiation – the conversion from one differentiated cell type to another, sometimes utilizing an undifferentiated intermediate. A classic example of transdifferentiation is the striking regeneration in adult newts after lens dissection. In this system, a new, functional lens emerges in transdifferentiation events from the dorsal, but not the ventral, pigmented iris tissue. These events also occur in cultured explants, and this system has been used to identify signals that can influence lens induction from dorsal or even ventral iris tissue54, 55. Another example of transdifferentiation was recently reported in which glucagon-producing alpha cells of the mouse pancreas were converted to insulin-producing beta cells, after massive diptheria toxin-induced lesions of beta-cells and provision of insulin56. Thus, an exciting revelation over the past few years has been the identification of more cell plasticity than previously suspected among regenerating tissues.
The newt limb has been intensely studied with respect to questions about the relative contributions of dedifferentiation and transdifferentiation during regeneration. For nearly 80 years, this system has been used to study how the limb blastema arises, including the extent to which dedifferentiation occurs and the multipotency or otherwise of the progeny of such events57, 58. These experiments have generally involved transplantation of tissues or cell populations that may or may not have contaminating cell types. While there is sufficient evidence to conclude that newt skeletal myofibers can fragment into proliferative myoblasts59, 60, the roles of dedifferentiation and/or transdifferentiation in the regeneration of limb structures are still subject to debate61.
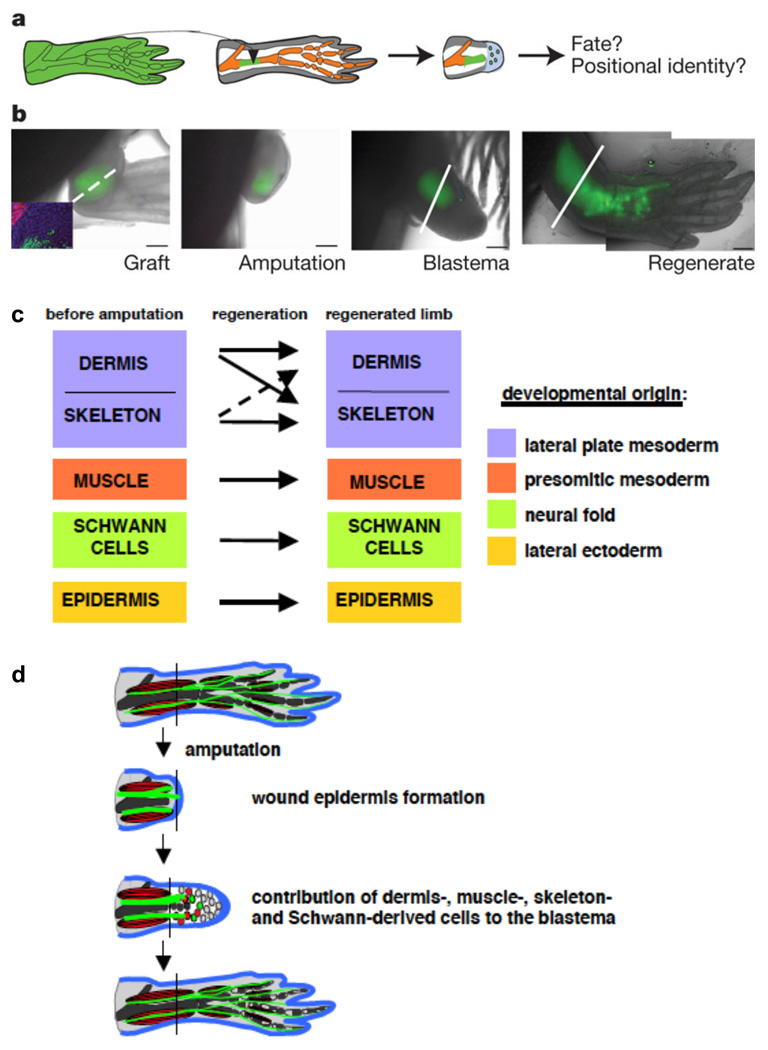
Axolotls have shorter generation times than newts and are more amenable to transgenic approaches. Recently, transgenic axolotls that express EGFP in all tissues62 have been used in combination with the elegant transplantation experiments possible in salamander embryos and adults to provide evidence in favor of lineage restriction with only limited transdifferentiation (Figure 1). That is, limbs with transplanted EGFP-labeled skeletal muscle tissue gave rise only to labeled skeletal muscle in the regenerated tissue, and transplanted labeled skeletal cartilage gave rise only to labeled cartilage62, 63. While these experiments did not address the extent to which dedifferentiation occurs, they are the most rigorous demonstration to date that a vertebrate appendage blastema need not be comprised of dedifferentiated, multipotent cells, but rather compartments of lineage-restricted cells.
Figure 1. Injured tissues retain lineages during axolotl limb regeneration.
A. Schematic of cartilage grafting from transgenic EGFP-expressing axolotl to wild-type recipient. After amputation, the tissue contributions of the donor graft and location in regenerated structures can be assessed.
B. Time course indicating the progression of EGFP-labeled donor tissue throughout the regeneration experiment. The amputation (dotted line) is made through an area containing a stable cartilage graft (green), and the graft and its derivatives are then visualized by whole-mount imaging throughout stages of regeneration. The Inset in the first image indicates that the EGFP-labeled cartilage graft does not co-label with a marker of skeletal muscle (red).
C. Summary of results from tissue-grafting experiments by Kragl and colleagues. Key tissue lineages like muscle, cartilage, Schwann cells, and epidermis remain restricted to their developmental origin and do not transdifferentiate to other lineages during limb regeneration.
D. Model of tissue contributions during blastema formation and limb regeneration, from ref 63. Blastemal cells arise from different tissue types but remain compartmentalized in the blastema. During regeneration, there is little lineage-switching, other than dermal cells; that is, blastemal cells retain their memory of origin as they are patterned into new limb structures. All figure panels reprinted with permission from 63.
What are the molecular mechanisms that underlie dedifferentation and trandifferentiation? Several findings indicate that the forced expression of fate-determining transcription factors can eventually wrest control of the developmental program of a cell type that has previously been committed to a specific lineage. Notable examples are the derivation of iPS cells from adult somatic cells48, the direct reprogramming of pancreatic beta cells from exocrine cells64, and the reprogramming of cardiac or dermal fibroblasts to cardiac muscle cells65. It remains to be determined whether instances of natural dedifferentiation require analogous developmental activity by transcription factors, although a recent report indicated upregulation of reprogramming factors like Sox2, Klf4, and c-Myc in regenerating newt limb and lens tissues66. If they do, information gained from dedifferentiation during limb regeneration in salamanders or heart regeneration in fish might be useful for attempts at optimizing reprogramming, or guiding subsequent contributions to regeneration, by mammalian cells. One absence in many if not all of these studies is a definitive marker of dedifferentiation. Presumably, intermediate states exist as a differentiated cell reverses its course toward a more plastic or proliferative form. If indeed so, the identification of expression signatures specific to these intermediate states would be of high value in understanding and further dissecting the process.
Finally, it should be noted that cellular sources can vary for different forms of injury to a given tissue. For instance, the endocrine beta cells and hepatocytes in the injured pancreas and liver, respectively, will reconstitute like cells after mechanical removal of tissue from these organs67, 68. However, alternative cells in either organ can differentiate into new parenchymal cells under certain conditions like ischemia or hepatotoxicity69, 70. Pancreatic endocrine alpha cells can acquire beta cell phenotypes like insulin production under conditions of extreme beta cell loss56. Thus, multiple experimental injury models are needed, as the key cells and signals involved in regenerative mechanisms will be best considered in the context of specific injuries.
Initiation and targeting of regeneration
In tissues that are competent for regeneration, signals must be released that identify the correct area for regenerative events to occur and direct nearby cellular sources to undergo regenerative programs. As described below, mechanisms that operate either locally or at a distance have been suspected, and in some cases identified, that stimulate regeneration. This is an important area of research, as these early signals have the potential to jumpstart the regenerative process.
Local regenerative stimuli
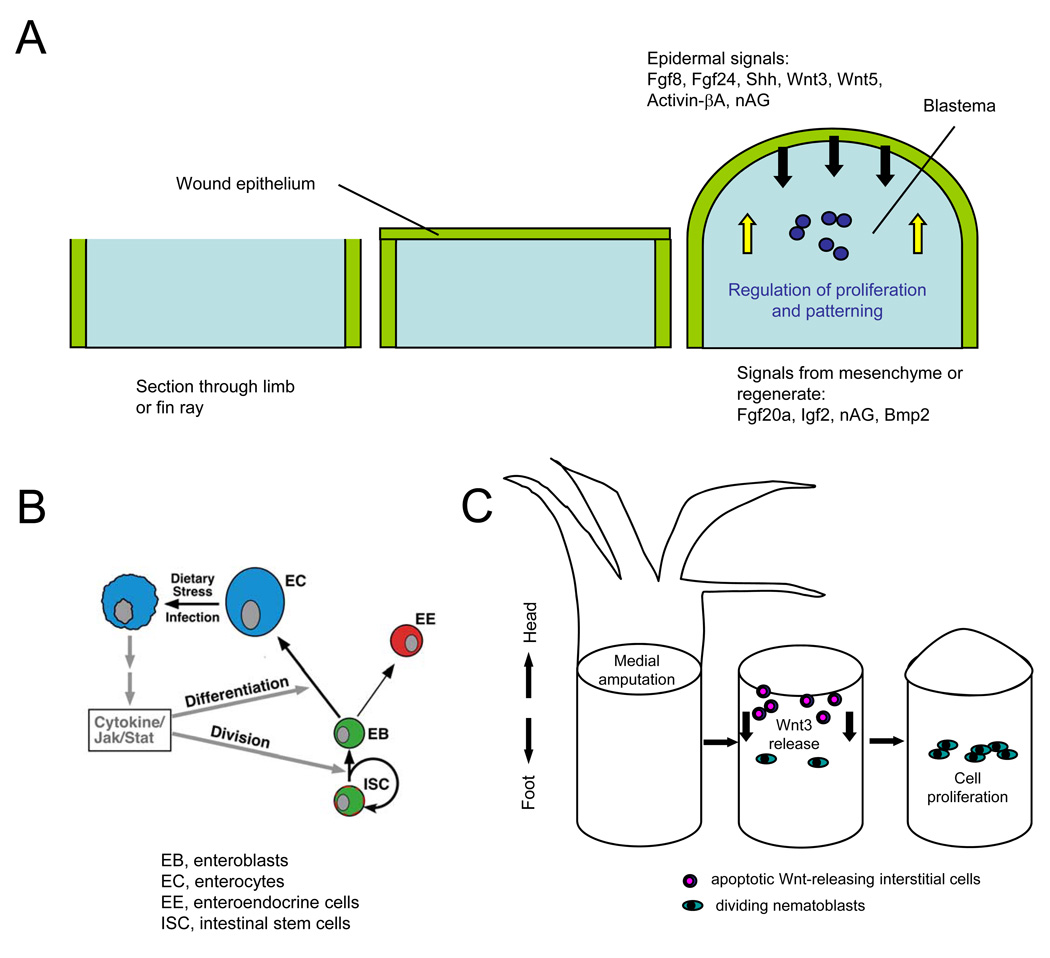
The simplest mechanism one might imagine for initiating regeneration is that a critical signal(s) is released locally upon injury that can stimulate regeneration in the spared tissues. This paradigm has been studied in many tissues from both invertebrate and vertebrate laboratory models. Limb regeneration in urodeles and fin regeneration in teleosts like zebrafish have been particularly attractive systems, as the high capacities for regeneration in these appendages starkly contrast with mammalian limbs, and because vertebrate limb bud development is a rich field of organogenesis that has provided several candidate signals71. For many years, we have known that amphibian limbs and teleost fins initiate regeneration by epidermal healing of the wound, within minutes to hours of trauma. As this regeneration epidermis matures into a multilayered structure, it begins to secrete factors critical for organizing the underlying mesenchyme into a blastema (Figure 2a). The steps resulting in blastema formation are specific to regenerative versus non-regenerative limbs, and thus key to regenerative capacity, and are analogous to how embryonic limb bud mesenchyme is organized under a developing apical ectodermal ridge in chick or mouse. A number of developmental factors have been identified in amphibian and fish appendages that are expressed very early after injury, and whose activity is critical for blastema formation (Figure 2a and 72). For example, in regenerating zebrafish fins these signals include Fgf20a, certain Wnt ligands, and Activinβ-A9, 73–75. While it is expected from studies to date that released signals like these will often recapitulate signals present in embryonic development of the same structures, understanding the regulation of their release and responses by adult cells that receive them will provide clues for whether and how a process like limb regeneration can be stimulated in adult mammals.
Figure 2. Signals initiating regeneration.
A. Signaling during blastema formation. Following amputation of a zebrafish fin or salamander limb, a wound epithelium quickly covers the appendage stump and matures into a key paracrine signaling structure for the blastema. Multiple factors are synthesized in the epidermis that are important for initiation and/or regulation of blastemal proliferation, while other factors appear to signal from underlying structures like the blastema or nerve to the epidermis.
B. Model for regenerative signaling in the Drosophila midgut. Enterocytes (EC) encountering stress or undergoing cell death release Unpaired cytokines to activate production of replacement enterocytes and enteroendocrine cells (EE) by intestinal stem cells (ISC) and enteroblast (EB) progenitors, through activating Jak/Stat signaling in these progenitor cell types. Reprinted with permission from 79.
C. Model for regenerative signaling by apoptotic cells after mid-gastric bisection in Hydra. Amputation causes apoptosis in cells near the plane of injury within 30 minutes, and concomitant Wnt3 release from those apoptotic cells. Wnt3 acts as a mitogen for interstial cells like nematoblasts, promoting regeneration by effects in addition to its role as a head organizer78.
Other mechanisms that stimulate local regeneration have recently emerged. Local cell stress or death make sense as signals for instructing local regenerative replacement, as the signals are likely to be restricted to areas of need. Recent findings have shown how apoptotic cells are assocated with and can even stimulate nearby regenerative events76–79 (Figure 2b). In the adult Drosophila midgut, Jiang and colleagues identified an elegant mechanism by which parenchymal enterocytes undergoing apoptosis or infection release Unpaired cytokines79, which activate nearby intestinal stem cells that divide and generate new enterocytes. In this way, a tissue with high turnover like intestinal epithelium has a robust mechanism for local regenerative replacement, during either normal homeostasis or injury and infection. An analogous mechanism can occur during Hydra head regeneration. Hydra rapidly regenerate their head or foot after bisection midway through the body column, but apoptosis of interstitial cells is focused in the anterior injury site. Here, as these cells die, they synthesize and release Wnt3, a factor that rapidly stimulates local cell proliferation (Figure 2c)78. Wnt3 produced by endodermal epithelial cells hours later is then also key for head formation80. Apoptosis is necessary and sufficient for activation of this type of regeneration, and has also been recognized as a regenerative requirement in tail regeneration in tadpoles77. Thus, signals released from dying cells may also represent instructive mechanisms for regeneration in vertebrates. It will be interesting to determine whether apoptosis also has roles during the prolonged process of regeneration; e.g. in other contexts described below.
Dynamic injury responses
While a freshly healed epidermis or a focus of dying cells provides an obvious source for initiating signals, regeneration can also involve organ- or organism-wide events. As indicated above, local skeletal muscle regeneration can be impacted by circulating factors whose efficacies change with age33. Similarly, exercise is known to stimulate neurogenesis from neural stem cells in the subventricular zone of adult rodents81. Thus, signals distant from the injured area can influence regenerative capacity.
Conversely, we also know that local injury can impact tissues distant from the trauma. In mammals, distal blood loss stimulates haematopoietic stem cell activity in marrow, through the action of cytokines and growth factors within niches. Similarly, partial hepatectomy activates compensatory hepatocyte hyperplasia in spared lobes, in part through released factors like IL-6, TNF-alpha, and HGF68. A surprising, organ-wide response is also displayed by the regenerating heart of zebrafish. Within days after a focal injury removing the apex of the ventricle, embryonic gene programs are activated in cell types throughout the chamber, not just adjacent to the wound82. Epicardial cells surrounding the entire chamber upregulate expression of the retinoic acid-synthesizing enzyme Raldh2, and subepicardial muscle cells adjacent to them activate regulatory sequences of the gata4 transcription factor gene53, 82. Activated programs localize to the injured and regenerating area in the following days, and these cells make key contributions to the renewed structures. It is unclear how signals are transmitted from injured to uninjured areas in this cardiac regeneration model, but it is possible that this sequence of events represents a process of searching for and identifying the injury site, which might be enhanced in a regenerative heart versus a non-regenerative (mammalian) heart.
Analogous regulatory phenomena are present in invertebrates. Decapitated planaria reset their body axes, involving animal-wide changes in expression of many released factors and signaling mediators (see the next section for further details). One of the more striking animal-wide effects occurs in Drosophila, which regenerate injured imaginal wing discs83–85. After X-ray irradiation, or mechanical or genetic ablation of disc tissue, animal development toward pupariation and eclosure is delayed as the disc regenerates86–88. Thus, there is some signal that senses local trauma and delays larval metamorphosis – an elegant mechanism for preserving synchronized development. Recent data from genetic studies indicate that this detection mechanism inhibits synthesis of the major metamorphosis signal, ecdysone, delaying pupariation89. Midbrain expression of the neuropeptide PTTH, which regulates ecdysone production, is inhibited by tissue damage, providing a likely mechanism of control. Retinoids were implicated as the systemic regulator of PTTH expression, as mutants in retinoid synthesis had attenuated delays in PTTH expression and development.
Over the next few years, one can predict to see increasing numbers of examples across phyla in which regeneration is not an isolated developmental event, but can control (and be controlled by) the physiology of the entire organ and/or animal. This may prove to add convenience or complexity in the context of regenerative medicine. In some cases, potential interventions need not be locally applied; in others, stimulating regeneration may trigger responses in unintended locations.
Control of proliferation and patterning
Once a tissue is competent for regeneration and the correct cell source is stimulated in the correct location, regeneration must be meticulously regulated such that only the appropriate structures are replaced. Like all examples of organogenesis, these events require mitogens and patterning signals, as well as some mechanism that senses scale to slow and eventually end the process.
Mitogens and patterning
It is clear that during adult regeneration, molecules important for embryonic development are called upon once more. As alluded to above, the large amount of mechanistic information yielded from studies of limb development over the past 15 years have revealed many candidate factors with potential roles during limb regeneration. For instance, factors like Sonic hedgehog, Fgfs, Wnts, retinoic acid, and BMPs that are key for patterning and outgrowth of the embryonic limb bud appear to exert similar effects on the regenerating adult limb or fin blastema72. In fact, we likely know more about basic signals for proliferation and patterning during regeneration than we do about other key events discussed elsewhere in this review.
However, regenerative growth of adult tissues is not strictly a repeat of embryonic development. For instance, the recent discovery of Anterior gradient (nAG) as a blastemal mitogen in the regenerating newt limb did not occur from comparisons with embryonic development19, 90. Indeed, there are key differences between embryonic development and regeneration that are worth pointing out. For instance, an injured organ may have an effect on other uninjured tissues, or indeed be affected by these tissues, as discussed earlier. However, only the injured organ is amenable to developmental cues that augment mass during regeneration, while uninjured tissues retain their size. By contrast, embryonic organs develop simultaneously and gain mass simultaneously as the animal grows, albeit by organ-specific mechanisms. In addition, the source of dividing cells is different: an adult organ regenerates from adult cells that have been homeostatically maintained for months or years, whereas embryonic tissues typically develop from fields of progenitor cells not so far removed in time from the fertilized one-cell animal. The issue of how the correct scale and shape are determined is one of the most fascinating and poorly understood aspects of regeneration. There must be a positional memory, by which adult cells retain the developmental ability to construct organ parts of the correct size and shape, replacing only that which was lost. Our limited understanding of this phenomenon is discussed below.
Positional memory
During embryonic patterning, cells decode complex, position-dependent signals to initiate molecular programs that control proliferation and differentiation. How cells interpret different concentrations and combinations of signals in developing systems has been heavily investigated 85, 91–95, with advances in understanding at the molecular level having been gained recently in Drosophila 96–99. During adult tissue regeneration, cells spared by injury must either remain competent to positional instructions even as a part of fixed, determined structures, or they must somehow maintain the instructions themselves.
Positional memory during regeneration is mainly considered in the context of appendages, which have obvious axes and patterns. Regulators of positional memory in appendages are expected to have two main characters: first, they should be present in a gradient or restricted pattern within the intact adult structure; second, their misexpression or blockade should affect the regenerative pattern. Surprisingly few factors to date have been implicated in control of positional memory during appendage regeneration. Retinoic acid (RA) treatment proximalizes amphibian limb regenerates, causing wrist-level amputations to sprout shoulder-level regenerates100. While these data are spectacular, the endogenous role of RA in positional memory is unclear, since there is no discernable PD gradient of RA in the intact limb, and loss-of-function experiments have not been successful. Other candidate factors include Prod1, a proposed receptor for nAG90, 101. Prod1 is induced by exogenous RA and expressed at slightly higher levels (~1.8-fold) in proximal intact limb regions as compared to distal regions102. While in vivo function is unclear, antibody knockdown of Prod1 in cultured blastemas blocks characteristic in vitro behavior of proximal blastemas, and blastema cells electroporated with excess Prod1 distribute proximally during regeneration as compared to control electroporations101, 103.
Much remains to be discovered about regulation of positional memory in adult tissues. While generating stem cells in plasticware is now routine, regenerative medicine of complex, patterned tissues like limbs requires major advances in understanding how to confer tissue-building instructions. That adult cells maintain positional information, even in mammals, is a relatively new concept. While most scientists might have assumed that skin fibroblasts isolated from different areas of the human body are molecularly the same, we know now through mRNA microarray studies that they are in fact distinct. Even when maintained in culture for several passages after isolation, the expression profiles of fibroblasts cluster among cells also taken from that anatomic site in different individuals, rather than with cells taken from different sites on an individual. One topographically controlled set of genes is the Hox genes. These genes are classic regulators of morphogenesis; thus, this finding suggests that the maintained expression of site-specific Hox codes in fibroblasts represents positional memory104. There is also evidence for positional memory distinguishing different subsets of adult bone cells. Adult murine mesoderm-derived bone and neural crest-derived bone were shown to heal injuries using progenitors of the same embryonic origins. However, mesoderm-derived adult progenitor cells transplanted to injured neural crest-derived bone did not contribute to healing105. Another piece of convincing evidence of positional memory has come from lineage analysis of neural stem cells from different regions within the ventricles of the adult mammalian brain. These cells may have a similar niche architecture and molecular profile, but they maintain positional information to generate distinct neurons, even in culture or if grafted to heterologous locations106. Thus, we are beginning to learn more about examples of positional memory within adult mammalian tissues.
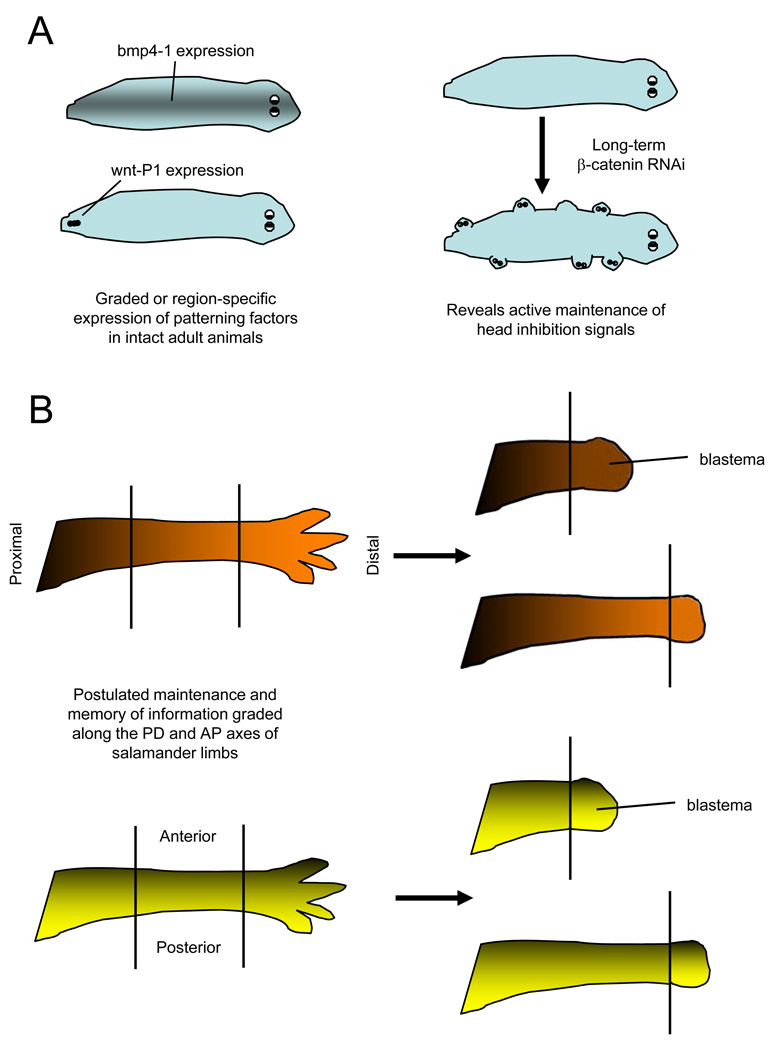
The positional memory displayed by planarian cells during regeneration may be related to these properties of mammalian cells. Intact planarians maintain graded or region-specific expression patterns of developmental signaling molecules that are also called upon during regeneration (Figure 3a). For instance, wnt-P1 expression is restricted to a small number of cells at the posterior midline of the intact animal107, and bmp4 expression exists in a rostral-to-caudal stripe of midline cells108. If extended RNA interference is used to disrupt such maintained signals in the absence of injury, the animal can take on monstrous forms. Among spectacular examples, knockdown of beta-catenin leads to emergence of ectopic heads from all body regions109, 110 (Figure 3a), and knockdown of the slit neural guidance cue causes ectopic nervous system structures to form111. Thus, positional memory in planarians is manifest in part by the active maintainence of patterning signals in gradients and region-specific distribution across the entire animal.
Figure 3. Models of positional memory in invertebrates and vertebrates.
A. Maintenance of patterning signals in planarians. (Left) bmp4-1, which regulates midline and dorsoventral patterning, is expressed in the midline region of intact animals108. wntP-1 regulates anteroposterior (AP) polarity during regeneration, and is normally expressed in a few cells in the tail of intact animals107. (Right) Knockdown of β-catenin causes head-like protrusions in ectopic locations, revealing homeostatic maintenance of the AP axis by this signaling molecule109, 110.
B. Model for contributions to positional memory in highly regenerative vertebrate tissues. Graded distribution of morphogenetic factors along the proximodistal (PD; top) or anteroposterior (AP; bottom) axis might also assist in recognition of positional identity during amphibian limb regeneration. Such factors would have a graded or region-specific distribution in the intact limb that helps maintain cell identity, and that can be quickly recovered in the regeneration blastema to help pattern the regenerate.
As mentioned earlier, active maintenance of developmental signals like Shh and Fgfs also occurs in the uninjured axolotl spinal cord and zebrafish tailfins27, 29. Factors like these may link somehow to genes that maintain bona fide positional memory across a field of adult cells. This may explain how the adult salamander spinal cord maintains a graded distribution of Hox gene expression along the anteroposterior axis of its spinal cord, giving each location along the spinal cord a distinct positional identity112. It seems reasonable to postulate that the adult cells of highly regenerative vertebrates like salamanders and fish may have a more ‘planarian’ profile than mammals, retaining positional memory in part by retaining the expression of embryonic morphogenetic stimuli or responders (Figure 3b).
Outlook
Regeneration research has lagged behind other fields such as embryology, immunology, and neuroscience in reaching the molecular age. This is largely because regeneration is an adult process that employs genes that were necessary for viable embryogenesis, and because the most spectacular regenerators have not been the most tractable genetic systems. The establishment of advanced tissue- and stage-specific tools in multiple species, for example based on control of recombinases like Cre, will allow access to regeneration and its mechanisms in the coming years. Exquisite tools are already available for regeneration studies in fruit flies and mice, where they are bearing excellent fruit. Similar tools are likely to continue to mature quickly in zebrafish, because of the large body of genetic researchers, and to emerge in other species like planarians and salamanders because of their attractive regenerative properties and increasing interest in community resources.
Furthermore, as it has permeated all fields of biological research, the continued accumulation of genome and transcriptome sequence information will aid future regeneration studies. Genome sequencing, especially of highly regenerative animals with large or understudied genomes like salamanders, may reveal gene regulatory sequences key to regenerative events. The ability to sequence genomes of individual animals will also make it easier to identify mutations that underlie regenerative defects. High-throughput deep sequencing will improve upon microarray technology, generating more sensitive mRNA expression profiles in regenerating tissues of organisms with or without fully annotated genomes. This work will reveal candidate genes that can be filtered from data from different tissues or different organisms, and may be used to home in on important differences in expressed genes that could contribute to regenerative capacity - for instance, adult cardiomyocytes from a regenerative species like zebrafish versus a non-regenerative species like mouse. Integration with characterization of epigenetic regulatory mechanisms and targets during regeneration will generate high-resolution molecular models of how regeneration is initiated and controlled. New information from these findings will then need to be synthesized to derive methods that encourage stem cell activity, build regenerative scaffolds, or impart enhanced regenerative capacity with biological factors.
Acknowledgements
I thank HHMI, NIGMS, NHLBI, AHA, AFAR, Pew Charitable Trusts, and the Whitehead Foundation for funding my laboratory’s research on regeneration. I apologize to colleagues in the field if discussion or depth was omitted due to space constraints.
References
- 1.Morgan TH. Experimental studies of the regeneration of Planaria maculata. Arch. Entw. Mech. Org. 1898;7:364–397. [Google Scholar]
- 2.Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 3.Gierer A, et al. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 4.Bosch TC. Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol. 2007;303:421–433. doi: 10.1016/j.ydbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Garza-Garcia AA, Driscoll PC, Brockes JP. Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integrative and Comparative Biology. 2010:1–8. doi: 10.1093/icb/icq022. [DOI] [PubMed] [Google Scholar]
- 6.Nechiporuk A, Poss KD, Johnson SL, Keating MT. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258:291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 7.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 8.Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. This paper describes the positional cloning of a mutation in fgf20a that disrupts zebrafish fin regeneration, delineating a key early signal in initiating regeneration.
- 10.Gonzalez-Quevedo R, Lee Y, Poss KD, Wilkinson DG. Neuronal regulation of the spatial patterning of neurogenesis. Dev Cell. 2010;18:136–147. doi: 10.1016/j.devcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishida T, Nakajima T, Kudo A, Kawakami A. Phosphorylation of Junb family proteins by the Jun N-terminal kinase supports tissue regeneration in zebrafish. Dev Biol. 2010;340:468–479. doi: 10.1016/j.ydbio.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama H, et al. Mesenchyme with fgf-10 expression is responsible for regenerative capacity in Xenopus limb buds. Dev Biol. 2000;219:18–29. doi: 10.1006/dbio.1999.9587. [DOI] [PubMed] [Google Scholar]
- 14.Beck CW, Christen B, Slack JM. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell. 2003;5:429–439. doi: 10.1016/s1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- 15.Adams DS, Masi A, Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134:1323–1335. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- 16.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin VP, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Illingworth CM. Trapped fingers and amputated finger tips in children. J Pediatr Surg. 1974;9:853–858. doi: 10.1016/s0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 21.Douglas BS. Conservative management of guillotine amputation of the finger in children. Aust Paediatr J. 1972;8:86–89. doi: 10.1111/j.1440-1754.1972.tb01793.x. [DOI] [PubMed] [Google Scholar]
- 22.Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- 23.Borgens RB. Mice regrow the tips of their foretoes. Science. 1982;217:747–750. doi: 10.1126/science.7100922. [DOI] [PubMed] [Google Scholar]
- 24.Yu L, et al. BMP signaling induces digit regeneration in neonatal mice. Development. 137:551–559. doi: 10.1242/dev.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han M, Yang X, Farrington JE, Muneoka K. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development. 2003;130:5123–5132. doi: 10.1242/dev.00710. [DOI] [PubMed] [Google Scholar]
- 26.Drenckhahn JD, et al. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cel. 2008;15:521–533. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005;132:3243–3253. doi: 10.1242/dev.01906. [DOI] [PubMed] [Google Scholar]
- 28.Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135:183–192. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- 29.Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 32.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. This paper indicates that a circulating factor in young mice can increase the normally reduced regenerative capacity of skeletal muscle in old mice.
- 34.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 36.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 37.Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Newmark PA, Sanchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 39.Baguna J, Salo E, Auladell C. Regeneration and pattern formation in planarians III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- 40.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 42.Buckingham M, Montarras D. Skeletal muscle stem cells. Curr Opin Genet Dev. 2008;18:330–336. doi: 10.1016/j.gde.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009;266:372–389. doi: 10.1111/j.1365-2796.2009.02158.x. [DOI] [PubMed] [Google Scholar]
- 44.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 45.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Driesch H. Studieren uber das Regulations vermogen der Organismen. 6. Die Restitution der Clavellina lepadiformis. Arch EntwMech. 1902;14:247–287. [Google Scholar]
- 48.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 49.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. This paper identifies heart regeneration in adult zebrafish and introduces this system as a new means to discover factors that block or enhance cardiac regeneration.
- 52.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grogg MW, et al. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature. 2005;438:858–862. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eguchi G, Abe SI, Watanabe K. Differentiation of lens-like structures from newt iris epithelial cells in vitro. Proc Natl Acad Sci U S A. 1974;71:5052–5056. doi: 10.1073/pnas.71.12.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorel F, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- 58.Dinsmore C. A history of regeneration research. Cambridge: Cambridge University Press; 1991. [Google Scholar]
- 59.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar A, Velloso CP, Imokawa Y, Brockes JP. The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol. 2004;2:E218. doi: 10.1371/journal.pbio.0020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrison JI, Loof S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sobkow L, Epperlein HH, Herklotz S, Straube WL, Tanaka EM. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev Biol. 2006;290:386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 63. Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. This paper uses transgenic salamander to provide evidence that the axolotl limb blastema is comprised of a heterogeneous population of lineage-restricted cells.
- 64.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ieda M, et al. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maki N, et al. Expression of stem cell pluripotency factors during regeneration in newts. Dev Dyn. 2009;238:1613–1616. doi: 10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 68.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 69.Xu X, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 70.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 71.Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- 72.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 73.Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 74.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 75.Kawakami Y, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006 doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pellettieri J, et al. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol. 2007;301:62–69. doi: 10.1016/j.ydbio.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chera S, et al. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev Cell. 2009;17:279–289. doi: 10.1016/j.devcel.2009.07.014. This paper indicates a mechanism by which head amputation triggers apoptosis and concomitant Wnt3 release from those apoptotic cells, stimulating animal regeneration.
- 79. Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. This paper describes an elegant mechanism for regeneration of the midgut epithelial tissue in Drosophila, in which stressed or dying cells stimulate stem cell activity and tissue regeneration.
- 80.Lengfeld T, et al. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 2009;330:186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 81.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 82.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 83.Kirby BS, Bryant PJ, Schneiderman HA. Regeneration following duplication of imaginal wing disc fragments of Drosophila melanogaster. Dev Biol. 1982;90:259–271. doi: 10.1016/0012-1606(82)90375-x. [DOI] [PubMed] [Google Scholar]
- 84.Bryant PJ. Regeneration and duplication in imaginal discs. Ciba Found Symp. 1975;0:71–93. doi: 10.1002/9780470720110.ch5. [DOI] [PubMed] [Google Scholar]
- 85.Bryant PJ. Pattern formation in the imaginal wing disc of Drosophila melanogaster: fate map, regeneration and duplication. J Exp Zool. 1975;193:49–77. doi: 10.1002/jez.1401930106. [DOI] [PubMed] [Google Scholar]
- 86.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell. 2009;16:797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simpson P, Berreur P, Berreur-Bonnenfant J. The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J Embryol Exp Morphol. 1980;57:155–165. [PubMed] [Google Scholar]
- 88.Hussey RG, Thompson WR, Calhoun ET. The Influence of X-Rays on the Development of Drosophila Larvae. Science. 1927;66:65–66. doi: 10.1126/science.66.1698.65. [DOI] [PubMed] [Google Scholar]
- 89.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol. 20:458–463. doi: 10.1016/j.cub.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. This study identified newt Anterior gradient as a blastemal mitogen released from nerves, giving an explanation for the reliance of amphibian limb regeneration on innervation.
- 91.French V. Pattern regulation and regeneration. Philos Trans R Soc Lond B Biol Sci. 1981;295:601–617. doi: 10.1098/rstb.1981.0163. [DOI] [PubMed] [Google Scholar]
- 92.Wolpert L. Positional information and pattern formation. Curr Top Dev Biol. 1971;6:183–224. doi: 10.1016/s0070-2153(08)60641-9. [DOI] [PubMed] [Google Scholar]
- 93.Wolpert L. One hundred years of positional information. Trends Genet. 1996;12:359–364. doi: 10.1016/s0168-9525(96)80019-9. [DOI] [PubMed] [Google Scholar]
- 94.Wolpert L. Positional information revisited. Development. 1989;107 Suppl:3–12. doi: 10.1242/dev.107.Supplement.3. [DOI] [PubMed] [Google Scholar]
- 95.French V, Bryant PJ, Bryant SV. Pattern regulation in epimorphic fields. Science. 1976;193:969–981. doi: 10.1126/science.948762. [DOI] [PubMed] [Google Scholar]
- 96.Baker NE. Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev. 2007;17:287–293. doi: 10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 97.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rogulja D, Irvine KD. Regulation of cell proliferation by a morphogen gradient. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 99.Piddini E, Vincent JP. Interpretation of the wingless gradient requires signaling-induced self-inhibition. Cell. 2009;136:296–307. doi: 10.1016/j.cell.2008.11.036. [DOI] [PubMed] [Google Scholar]
- 100.Maden M. Vitamin A and pattern formation in the regenerating limb. Nature. 1982;295:672–675. doi: 10.1038/295672a0. [DOI] [PubMed] [Google Scholar]
- 101.da Silva SM, Gates PB, Brockes JP. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell. 2002;3:547–555. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 102.Kumar A, Gates PB, Brockes JP. Positional identity of adult stem cells in salamander limb regeneration. C R Biol. 2007;330:485–490. doi: 10.1016/j.crvi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 103.Echeverri K, Tanaka EM. Proximodistal patterning during limb regeneration. Dev Biol. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 104.Chang HY, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leucht P, et al. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development. 2008;135:2845–2854. doi: 10.1242/dev.023788. [DOI] [PubMed] [Google Scholar]
- 106.Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- 107.Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 109.Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 110.Gurley KA, Rink JC, Sanchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cebria F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nicolas S, Papillon D, Perez Y, Caubit X, Le Parco Y. The spatial restrictions of 5'HoxC genes expression are maintained in adult newt spinal cord. Biol Cell. 2003;95:589–594. doi: 10.1016/j.biolcel.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 113. Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. This study employed a powerful, large-scale RNA interference approach to systematically explore gene requirements during planarian regeneration.
- 114.Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 115.Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]